Zelditch M.L. (и др.) Geometric Morphometrics for Biologists: a primer
Подождите немного. Документ загружается.

chap-07 4/6/2004 17: 24 page 158
158 GEOMETRIC MORPHOMETRICS FOR BIOLOGISTS
X
1
X
1
X
2
X
2
X
3
X
3
(A)
(B)
(C)
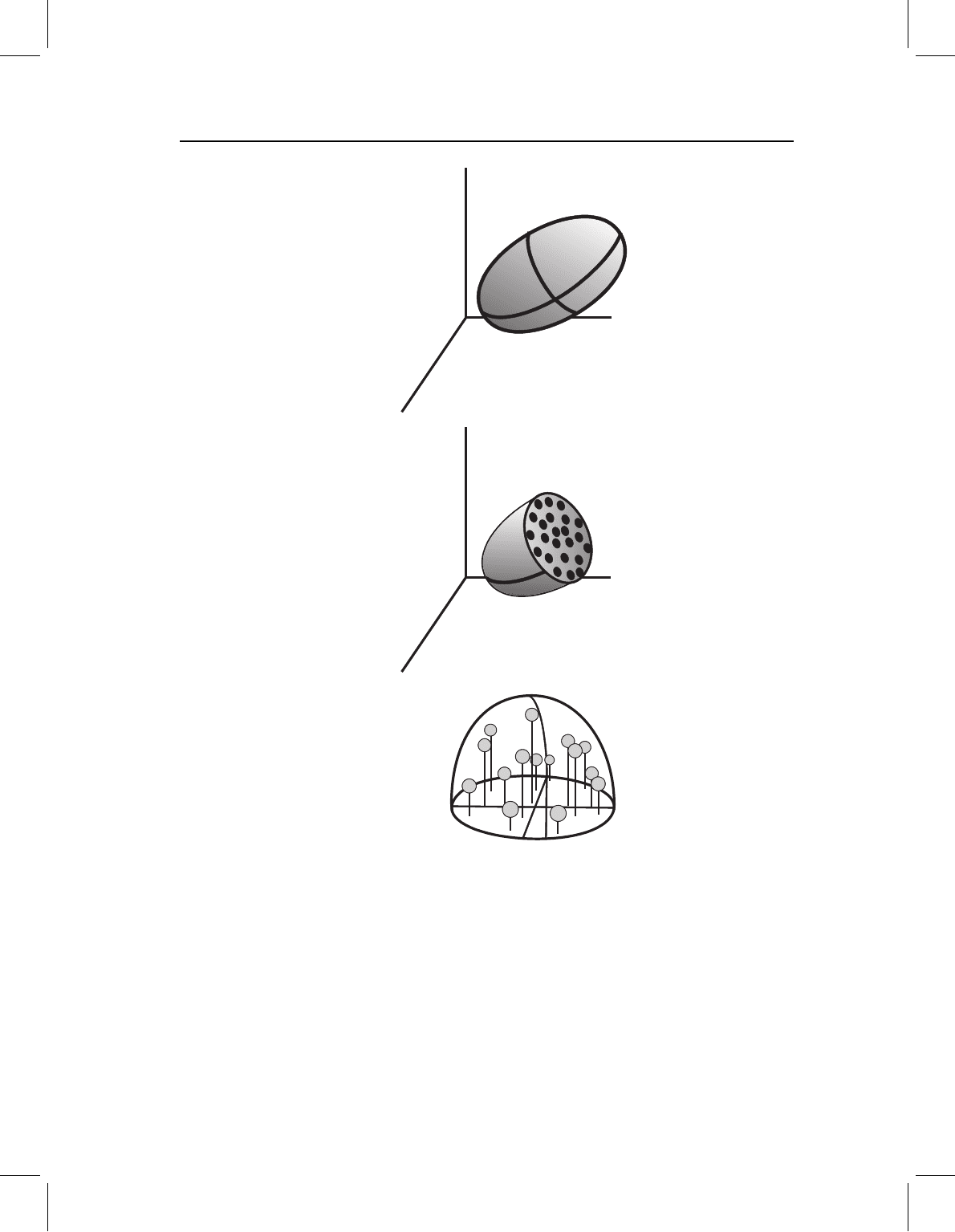
Figure 7.2 Graphical representation of PCA on three original variables (X
1
, X
2
, X
3
). (A) The
distribution of individual specimens on the three original axes is summarized by a three-dimensional
ellipsoid; (B) the three-dimensional ellipsoid is cut by a plane passing through the sample centroid
and perpendicular to the longest axis (PC1) at its midpoint, showing the distribution of individuals
around the longest axis in the plane of the section; (C) the upper half of the ellipsoid in B has been
rotated so that the cross-section is in the horizontal plane. Perpendicular projections of all individuals
(from both halves) onto this plane are used to solve for the second and third PCs.
first principal component (PC1). In an idealized case like that shown in Figure 7.1A, the line
we seek is approximately the line through the two cases that have extreme values on both
variables. Real data rarely have such convenient distributions, so we need a criterion that
has more general utility. If we want to maximize the variance that the first axis describes,

chap-07 4/6/2004 17: 24 page 159
ORDINATION METHODS 159
then we also want to minimize the variance that it does not describe – in other words, we
want to minimize the sum of the squared distances of points away from the line (Figure
7.1C). (Note: the distances that are minimized by PCA are not the distances minimized in
conventional least-squares regression analysis – see Chapter 10.)
The next step is to describe the variation that is not described by PC1. When there are
only two original variables this is a trivial step; all of the variation that is not described by
the first axis of the ellipse is completely described by the second axis. So, let us consider
briefly the case in which there are three observed traits: X
1
, X
2
and X
3
. This situation is
unlikely to arise in optimally superimposed landmark data, but it illustrates a generalization
that can be applied to more realistic situations. As in the previous example, all traits
are normally distributed and no trait is independent of the others. In addition, X
1
has
the largest variance and X
3
has the smallest variance. A three-dimensional model of this
distribution would look like a partially flattened blimp or watermelon (Figure 7.2A). Again
PC1 is the direction in which the sample has the largest variance (the long axis of the
watermelon), but now a single line perpendicular to PC1 is not sufficient to describe the
remaining variance. If we cut the watermelon in half perpendicular to PC1, the cross-
section is another ellipse (Figure 7.2B). The individuals in the section (the seeds in the
watermelon) lie in various directions around the central point, which is where PC1 passes
through the section. Thus, the next step of the PCA is to describe the distribution of data
points around PC1, not just for the central cross-section, but also for the entire length of
the watermelon.
To describe the variation that is not represented by PC1, we need to map, or project,
all of the points onto the central cross-section (Figure 7.2C). Imagine standing the halved
watermelon on the cut end and instantly vaporizing the pulp so that all of the seeds drop
vertically onto a sheet of wax paper, then repeating the process with the other half of
the watermelon and the other side of the paper. The result of this mapping is a two-
dimensional elliptical distribution similar to the first example. This ellipse represents the
variance that is not described by PC1. Thus, the next step of the three-dimensional PCA
is the first step of the two-dimensional PCA – namely, solving for the long axis of a
two-dimensional ellipse, as outlined above. In the three-dimensional case, the long axis
of the two-dimensional ellipse will be PC2. The short axis of this ellipse will be PC3, and
will complete the description of the distribution of seeds in the watermelon. By logical
extension, we can consider N variables measured on some set of individuals to represent
an N-dimensional ellipsoid. The PCs of this data set will be the N axes of the ellipsoid.
After the variation in the original variables has been redescribed in terms of the PCs, we
want to know the positions of the individual specimens relative to these new axes (Figure
7.3). As shown in Figure 7.3A, the values we want are determined by the orthogonal pro-
jections of the specimen onto the PCs. These new distances are called principal component
scores. Because the PCs intersect at the sample mean, the values of the scores represent
the distances of the specimen from the mean in the directions of the PCs. In effect, we are
rotating and translating the ellipse into a more convenient orientation so we can use the
PCs as the basis for a new coordinate system (Figure 7.3B). The PCs are the axes of that
system. All this does is allow us to view the data from a different perspective; the positions
of the data points relative to each other have not changed.
As suggested by Figure 7.4, we could compute an individual’s score on a PC from the
values of the original variables that were observed for that individual and the cosines of
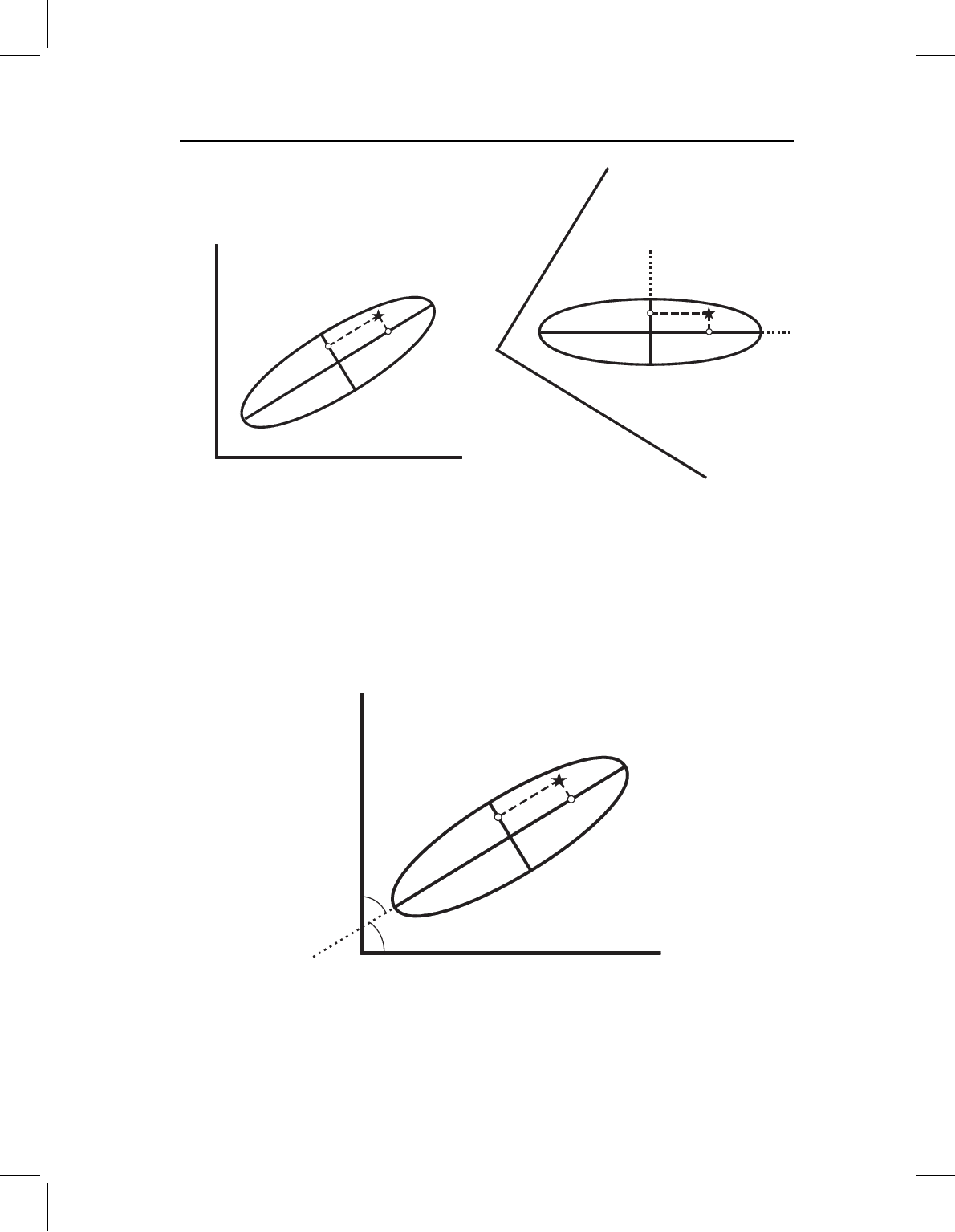
chap-07 4/6/2004 17: 24 page 160
160 GEOMETRIC MORPHOMETRICS FOR BIOLOGISTS
(A) (B)
X
1
X
2
PC2
PC1
S
1
S
2
X
1
X
2
S
1
S
2
PC2
PC1
Figure 7.3 Graphical interpretation of PC scores. (A) The star is the location of an individual in
the sample. Perpendiculars from the star to PCs indicate the location of the star with respect to those
axes. The distances of points S
1
and S
2
from the sample centroid (intersection of PC1 and PC2)
are the scores of the star on PC1 and PC2. (B) The figure in part A has been rotated so that PCs
are aligned with the edges of the page. The PCs will now be used as the reference axes of a new
coordinate system; the scores on these axes are the location of the individual in the new system. The
relationships of the PC axes to the original axes has not changed, nor has the position of the star
relative to either set of axes.
X
1
X
2
PC2
PC1
a
2
a
1
S
2
S
1
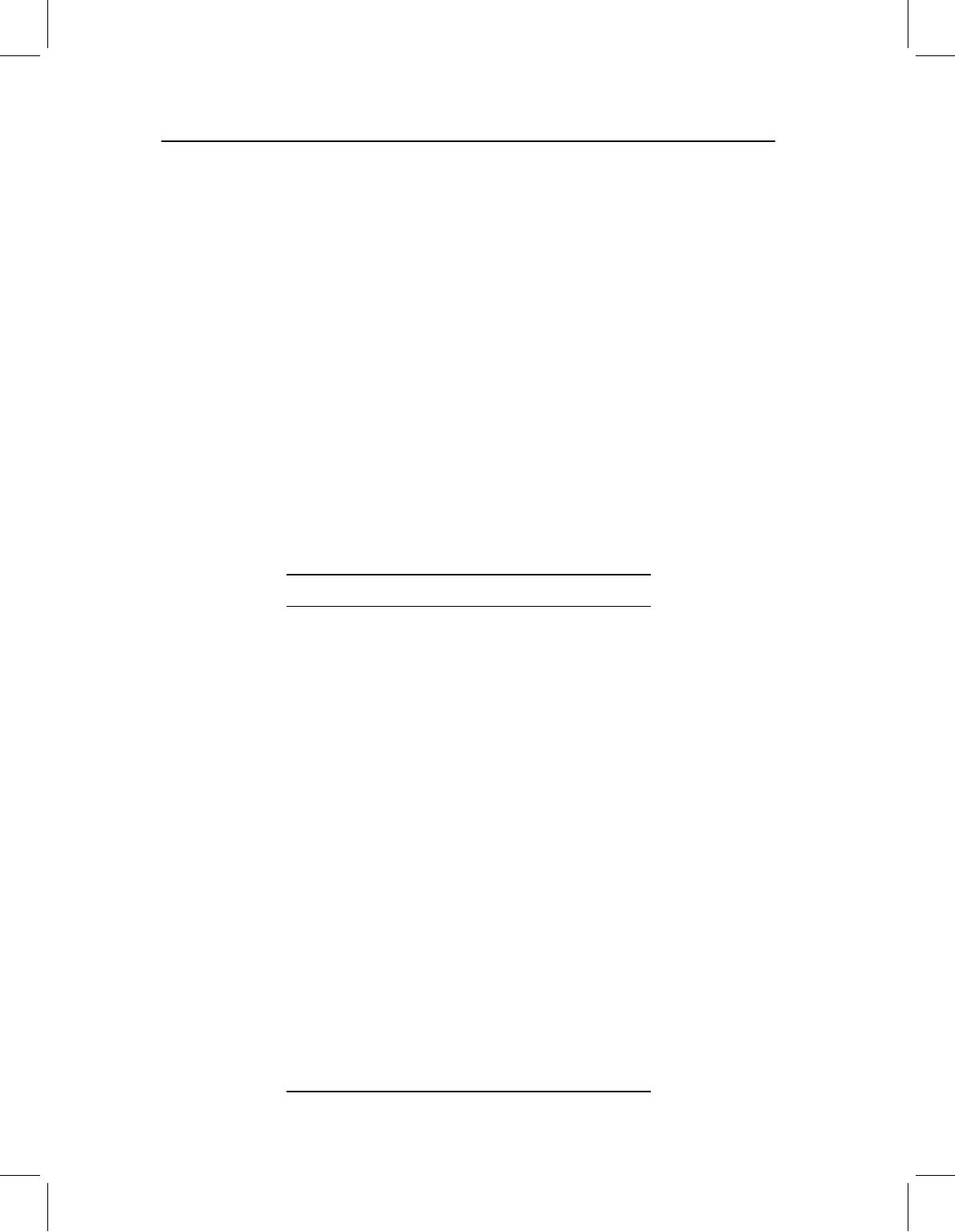
Figure 7.4 Graphical interpretation of PC scores, continued. The angles α
1
and α
2
indicate the
relationship of PC1 to the original axes X
1
and X
2
. Thus, S
1
can be computed from the coordinates
of the star on X
1
and X
2
and the cosines of the angles between PC1 and the original axes. S
2
can be
computed from the coordinates of the star on X
1
and X
2
and the cosines of the angles between PC2
and the original axes.

chap-07 4/6/2004 17: 24 page 161
ORDINATION METHODS 161
the angles between the original variables and the PCs. In our simple two-dimensional case,
the new scores, Y, could be calculated as:
Y
1
= A
1
X
1
+A
2
X
2
(7.1)
where A
1
and A
2
are the cosines of the angles α
1
and α
2
and the values of individuals
on X
1
and X
2
are the differences between them and the mean, not the observed values of
those variables.
It is important to bear in mind for our algebraic discussion that Equation 7.1 represents a
straight line in a two-dimensional space. Later we will see equations that are expansions of
this general form and represent straight lines in spaces of higher dimensionality. So, in case
the form of Equation 7.1 is unfamiliar, the next few equations illustrate the simple conver-
sion of this equation into a more familiar form. First, we rearrange the terms to solve for X
2
:
Y
1
−A
1
X
1
= A
2
X
2
(7.2)
A
2
X
2
=−A
1
X
1
+Y
1
(7.3)
X
2
=
−A
1
X
1
A
2
+
Y
1
A
2
(7.4)
Then we make two substitutions (M =−A
1
/A
2
and B =Y
1
/A
2
) to produce:
X
2
= MX
1
+B (7.5)
Thus the formula for the PC is, indeed, the formula for a straight line.
Algebraic description of PCA
We begin this description of PCA by repeating the starting conditions and the constraints
we want to impose on the new variable. We have a set of observations of P traits on N
individuals, where P is the number of shape variables (not the number of landmarks). The
data comprise P variances and P(P −1)/2 covariances in the sample. We want to compute a
new set of P variables (PCs) with variances that sum to the same total as that computed from
the variances and covariances of the original variables, and we also want the covariances
of all the PCs to be zero. In addition, we want PC1 to describe the largest possible portion
of variance, and we want each subsequent PC to describe the largest possible portion of
the variation that was not described by the preceding components.
The full set of observations can be written as the matrix X:
X =
X
11
X
12
X
13
... X
1P
X
21
X
22
X
23
... X
2P
X
31
X
32
X
33
... X
3P
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
X
N1
X
N2
X
N3
... X
NP
(7.6)
where X
NP
is the value of the Pth coordinate in the Nth individual. We can also think of
this as a P-dimensional space with N points plotted in that space – just a multi-dimensional
version of the simplistic examples presented in the previous section.

chap-07 4/6/2004 17: 24 page 162
162 GEOMETRIC MORPHOMETRICS FOR BIOLOGISTS
Our problem is to replace the original variables (X
1
, X
2
, X
3
, ...X
P
), which are the
columns of the data matrix, with a new set of variables (Y
1
, Y
2
, Y
3
, ...Y
P
), the PCs
that meet the constraints outlined in the first paragraph of this section. Each PC will be a
straight line through the original P-dimensional space, so we can write each Y
j
as a linear
combination of the original variables:
Y
j
= A
1j
X
1
+A
2j
X
2
+···+A
Pj
X
P
(7.7)
which can be expressed in matrix notation as:
Y
j
= A
T
j
X (7.8)
where A
T
j
is a vector of constants {A
1j
, A
2j
, A
3j
...A
Pj
}. (The notation A
T
j
refers to the
transpose, or row form, of the column matrix A
j
.) All this means is that the new values of
the individuals, their PC scores, will be computed by multiplying their original values (listed
in matrix X) by the appropriate values A
T
j
of and summing the appropriate combinations
of multiples. Now we can see that our problem is to find the values of A
T
j
that satisfy the
constraints outlined above.
The first constraint we will address is the requirement that the total variance is not
changed. Variance is the sum of the squared distances of individuals from the mean, so
this is equivalent to requiring that distances in the new coordinate system are the same as
distances in the original coordinate system. The total variance of a sample is given by the
sample variance–covariance matrix S:
S =
s
11
s
12
s
13
... s
1P
s
21
s
22
s
23
... s
2P
s
31
s
32
s
33
... s
3P
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
s
P1
s
P2
s
P3
... s
PP
(7.9)
in which s
ii
is the sample variance observed in variable X
i
, and s
ij
(which is equal to s
ji
)is
the sample covariance observed in variables X
i
and X
j
.
We can meet the requirement that the total variance is unchanged by requiring that each
PC is a vector of length one. If we multiply matrix X by a vector of constants as indicated
in Equation 7.8, the variance of the resulting vector Y
j
will be:
Var(Y
j
) = Var
A
T
j
X
= A
T
j
SA
j
(7.10)
Thus the constraint that variance is unchanged can be formally stated as the requirement
that the inner product or dot product of each vector A
T
j
with itself must be one:
A
T
j
A
j
= 1 =
p
k=1
A
2
kj
(7.11)
This means that the sum of the squared coefficients will be equal to one for each PC.
Substituting Equation 7.11 into Equation 7.10 yields Var(Y
j
) =S, demonstrating that the
constraint has been met.

chap-07 4/6/2004 17: 24 page 163
ORDINATION METHODS 163
The next constraint is the requirement that principal component axes have covariances
of zero. This means that the axes must be orthogonal. More formally stated, this constraint
is the requirement that the dot product of any two axes must be zero. For the first two
PCs, the constraint is expressed as:
A
T
j
A
2
= 0 =
A
1i
A
2i
(7.12)
The general requirement that the products of corresponding coefficients must be zero for
any pair of PCs is expressed as:
A
T
i
A
j
= 0(7.13)
The requirements imposed by Equations 7.11 and 7.13 indicate that we are solving for
an orthonormal basis. A basis is the smallest number of vectors necessary to describe a
vector space (a matrix). An orthogonal basis is one in which each vector is orthogonal
to every other, so that a change in the value of one does not necessarily imply a change
in the value of another – in other words, all the variables are independent, or have zero
covariance (Equation 7.13) in an orthogonal basis. An orthonormal basis is an orthogonal
basis in which each axis has the same unit length. This very particular kind of normality
was imposed by the first requirement (Equation 7.11). In an orthonormal basis, a distance
or difference of one unit on one axis is equivalent to a difference of one unit on every other
axis; consecutive steps of one unit on any two axes would describe two sides of a square.
So far, we have defined important relationships among the values of A
T
i
. There is an infi-
nite number of possible orthonormal bases that we could construct to describe the original
data. The third constraint imposed above defines the relationship of the new basis vectors
to the original vector space of the data. Specifically, this constraint is the requirement that
the variance of PC1 is maximized, and that the variance of each subsequent component is
maximized within the first two constraints.
We begin with the variance of PC1. From Equation 7.10 we know that:
Var(Y
1
) = Var
A
T
1
X
= A
T
1
SA
1
(7.14)
The matrix S can be reduced to:
=
λ
1
00... 0
0 λ
2
0 ... 0
00λ
3
... 0
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
000... λ
p
(7.15)
where each λ
i
is an eigenvalue, a number that is a solution of the characteristic equation:
S −λ
i
I = 0(7.16)

chap-07 4/6/2004 17: 24 page 164
164 GEOMETRIC MORPHOMETRICS FOR BIOLOGISTS
In the characteristic equation, I is the P ×P identity matrix:
I =
100... 0
010... 0
001... 0
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
000... 1
(7.17)
If each original variable in the data matrix X has a unique variance (cannot be replaced by a
linear combination of the other variables), then each λ
i
has a unique value greater than zero.
Furthermore, the sum of the eigenvalues is equal to the total variation in the original data.
For each eigenvalue, there is a corresponding vector A
i
, called an eigenvector, such that:
SA
i
= λ
i
A
i
(7.18)
This must be true, because we have already required:
S −λ
i
I = 0(7.19)
Therefore:
(S −λ
i
I)A
i
= 0(7.20)
which can be rearranged to:
SA
i
= λ
i
A
i
(7.21)
Thus, the eigenvectors are a new set of variables with variances equal to their eigenvalues
and covariances equal to zero. Because the covariances are zero, the eigenvectors satisfy the
constraint of orthogonality. Eigenvectors usually do not meet the constraint of normality
(A
T
i
A
i
=1), but this can be corrected simply by rescaling. Accordingly, the rescaled eigen-
vectors are the PCs, which comprise an orthonormal basis for the variance–covariance
matrix S.
All that remains is to order the eigenvectors so that the eigenvalues are in sequence
from largest to smallest. We can now show that the variance of PC1 is the first and largest
eigenvalue. From Equation 7.10 we have Var(Y
j
) =A
T
j
SA
j
, and from Equation 7.18 we
have SA
i
=λ
i
A
i
. Putting these together, we get:
Var(Y
1
) = A
T
1
λ
1
A
1
(7.22)
We can rearrange this to:
Var(Y
1
) = λ
1
A
T
1
A
1
(7.23)
which simplifies to λ
1
because we have already imposed the constraint that (A
T
1
A
1
=1).
A formal proof that principal components are eigenvectors of the
variance–covariance matrix
This is the derivation as presented by Morrison (1990). Let us suppose that we have a
set of measures or coordinates X =(X
1
, X
2
, X
3
...X
P
), and we want to find the vector
A
1
=(A
11
, A
21
, A
31
...A
P1
) such that:
Y
1
= A
11
X
1
+A
21
X
2
+A
31
X
3
+···+A
P1
X
P
(7.24)

chap-07 4/6/2004 17: 24 page 165
ORDINATION METHODS 165
We would like to maximize the variance of Y
1
:
s
2
Y
1
=
P
i=1
P
j=1
A
i1
A
j1
s
ij
(7.25)
where s
ij
is the element on the ith row and jth column of the variance–covariance matrix
S of the observed specimens. We can write the variance of Y
1
in matrix form as:
s
2
Y
1
= A
T
1
SA
1
(7.26)
Now we seek to maximize s
2
Y
1
subject to the constraint that A
1
has a magnitude of one,
which means that (A
T
1
A
1
=1). To do this, we introduce a term called a Lagrange multiplier
λ
1
, and use it to form the expression:
s
2
Y
1
+λ
1
1 −A
T
1
A
1
(7.27)
which we seek to maximize with respect to A
1
. Therefore, we take this new expression for
the variance of Y
1
and set its partial derivative with respect to A
1
to zero:
∂
∂A
1
s
2
Y
1
+λ
1
(1 −A
T
1
A
1
)
= 0(7.28)
Using Equation 7.26, we can expand the expression for the partial derivative to:
∂
∂A
1
A
T
1
SA
1
+λ
1
(1 −A
T
1
A
1
)
= 0(7.29)
which we now simplify to:
2(S −λ
1
I)A
1
= 0(7.30)
where I is the P ×P identity matrix. Because A
1
cannot be zero, Equation 7.30 is a vec-
tor multiple of Equation 7.16, the characteristic equation. In Equation 7.30, λ
1
is the
eigenvalue and A
1
is the corresponding eigenvector.
Given Equation 7.30, we can also state that:
(S −λ
1
I)A
1
= 0(7.31)
This can be rearranged as:
SA
1
−λ
1
IA
1
= 0(7.32)
and simplified to:
SA
1
−λ
1
A
1
= 0(7.33)
and further rearranged so that:
SA
1
= λ
1
A
1
(7.34)
This leads to the following substitutions and rearrangements of Equation (7.26):
s
2
Y
1
= A
T
1
SA
1
= A
T
1
λ
1
A
1
= λ
1
A
T
1
A
1
= λ
1
(7.35)
Thus, the eigenvalue λ
1
is the variance of Y
1
.

chap-07 4/6/2004 17: 24 page 166
166 GEOMETRIC MORPHOMETRICS FOR BIOLOGISTS
Interpretation of results
As we stated above, PCA is nothing more than a rotation of the original data; it is simply
a descriptive tool. The utility of PCA lies in the fact that many (if not all) of the features
measured in a study will exhibit covariances because they interact during, and are influ-
enced by, common processes. Below, we use an analysis of jaw shape in a population of
tree squirrels to demonstrate how PCA can be used to reveal relationships among traits.
Fifteen landmarks were digitized on the lower jaws of 31 squirrels (Figure 7.5). These
landmarks capture information about the positions of the cheek teeth (2–5), the incisor
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
Figure 7.5 Outline drawing of the lower jaw of the fox squirrel, Sciurus niger, showing the locations
of 15 landmarks.
0.0 0.1 0.2 0.3
⫺0.1
⫺0.2
⫺0.3
⫺0.4 ⫺0.3 ⫺0.2 ⫺0.1
0.0
0.1
0.2
X
Y
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
Figure 7.6 Plot of landmark coordinates of 31 S. niger jaws after partial Procrustes superimposition.
The locations of landmark 6 in all 31 specimens are enclosed by an ellipse. Similar ellipses could be
drawn for each landmark.
chap-07 4/6/2004 17: 24 page 167
ORDINATION METHODS 167
(1, 14 and 13), muscle attachment areas (6, 9–12, 15) and the articulation surface of the
jaw joint (7 and 8). The 31 specimens include 23 adults and 8 juveniles (individuals lacking
one or more of the adult teeth).
Figure 7.6 shows the landmark configurations of all 31 specimens, after partial Pro-
crustes superimposition. This plot does not tell us much beyond the fact that there is
shape variation in the sample. We can infer from the areas of the scatters for individual
landmarks that there is not much variation in the relative positions of the cheek teeth. In
contrast, many of the ventral landmarks have noticeably larger scatters, suggesting that
their positions relative to the teeth are more variable.
To obtain more precise information about the pattern of shape variation, the 31 sets of
landmark coordinates are converted into shape variables (see Chapter 6 for review), and
these shape variables are subjected to PCA. The 15 landmarks yield 26 shape variables,
so there are 26 PCs, and 26 scores for each specimen (its score on each component). The
output from PCA consists of the list of coefficients describing the PCs, the variance of each
component and its percentage of the total variance, and the scores of each specimen on
each component.
As shown in Table 7.1, each PC has progressively less variance. Many of the components
represent such a small proportion of the total variance that it is reasonable to ask whether
Table 7.1 Eigenvalues from PCA of squirrel jaws
PC Eigenvalues % of total variance
1 1.13 ×10
−3
51.56
2 2.15 ×10
−4
9.83
3 1.64 ×10
−4
7.49
4 1.36 ×10
−4
6.22
5 1.16 ×10
−4
5.32
6 9.52 ×10
−5
4.36
7 7.18 ×10
−5
3.28
8 5.45 ×10
−5
2.49
9 4.49 ×10
−5
2.05
10 3.58 ×10
−5
1.64
11 3.25 ×10
−5
1.49
12 2.36 ×10
−5
1.08
13 1.79 ×10
−5
0.82
14 1.37 ×10
−5
0.63
15 9.83 ×10
−6
0.45
16 9.31 ×10
−6
0.43
17 6.87 ×10
−6
0.31
18 3.72 ×10
−6
0.17
19 3.06 ×10
−6
0.14
20 2.17 ×10
−6
0.10
21 1.66 ×10
−6
0.08
22 7.04 ×10
−7
0.03
23 5.37 ×10
−7
0.02
24 3.62 ×10
−7
0.02
25 1.15 ×10
−7
0.01
26 5.02 ×10
−8
<0.01
