Seuront L. Fractals and Multifractals in Ecology and Aquatic Science
Подождите немного. Документ загружается.

64 Fractals and Multifractals in Ecology and Aquatic Science
the positive kurtosis shows a distribution that is more peaked than expected in the case of normal-
ity (g
2
= 1.83). As organisms foraging for food (for example, the amphipod C. arenarium; see
Section 3.2.3) are likely to actively select areas of high chlorophyll a concentration, they may take
advantage of positively skewed distribution. Equation (3.22) was then applied to the distribution pat-
tern of chlorophyll a concentration higher than a given threshold (such as C > 8.85 mg Chl.a m
–2
;
Figure 3.20B) to investigate the relative advantages that foraging organisms may have to actively select
a specic range of food concentrations. The whole plot has size 15 × 15 in plot units, and each pixel
represents a surface of 6.67 × 6.67 cm in the eld. The log-log plot of m(d) vs. d exhibits a very clear
scaling behavior, resulting in the mass dimension D
m
= 1.82 for C > 8.85 mg Chl.a m
−2
(Figure 3.20C).
More generally, the fractal dimensions D
m
related to chlorophyll a concentrations ranging from 1.9 to
26.6 mg Chl.a m
−2
clearly decrease with increasing chlorophyll a concentration (Figure 3.21).
3.2.4.2.4 Ecological Interpretation
Low chlorophyll densities (C ≤ 6.65 mg Chl.a m
−2
) are characterized by high fractal dimensions,
D
m
= 1.89 ± 0.01. Such high dimensions (the maximum value that D
m
can reach is D
m
= 2.00) char-
acterize very complex processes where short-range, local variability is highly developed and tends to
obfuscate long-range trends; the variable is more evenly or regularly distributed (that is, less struc-
tured) in space. In other words, this indicates that the variation within a sampling unit is equal to
the variation among the sampling units. Alternatively, the dimensions D
m
related to high chlorophyll
concentrations (C ≤ 6.65 mg Chl.a m
−2
) are very low (D
m
= 1.03 ± 0.02) and cannot be distinguished
from the lower D
m
value (D
m
= 1). This indicates that the variability of high microphytobenthos
concentrations is characterized by the so-called random point pattern (Li 2000) (see Figure 2.9D).
Finally, patches corresponding to intermediate chlorophyll a concentrations (7.60 ≤ C ≤ 21.85 mg
Chl.a m
−2
) are characterized by decreasing fractal dimensions D
m
from D
m
= 1.85 to 1.07. According
to the optimal foraging theory (Pyke 1984), organisms are expected to optimize the energy required
to capture a given amount of food. This is particularly relevant here as food availability changes
depending on its fractal dimension. A low fractal dimension relates to a smooth and predictable dis-
tribution of food items gathered in a small number of patches. In contrast, a high fractal dimension
means rough, fragmented, space-lling and less predictable distribution. When a predator has no
2.0
Mass Dimension D
m
1.8
1.6
1.4
1.2
1.0
3.8 7.6 11.4 15.2 19.0
C
22.8 26.6
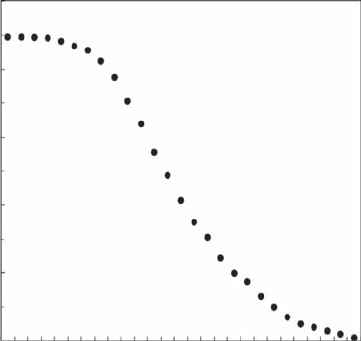
Figure 3.21 Mass fractal dimension D
m
estimated from discrete patterns of increasing microphytobenthos
chlorophyll a concentrations. The dimension D
m
is estimated as the slope of the log-log plot of m(d ) vs. d
(Equation 3.20). (See Figure 3.20C.)
2782.indb 64 9/11/09 12:04:55 PM

Self-Similar Fractals 65
detection ability, prey distributions with high dimensions should be more efcient, as food avail-
ability (which is here equivalent to the predator–prey encounter rates) becomes proportional to the
searched volume as fractal dimension increases. When a predator can remotely detect food items,
prey distributions with low fractal dimension should be relatively better. Given the sensory abilities
of intertidal organisms preying on microphytobenthos (see, for example, Erlandson and Kostylev
1995; Hutchinson et al. 2007), in the specic case studied here, microphytobenthos grazers should
then derive maximum benet from high concentration patches.
3.2.4.3 comparing the mass dimension D
m
to other Fractal dimensions
From Equations (3.9) and (3.10), and from Equations (3.19) and (3.20), it is readily seen that for
strictly self-similar mathematical fractals, such as the Cantor dust (Figure 3.3) and the Sierpinski
carpet and gasket (Figure 2.8), the mass dimension D
m
is the same as the Hausdorff dimension D
H
,
and strictly speaking to any other fractal dimension related to Equations (3.9) and (3.10) in this
section. However, for real-world fractals, there are signicant differences. The mass dimension can
nevertheless still be directly compared to the box dimension, D
b
.
Consider the mass of occupied pixels in a window (that is, box) of size (d ) as:
m
N
N
i
Oi
Ti
()
()
()
δ
δ
δ
=
(3.23)
where
m
i
()
δ
is the mass of the occupied pixels
N
O
()
δ
, and
N
Ti
()
δ
the total number of pixels in the
ith box of size d. The average mass,
m()
δ
, in
N()
δ
boxes of size d is then:
m
N
m
i
i
N
()
()
()
()
δ
δ
δ
δ
=
=
∑
1
1
(3.24)
Now, rethinking Equation (3.23) in terms of probabilistic arguments leads to expressing count fre-
quencies as probability density function p(d) following:
pm
i
i
N
() ()
δδ
==
=
∑
1
1
(3.25)
One may note here that for a given value of d, the mass m(d ) is expressed as the rst moment (that
is, the mean) of the probability distribution as:
mNp
Oi
i
N
() ()
δδδ
()=
=
∑
1
(3.26)
Using Equations (3.25) and (3.24) can be equivalently thought of as
mN() /(),
δδ
=1
leading to
rewrite Equation (3.22) as:
Nk
D
m
()
δδ
=
−
(3.27)
which is fully similar to the power-law relationship used to estimate the box dimension (
Nk
D
b
()
δδ
=
−
),
leading to:
DD
mb
=
(3.28)
2782.indb 65 9/11/09 12:05:03 PM
66 Fractals and Multifractals in Ecology and Aquatic Science
and using Equation (3.17) to
D
m
= D
b
= D
c
(3.29)
The mass and box dimensions estimated for the distribution pattern of Corophium arenarium
shown in Figure 3.17A are respectively D
m
= 1.29 ± 0.02 and D
b
= 1.28 ± 0.01, and cannot statisti-
cally be distinguished (p > 0.05) from the cluster dimension estimated in Section 3.2.3.2, D
c
= 1.28 ±
0.02. Similarly, the mass, box, and cluster dimensions estimated from the microscale distribution
patterns of microphytobenthos chlorophyll biomass (Figure 3.20A) cannot be statistically distin-
guished whatever the chlorophyll a concentration C considered.
3.2.5 in F o r m a T i o n di m E n s i o n , D
i
3.2.5.1 theory
The information dimension, D
i
, can be conceptually related to the box dimension D
b
and
the cluster dimension D
c
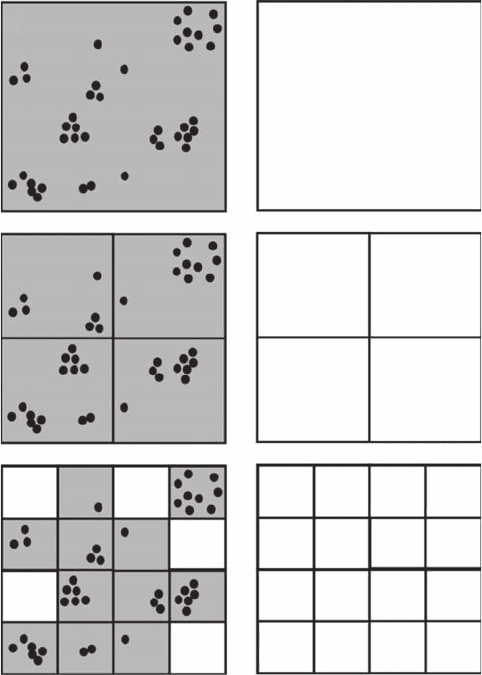
, because it is based on a count of occupied boxes of varying size d
(Figure 3.22). However, in the box and the cluster dimension estimates, a box is counted as occu-
pied (Figure 3.22A,B,C) and enters the calculations of N(d) (see Equations 3.12 and 3.14), regardless
A
D
B
E
FC
41
7
1014
10
1
9
3
31
6
3
6
621
Figure 3.22 Schematic illustration of three successive steps of the box-dimension procedure, or equiva-
lently the cluster dimension procedure (A, B, C) and information dimension procedure (D, E, F) using three
different characteristic box sizes: d
1
(A, D), d
2
(B, E), and d
3
(C, F) dened as d
1
= 2d
2
= 4d
3
.
2782.indb 66 9/11/09 12:05:05 PM

Self-Similar Fractals 67
of whether it contains one point or a relatively large number of points. The information dimen-
sion provides more details, as the number of points n
i
within each occupied box are counted
(Figure 3.22D,E,F), and is expressed as the relative frequency f
i
:
f
n
N
i
i
=
(3.30)
where
N
is the total number of points in the set
Σ
i
N
i
f
=
=
1
1
()
δ
, and
N()
δ
is the number of occupied
boxes of size d. A weight is then assigned to each box; the boxes containing a greater number of
points count more than boxes with fewer points. The information entropy, or Shannon entropy
H()
δ
at a scale d is subsequently dened as:
Hff
ii
i
N
() log
()
δ
δ
=−
=
∑
1
(3.31)
Consider now a uniformly distributed point pattern in one-, two-, and three-dimensional spaces.
The number of points
n
i
and the frequencies
f
i
are thus proportional to
δ
−1
,
δ
−2
, and
δ
−3
and to
δ
1
,
δ
2
, and
δ
3
, respectively. Equations (3.12) and (3.14) are subsequently rewritten as;
HkD
E
() loglog
δδ
=−
(3.32)
where
k
is a constant. For nonuniformly distributed point patterns, the information dimension
D
i
is subsequently dened as:
HkD
i
() loglog
δδ
=−
(3.33)
where
k
is still a constant.
3.2.5.2 comparing the information dimension D
i
to other Fractal dimensions
Although the above-mentioned arguments lead us to consider the information dimension as a gen-
eralization of the box dimension, these dimensions are nevertheless not equal. Consider a uni-
form point pattern embedded in a
D
E
-dimensional space. All the frequencies
f
i
are equal, each
fN
i
=1/ ()
δ
and Equation (3.31) leads to
HN() log()
δδ
=
, that is, the maximum value of
H()
δ
. All
the other (that is, smaller) values of
H()
δ
thus quantify the nonuniformity of the point pattern or
alternatively correct the dimension estimate by giving less weight to the boxes that contain relatively
fewer points. In other words, if one denes the information dimension as:
D
H
i
=
→
lim
()
log( /)
δ
δ
δ
0
1
(3.34)
and goes back to the formulation of the box dimension (Equation 3.15), it comes that:
DD
ib
=
(3.35)
for uniform point patterns, and
DD
ib
<
(3.36)
for nonuniform point patterns. The validity of Equations (3.33) and (3.34) is illustrated by the infor-
mation dimension analysis of the microscale distribution of the amphipod Corophium arenarium
2782.indb 67 9/11/09 12:05:21 PM
68 Fractals and Multifractals in Ecology and Aquatic Science
(Section 3.2.3; Figure 3.17A) and the related simulated uniform point pattern (Section 3.2.3)
(Figure 3.17B). As predicted by Equation (3.35), the information and box dimensions of the uni-
form point pattern cannot be statistically distinguished (p > 0.05). In turn, the information dimen-
sion D
i
is signicantly different from the box dimension D
b
for the point pattern of C. arenarium
(p < 0.01). Farmer et al. (1983) rened the denitions of both box and information dimensions,
stating that D
b
is a “metric dimension” (that is, it depends only on metric scaling properties, d),
while
D
i
is a “probabilistic dimension” (that is, it depends on both metric, d, and probabilistic,
f
i
,
properties).
3.2.6 co r r E l a T i o n di m E n s i o n , D
c o r
3.2.6.1 theory
The correlation dimension is well adapted to the characterization of spatial clustering in point pat-
terns and was initially introduced to characterize the dimension of strange attractors (see Section
6.1.3). This method, widely used in empirical analyses of dynamical systems (Grassberger and
Procaccia 1983) and in cosmology (McCauley 2001), has, to our knowledge, never been used in
ecology. A generalization of this method to the stochastic process has been applied by Ibanez (1986)
and Seuront (1999, 2004) to plankton transects and time series, respectively (see Chapter 6). The
correlation function of a point pattern, usually referred to as the correlation integral
C()δ
, is calcu-
lated as (Hentschel and Procaccia 1983):
C
N
C
i
i
N
() ()
δδ
=
=
∑
1
1
(3.37)
where
Cd
i
N
ij
N
ij
() ()
,
δθδ
=−
≠=
1
1
Σ
is the number of distinct pairs of points in a circle (or equivalently
a sphere if the point pattern is embedded in three dimensions) of radius
δ
,
centered on the ith of
N points,
d
ij,
is the Euclidean distance between the ith and the jth points (
dxx
ij ij,
||=−
), and q(x)
is the Heaviside function, dened as
θ
()x = 0
and
θ
()x = 1
for
x < 0
and
x ≥ 0
, respectively. It is
possible to take 0 < d < 1. This means that the original dimensional variable d for each point has
been rescaled by dividing it by d
max,i
, where d
max,i
is the value of the unscaled variable d for which
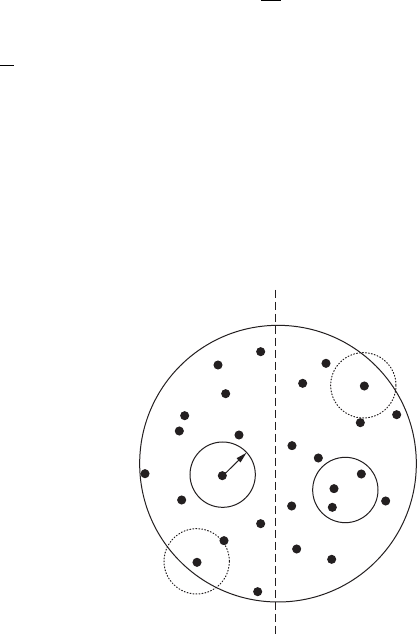
the radius d, centered on point i, just touches the boundary of the data set (Figure 3.23).
δ
Figure 3.23 A point pattern, illustrating which clusters are used (solid circles) and not used (dashed circles)
in the search for scale invariance.
2782.indb 68 9/11/09 12:05:30 PM

Self-Similar Fractals 69
Equation (3.37) can be rewritten more simply as:
C
N
N
ij
()
,
δ
=
1
2
(3.38)
where
N
is the total number of pairs of points in the set, and
N
ij,
is the number of pairs with
d
ij,
.<
δ
In other words, the correlation integral
C()
δ
represents the probability that the distance
between a pair of randomly chosen points will be less than a distance d
apart. In the case of a uni-
form point pattern, the correlation integral scales with the distance d
as:
C(d ) = kd
D
E
(3.39)
while for nonuniform distributions, C(d) is given by:
C(d ) = kd
D
cor
(3.40)
where
k
is a constant and the exponent
D
cor
is the correlation dimension, estimated as the slope of
the log-log plot of C(d ) vs. r.
3.2.6.2 comparing the correlation dimension D
cor
to other Fractal dimensions
Considering the correlation dimension as:
D
C
cor
=
→
lim
log()
log( )
δ
δ
δ
0
(3.41)
leads to:
DDD
corib
==
(3.42)
for uniform point patterns, and it is seen from the comparison of Equations (3.13), (3.32), and (3.39) that:
DDD
corib
<<
(3.43)
for nonuniform distributions. The correlation dimension D
cor
, information dimension D
i
, and box
dimension D
b
returned by respectively applying Equations (3.36), (3.29), and (3.12) to the microscale
point pattern of the amphipod Corophium arenarium (see Section 3.2.3, Figure 3.17A) and the
related uniform point pattern simulated with the same number of data points (see Section 3.2.3,
Figure 3.17B). As predicted by Equation (3.42), the correlation, information, and box dimensions of
the uniform point pattern are not statistically different ( p > 0.05) (Figure 3.24). On the other hand,
the three dimensions D
cor
, D
i
, and D
b
are signicantly different (p < 0.05) for the point pattern
of C. arenarium, with D
cor
= 1.15 ± 0.01, D
cor
= 1.22 ± 0.02, and D
cor
= 1.28 ± 0.02 (Figure 3.24),
hence verifying Equation (3.43). Chapter 5 shows in detail how the correlation dimension D
cor
, the
information dimension D
i
, and the box dimension D
b
can also be related using a generalization of
the entropy formulation introduced in Equation (3.31).
3.2.7 ar E a -pE r i m E T E r di m E n s i o n s
Area-perimeter methods have generally been used to estimate the fractal dimension of objects
coded as digitized images. Consider as an example a landscape consisting of a set of vegetation
patches (Figure 3.25). These patches can be either monospecic (Figure 3.25A,B) or plurispecic
(Figure 3.25C). Area-perimeter dimension methods can be used to determine the fractal dimen-
sion of a set of patches as a function of the complexity of their boundaries or their space-lling
2782.indb 69 9/11/09 12:05:36 PM

70 Fractals and Multifractals in Ecology and Aquatic Science
character and also as a function of the quality of the surrounding patches. Three approaches are
thus possible:
Perimeter-based• , to determine the extent that a patch perimeter lls the plane
Area-based• , to determine the extent that a patch lls the plane
Landscape-based• , to compare patch complexity in an environment characterized by
patches of different types
Each of the three methods described in this section measures a fractal dimension, but it will be
shown that their application and interpretation are quite different.
3.2.7.1 Perimeter dimension, D
p
The perimeter dimension (D
p
) method basically provides a measure of the perimeter-area ratio for
a patch or a set of patches (Figure 3.25A). Consider an “ideal” circular patch (for example, Okubo 1980)
in a two-dimensional space. The area A and the perimeter P are related as:
PrrA A
Ar
P
P
== ≈
==≈
2
4
2
2
2
ππ
π
π
(3.44)
where r is the radius of the patch, and “
≈
” means proportionality. In case of a non-Euclidean patch
structure, Equation (3.44) leads to the following perimeter-area relationship:
PkA
D
p
=
/2
(3.45)
where k is a constant, the area A is the number of pixels needed to cover a given object, the perimeter
P is the number of pixel edges, and D
p
is the perimeter dimension. For a single patch, the perimeter
dimension is thus simply written as:
DPA
p
= 2log /log
(3.46)
2.0
1.8
1.6
Fractal Dimension
1.4
1.2
1.0
D
corr
D
i
D
b
Figure 3.24 Correlation dimension D
cor
, information dimension D
i
, and box dimension D
b
estimated for the
microscale point pattern of the amphipod Corophium arenarium (see Figure 3.16) and a simulated uniform
point pattern with the same number of data points.
2782.indb 70 9/11/09 12:05:40 PM
Self-Similar Fractals 71
A
B
n
n
n
n
n
C
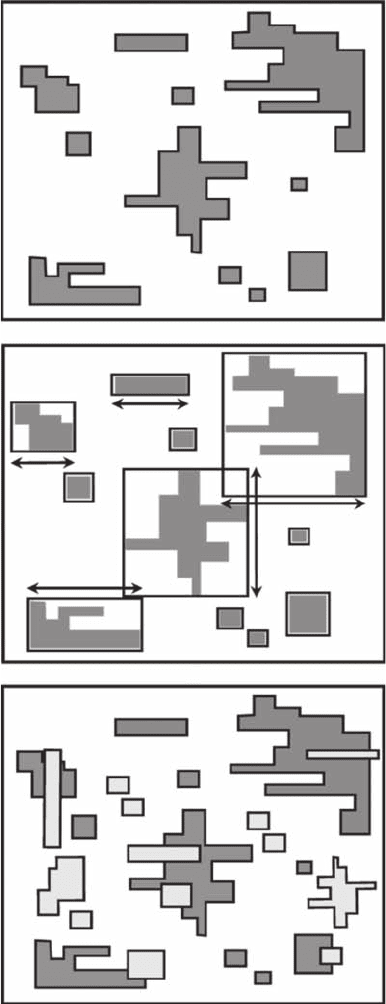
Figure 3.25 Area-perimeter dimensions. (A) Perimeter dimension D
p
is based on estimates of patch areas
(light gray) and perimeter (indicated by a black solid line). (B) Area dimension D
a
is based on estimates
of patch areas and largest horizontal or vertical dimension (n). (C) Landscape/seascape dimension D
s
is
a generalization of the perimeter dimension to patches of different species, here shown by different gray
intensities.
2782.indb 71 9/11/09 12:05:42 PM

72 Fractals and Multifractals in Ecology and Aquatic Science
Alternatively, the slope of the log-log area-perimeter plot for a set of objects gives a fractal dimen-
sion (Burrough 1986). Patterns characterized by perfectly circular patches (that is, a low perimeter-
area ratio) have a fractal dimension
D
p
=1
, while those containing highly convoluted patches (that
is, a high perimeter-area ratio) have a fractal dimension
D
p
→2
. This method is thus well adapted
to studies focusing on ecotonal boundaries (edges).
However, a digitized image of any fractal object is Euclidean by virtue of its projection onto a
grid, and thus leads to biased estimates of
D
p
; see, for example, Falconer (1993), and Section 7.2.1.
As a remedial procedure, the expression of the perimeter P in Equation (3.45) needs to be modied
as (Olsen et al. 1993):
PPA
1
1
4
21=+−[()]
(3.47)
The ratio 1:4 is determined as the proportionality constant for pixelized systems, and
21()A−
is
the maximum value of the perimeter fractal dimension
D
p
of an object of area
A
considered on
a square grid. The corrected perimeter
P
1
should then be used instead of
P
when estimating
D
p
from Equation (3.45). Some additional limitations related to the representation of digital images are
discussed in Section 7.2.1.
3.2.7.2 area dimension, D
a
To quantify the space-lling characteristics of a patch (Figure 3.25B), Voss (1988) modied the stan-
dard box-counting framework (see Section 3.2.2) and suggested that the fractal dimension of a patch
can be instead measured as D
a
= log A/log L, where L is the maximum of the row and column lengths
of the pixelized patch. Square patches of size n
(Ln=
,
An=
2
) completely ll a two-dimensional
space (that is,
Dnn
a
2==log/log
2
), while for a “rectangular” patch of length
n
and width 1
(
Ln=
,
An=
),
Dnn
a
==log/log1
. The corresponding length-area relationship is written as:
A = kL
D
a
(3.48)
where k is a constant, and
D
a
the area dimension. This method returns high fractal dimensions for
patches that best ll their embedding space (that is, circular patches). Thus, in studies focusing on
acquisition and retention of space—for instance, in the framework of invading species (Kooi and
Kooijman 2000)—a circular patch (where the area-to-edge ratio is high) would be more likely to
retain space than a thin, convoluted patch (that is, where the area-to-edge ratio is low).
3.2.7.3 landscape/seascape dimension, D
s
This framework is a generalization of the perimeter-area relationship provided above to a set of
patches of different types (for example, patches of different seagrass species; Figure 3.25C) to esti-
mate the extent to which the perimeter of a given patch interacts with neighboring patch types.
The related measure of fractal dimension, the so-called landscape/seascape dimension D
s
, can be
estimated using a modied perimeter length P
m
(Olsen et al. 1993):
P
m
= P + [2(A − 1)(N/(N − 1))]
(3.49)
where P and A are respectively the patch perimeter and area, N is the number of adjacent patch
types, and N
t
is the number of all patch types in the landscape/seascape. The related fractal dimen-
sion for the seascape is subsequently estimated by substituting Equation (3.48) into the perimeter-
area relationship given by Equation (3.45). This method can be extremely useful to estimate the
impact of intra- and interspecic competition for space and resources—for instance, in the frame-
work of zoobenthic community establishment and maintenance (Boström and Bonsdorff 2000;
Hovel et al. 2002).
2782.indb 72 9/11/09 12:05:52 PM

Self-Similar Fractals 73
3.2.7.4 Fractal dimensions, areas, and Perimeters
Although the perimeter and the area fractal dimensions introduced in Section 3.2.7 specically
describe the relationships between the perimeter P and the area A of a patch, they can more generally
be used to quantify the structure of any rough object such as proteins, biological aggregates obtained
from natural environments (for example, marine snow) or bioreactors (for example, bioocculated
microbial aggregates generated by the activated sludge process), inorganic colloidal aggregates (for
example, clays, alum, ferric hydroxides), and growth patterns of inorganic and organic systems (for
example, cluster formation, dendritic growth, diffusion-limited aggregation). However, it appears
that many fractal relationships involving the perimeter or the area of an object signicantly differ
from the concepts introduced above. These relationships are reviewed hereafter and discussed in
relation to the related fractal dimensions.
3.2.7.4.1 Fractal Structure of Surfaces
3.2.7.4.1.1 On the Fractal Surface Dimension of Proteins
The characteristic roughness and corrugation of protein surfaces are of extreme biological relevance
in their function, including (1) the association of different subunits; (2) the recognition, diffusion,
and binding of a ligan; and (3) the release of products. Typically, the surface areas of proteins is
dened by the area accessible to a probe sphere (Lee and Richards 1971; Richards 1977) and the
related fractal surface dimension D
s
, estimated as
D
dA
dR
s
p
=−2
log
log
(3.50)
or equivalently
AkR
P
D
s
=
−2
(3.51)
where k is a constant, A the molecular surface area, and R
P
the probe radius (Lewis and Rees 1985).
Note that Equation (3.51) is the strict equivalent of Equation (3.6), and consequently of the box-
counting dimension D
b
described in Section 3.2.2. This approach has been used to investigate the
fractal surface dimension of three enzymes (lysozyme, ribonuclease A, and superoxide dismutase),
which was D
b
= 2.44 on average for scales ranging from 0.1 and 0.35 nm (Lewis and Rees 1985).
The variation in D
s
over the protein surfaces revealed high fractal dimensions (D
s
> 2.5) for surface
regions of lysozyme characterized by elevated reactivity, while lower D
s
values (D
s
∈[2.3 − 2.5])
were obtained from regions located near the activity surface of the enzyme (Lewis and Rees 1985).
The region of greatest surface roughness corresponded to the dimmer interface and to the subunit
interface in superoxide dismutase and ribonuclease A, respectively. Regions involved in the forma-
tion of tight complexes and permanent binding (for example, interfaces between subunits, antibody-
combining regions) appeared to be more irregular than average (D
s
> 2.4). In contrast, regions of
proteins that interact transiently with ligans and cannot tolerate formation of stable complexes (for
example, active sites) appear to be smoother than average. Despite the very narrow range of scales
used to estimate the fractal dimensions and in the absence of any discussion related to the obvious
changes in the values of D
s
with the size of the probe (Lewis and Rees 1985; see their Figure 2b),
this work indicates that increased roughness favors strong bounds, thus relating the fractal structure
to specic functions and suggesting that fractal dimension could actually be used to predict protein
functional sites, as functional surfaces are much rougher than protein surfaces in general (Pettit and
Bowie 1999). Note that Equation (3.50) has further been modied to account for statistical errors
resulting from local variations in roughness at specic sites on a protein surface as
f
dA
dR
i
j
j
P
=−
∑
2
log
log
(3.52)
2782.indb 73 9/11/09 12:05:55 PM
