Jones M., Fleming S.A. Organic Chemistry
Подождите немного. Документ загружается.

23.4 Peptide Chemistry 1209
mild acid
(deprotection)
1. DCC
removal of tBoc
protecting group
DCC-mediated
amino acid coupling
COOH
R
tBoc
O
..
..
R
..
O
..
..
NH
2
R
..
O
..
..
NH
2
R
..
O
..
..
N
N
R
H
H
..
..
O
..
..
O
..
..
R
O
..
..
N
H
..
tBoc
tBoc
R
2.
R
N
N
H
H
..
..
O
..
..
tBoc
R
O
..
..
O
..
..
O
..
..
O
..
..
N
H
tBoc
..
N
H
O
..
..
O
..
..
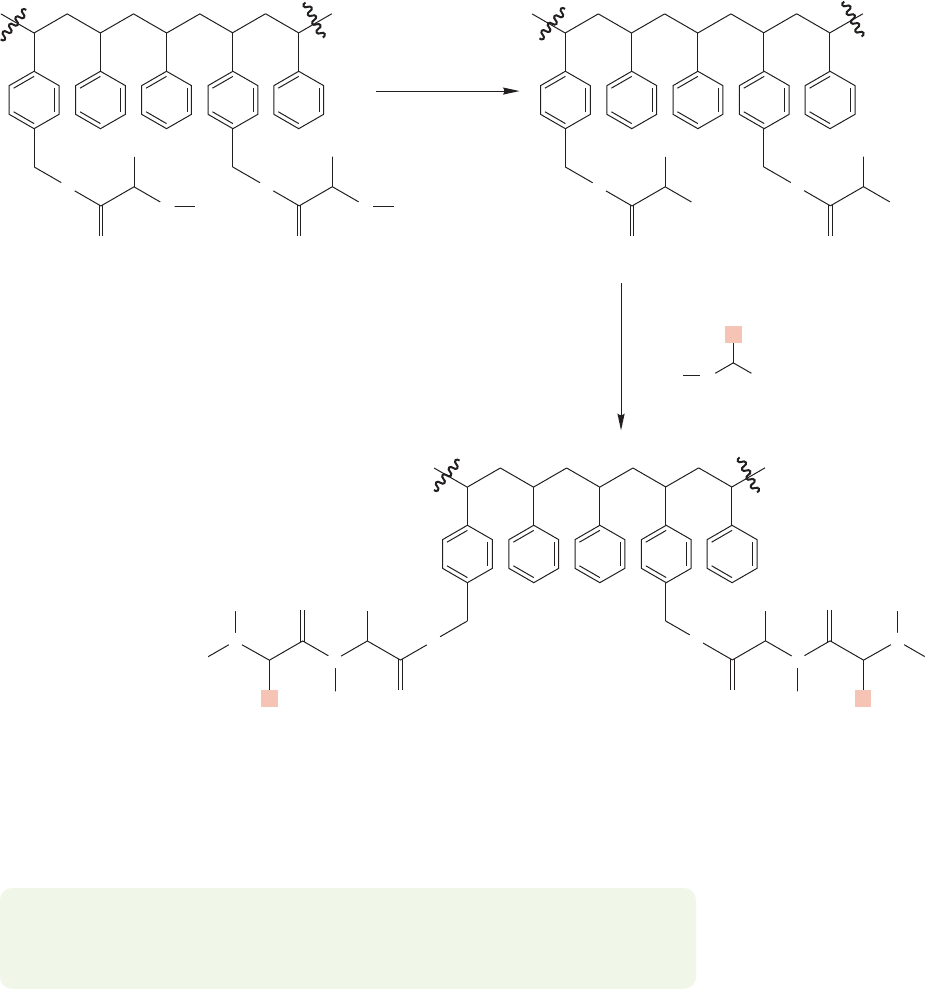
FIGURE 23.52 The steps of peptide synthesis can now be carried out on this immobilized
amino acid.
The tBoc group is then removed with mild acid hydrolysis, and a new, tBoc-
protected amino acid is attached to the bound amino acid through DCC-mediated
coupling.The multiple steps of peptide synthesis using various amino acids can now
be carried out on this immobilized amino acid (Fig. 23.52).
PROBLEM 23.22 What is the function of the benzene ring of polystyrene? Could
any ring-containing polymer have been used? Would cyclohexane have done as
well, for example?
This procedure can be repeated as many times as necessary. The growing pep-
tide chain cannot escape, because it is securely bound to the polystyrene; reagents
can be added and by-products can be washed away after each reaction.The final step
1210 CHAPTER 23 Amino Acids and Polyamino Acids (Peptides and Proteins)
The first relatively primitive homemade Merrifield machine managed to pro-
duce a chain of 125 amino acids in an overall 17% yield, a staggering accomplish-
ment.Merrifield was quite rightly rewarded with the chemistry Nobel prize in 1984.
4
In this case, “natural” is an artificial distinction, almost as foolish as labels describing natural as opposed to
artificial vitamin C. The Merrifield procedure is no less natural than what we are about to describe. The lab-
oratory synthesis is not unnatural, it’s just human-mediated.
N
NH
2
R
H
..
..
..
..
O
..
..
O
..
..
..
..
..
..
R
HF
N
N
R
H
H
..
..
O
O
..
..
..
..
O
..
..
R
N
NH
3
R
H
..
O
..
..
O
..
..
R
N
N
R
+
H
H
..
..
–
OOC
O
..
..
R
CH
2
F
+
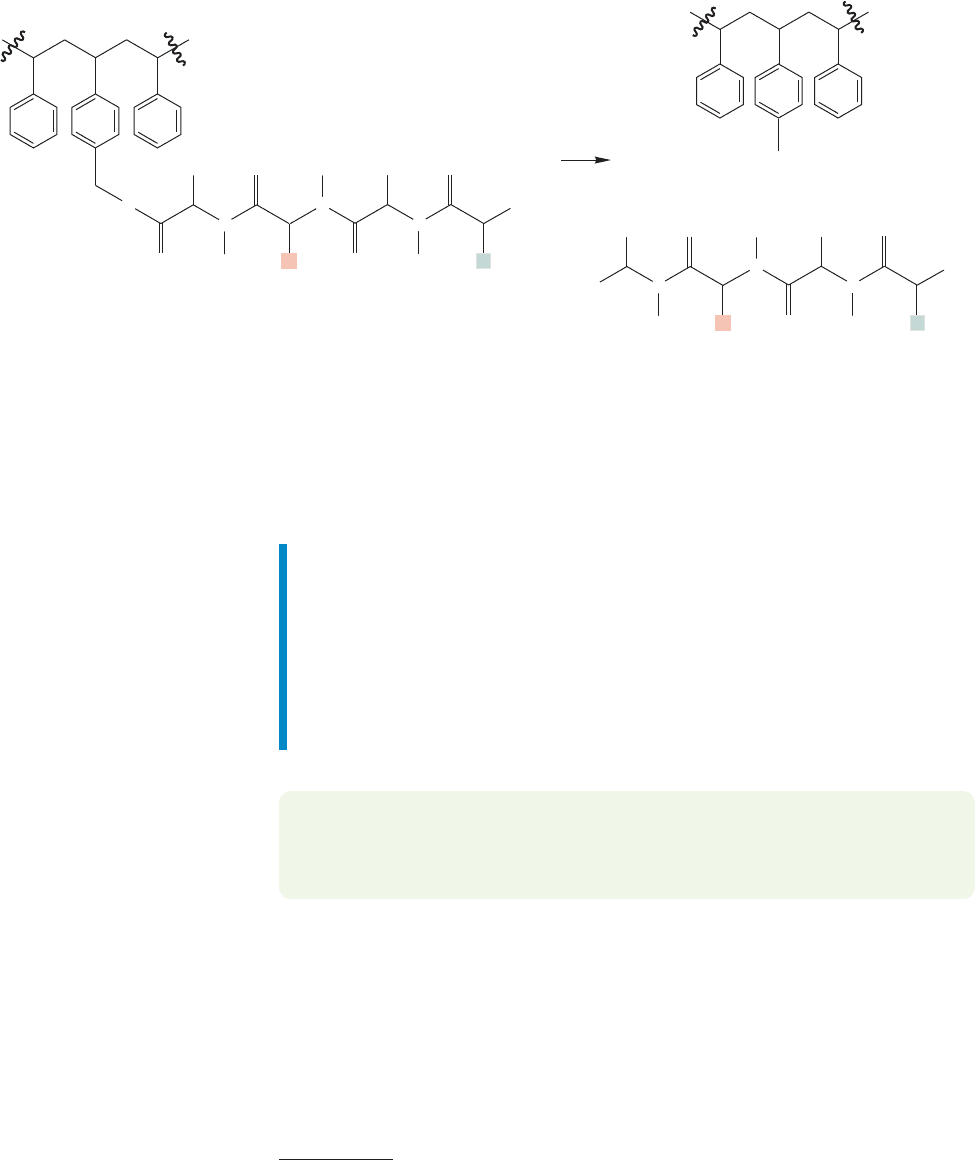
FIGURE 23.53 When synthesis is complete, the polypeptide can be detached from the polymer backbone by reaction with HF.
is detachment of the peptide from the polystyrene resin, usually through reaction
with hydrogen fluoride or another acid (Fig. 23.53).
Summary
Proteins have primary, secondary, tertiary, and quaternary structure.The primary
structure of a peptide can be determined by using a combination of enzymes and
small molecule reagents.There are several analytical tools that can also be applied
to this task.Peptides can be synthesized by employing protecting groups and acti-
vating reagents.The common procedures are: amide formation with tBoc or Cbz
for amine protection, esterification with ethanol for carboxylic acid protection,
and acid activation with DCC.
PROBLEM 23.23 Devise a synthesis of the dipeptide starting from the
amino acids leucine, alanine, and any other reagents described in this chapter.
Mechanisms are not necessary.
Leu
.
Ala
23.5 Nucleosides, Nucleotides, and Nucleic Acids
In Section 23.4c, the synthesis of proteins carried out by humans was described. In
the natural world
4
quite another technique is used, and the uncovering of how it
works is one of the great discoveries of human history.
In order to understand the biological synthesis of proteins, we must first learn
about the basic building block called a nucleoside. A nucleoside is a β-glycoside
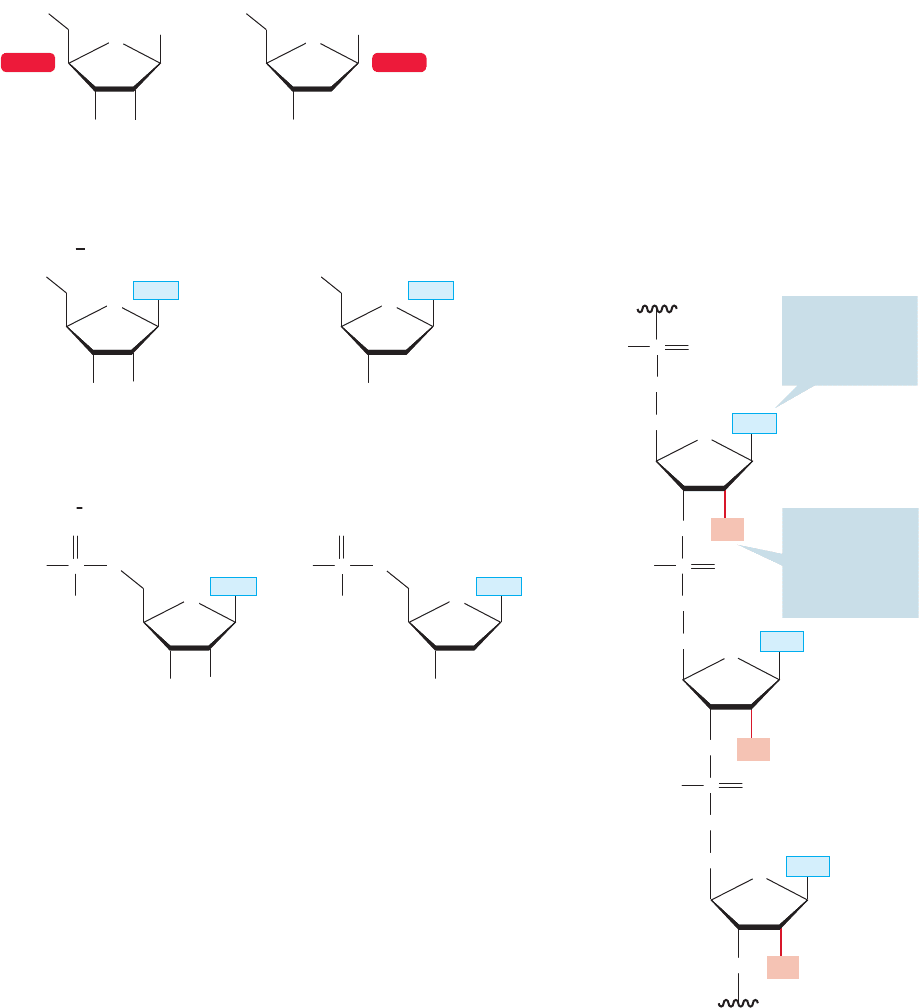
23.5 Nucleosides, Nucleotides, and Nucleic Acids 1211
1´
2´
3´
4´
5´
1´
2´
3´
4´
5´
O
OH
OH
HO
Base
O
OH
HO
Base
Nucleotides
–
–
O
O
O
O
P
–
–
O
O
O
O
P
O
OH
OH
HO
HO
1´
2´
3´
4´
5´
1´
2´
3´
4´
5´
Nucleosides
A ribonucleoside A deoxyribonucleoside
Base
O
OH
HO
HO
Base
––
Sugars
-Ribofuranose
-2-Deoxyribofuranose
(no oxygen at the 2-position)
O
OH
OH
HO
HO
1
2
3
4
5
O
OH
HO
HO
WEB 3DWEB 3D
The nucleic acids, deoxyribonucleic acid (DNA) and ribonucleic acid (RNA),
are polymers of nucleotides. The sugars in both nucleic acids, ribose in RNA and
deoxyribose in DNA,are linked to each other through phosphoric acid groups attached
to the 5¿ position of one sugar and the 3¿ position of the other (Fig.23.55). Each sugar
carries a heterocyclic base attached to the 1¿ carbon.
One of the striking features of this replication machine, for that is exactly what
we have begun to describe, is the economy with which it operates. So far, we have
phosphoric acid and two sugars, ribose and deoxyribose, held together in poly-
meric fashion. One might be pardoned for imagining that all manner of bases
are attached to this backbone, and that the diversity of the world of living things
CH
2
Base
1´
2´
3´
4´
5´
..
..
–
O
O
P
..
..
..
..
..
CH
2
Base
1´
2´
3´
4´
5´
..
–
O
O
P
..
..
..
..
..
OH
CH
2
Base
1´
2´3´
4´
5´
..
..
–
O
O
O
P
..
..
..
..
..
..
..
..
..
O
..
..
O
O
..
O
O
O
..
..
O
..
..
O
..
..
..
..
OH
..
..
OH
..
..
This OH at the
2' position is
present in RNA,
but absent in
DNA
These bases
can be different
in RNA and
DNA
FIGURE 23.55 Nucleic acids (DNA
and RNA) are polymeric collections
of nucleotides.
(p. 1148) formed between a sugar and a heterocyclic molecule (referred to as the
base). A ribonucleoside is a nucleoside that has ribose as the attached sugar and
a deoxyribonucleoside has deoxyribose as the sugar. Nucleosides that are phos-
phorylated at the 5¿ position are called nucleotides (Fig. 23.54). The atoms of
the sugar are given the primed numbers (e.g., 5¿) because the atoms of the het-
erocyclic base are numbered first.
FIGURE 23.54 The sugars ribose and deoxyribose, and the
corresponding nucleosides and nucleotides.
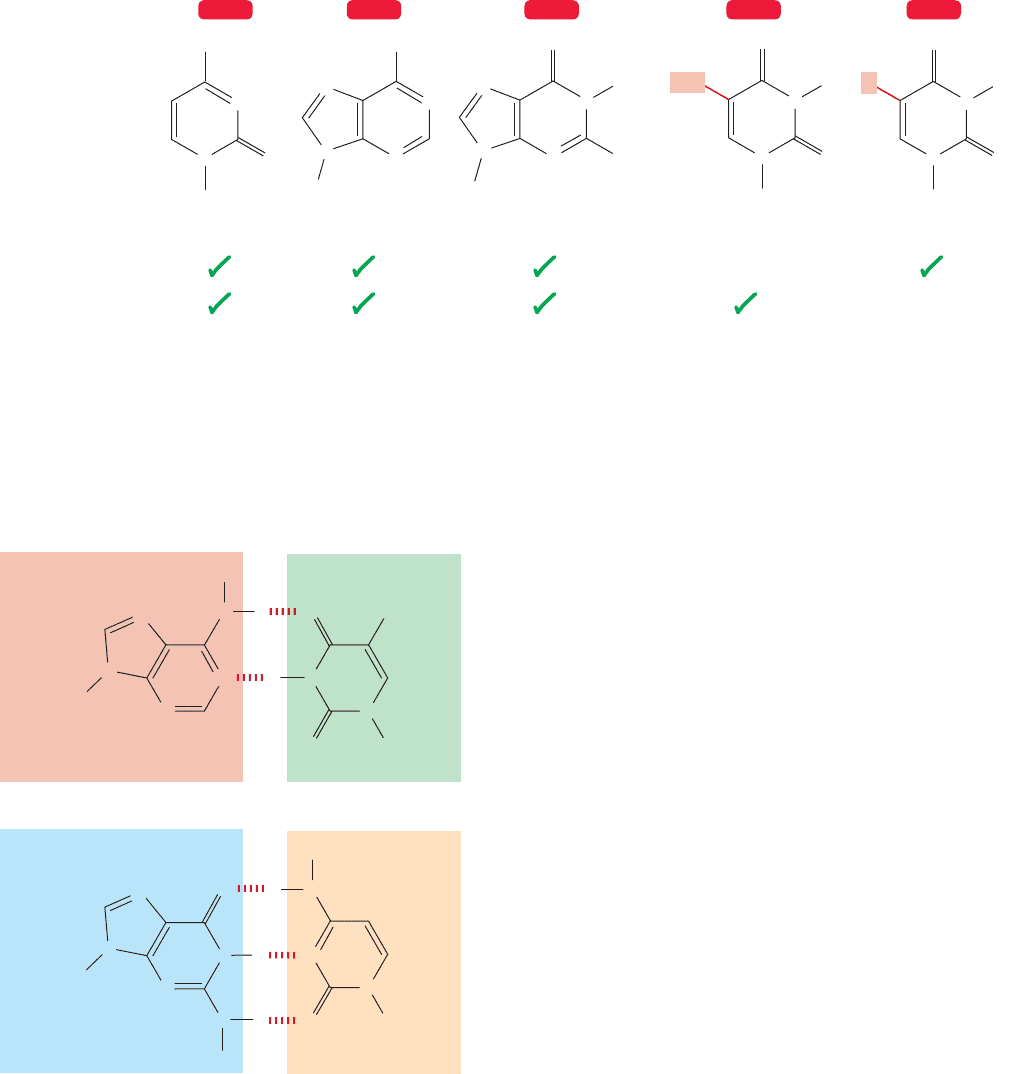
1212 CHAPTER 23 Amino Acids and Polyamino Acids (Peptides and Proteins)
In RNA
In DNA
Cytosine (C)
..
..
..
..
..
N
N
NH
2
O
H
Adenine (A)
..
..
..
..
N
N
..
N
NH
2
N
H
Guanine (G)
..
N
N
..
..
..
N
..
N
H
O
..
..
NH
2
H
Thymine (T)
..
..
..
N
N
O
H
..
O
..
..
H
H
3
C
WEB 3D WEB 3D WEB 3D WEB 3D
Uracil (U)
..
..
..
N
N
O
H
..
O
..
..
H
H
WEB 3D
FIGURE 23.56 The five bases present in RNA and DNA are shown.Three are common to both kinds of nucleic acid.
results from the introduction of detail in this way. But that idea would be wrong.
In fact, there are only five bases used in DNA and RNA, and three of them
(cytosine, adenine, and guanine) are common to both kinds of nucleic acid. The
other two, thymine which is in DNA and uracil which is in RNA, differ only in
the presence or absence of a single methyl group (Fig. 23.56). Not much diver-
sity there.
The bases are written in shorthand by using a single letter, C, A, G,T, or U.The
introduction of detailed information must come in another way.
Notice the similarities between these nucleic acid polymers and the protein poly-
mers.In the polyamino acids,the units are linked by amide bonds.In DNA and RNA
the nucleotides are attached by phosphates. The information
in proteins is carried by the side chains, the R groups, whose
steric and electronic demands determine the higher order
structures—secondary, tertiary, and quaternary—of these mol-
ecules. The information in DNA and RNA is carried by the
bases.There are only 20 common R groups for amino acids,but
there are even fewer bases—a mere four for each nucleic acid.
Nevertheless, higher ordered structures are also found in the
nucleic acids. Our next task is to see how this comes about.
A crucial observation was made by Erwin Chargaff
(1905–2002) who noticed in 1950 that in many different
DNAs the amounts of the bases adenine (A) and thymine (T)
were always equal, and the amount of guanine (G) was always
equal to the amount of cytosine (C). The four bases of
DNA seemed to be found in pairs.This observation led to the
reasonable notion that the members of the pairs must be asso-
ciated in some way, and to the detailed picture of hydrogen-
bonded dimeric structures of base pairs in which A is always
associated with T and G with C. These combinations are the
result of especially facile hydrogen bonding; the molecules fit
well together (Fig. 23.57).
The existence of base pairs shows how the order of bases
on one strand of DNA can determine the structure of anoth-
er strand that is hydrogen bonded to the first. For a sequence
A
T
G
C
..
..
..
N
N
..
..
N
H
H
H
H
CH
3
N
H
N
..
..
Deoxyribose
C(1')
Deoxyribose
C(1')
..
..
N
N
N
N
N
H
N
..
Deoxyribose
C(1')
Deoxyribose
C(1')
N
H
H
N
..
..
N
N
..
..
O
..
..
O
..
..
O
..
..
..
O
..
..
FIGURE 23.57 Adenine–thymine and guanine–cytosine form
base pairs through effective hydrogen-bond formation.
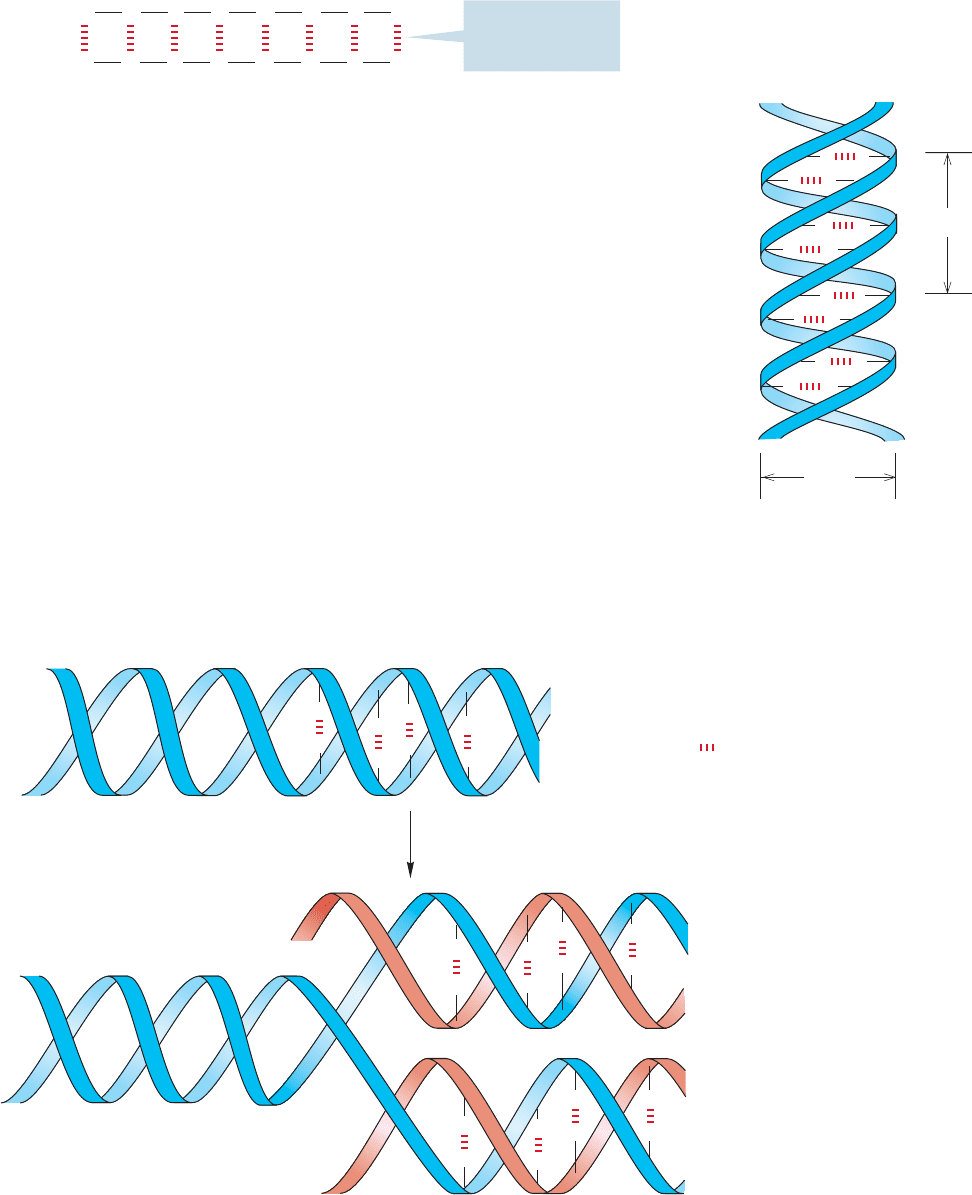
23.5 Nucleosides, Nucleotides, and Nucleic Acids 1213
FIGURE 23.58 The sequence of nucleotides on one polymer determines
the sequence in another through formation of hydrogen-bonded base
pairs, C-G and A-T.
of bases C C A T G C T A, the complementary sequence must be G G T A C G
A T in order to form the optimal set of hydrogen-bonded base pairs (Fig. 23.58).
~34 A
⬚
~20
AT
TA
GC
GC
CG
CG
A
TA
T
A
⬚
FIGURE 23.59 The famous double
helix of DNA is held together by
hydrogen bonds between base pairs.
A
T
unwind blue strands
New (red) strands
are assembled in an order
determined by the original
sequences of nucleotides
Two strands of DNA in a
double helix held together by
hydrogen bonds between
base pairs
C
A
T
A
T
A
T
A
T
G
C
G
C
G
C
G
C
A
T
G
C
G
FIGURE 23.60 The DNA replication machine works by first unraveling to form single-stranded sections. Assembly of
new strands is determined by the original base sequence. Where there is an A, a T must be added; a G requires a C.
C C A T G C T A
GGTACGAT
Hydrogen-bonded
base pairs as in
Figure 23.57
This notion was also instrumental in the realization by James D. Watson (b. 1928)
and Francis H. C. Crick (1916–2004), with immense help from X-ray diffraction pat-
terns determined by Rosalind Franklin (1920–1958) and Maurice Wilkins (1916–2004),
that the structure of DNA was a double-stranded helix in which the two DNA chains
were held together in the helix by A-T and G-C hydrogen-bonded base pairs. The
diameter of the helix is about 20 Å, and there are 10 nucleotides per turn of the helix.
Each turn is about 34 Å long (Fig. 23.59). The DNA chain is our genetic code.
We still have two outstanding problems: (1) Where does the density of infor-
mation come from? How is the seemingly inadequate pool of five bases translated
into the wealth of information that must be transferred in the synthesis of proteins
containing specific sequences made from the 20 amino acids? (2) What is the mech-
anism of the information transferal? The second of these two questions is the eas-
ier to tackle, so we will start there.
The replication of DNA involves partial unwinding of the double helix to yield two
templates for the development of new DNA strands.The order of bases in each strand
determines the structure of the new strand.Where there is a T,an A must appear,where
there is a G, a C must attach, and so on. Each strand of the original DNA must induce the
formation of an exact duplicate of the strand to which it was originally attached (Fig. 23.60).
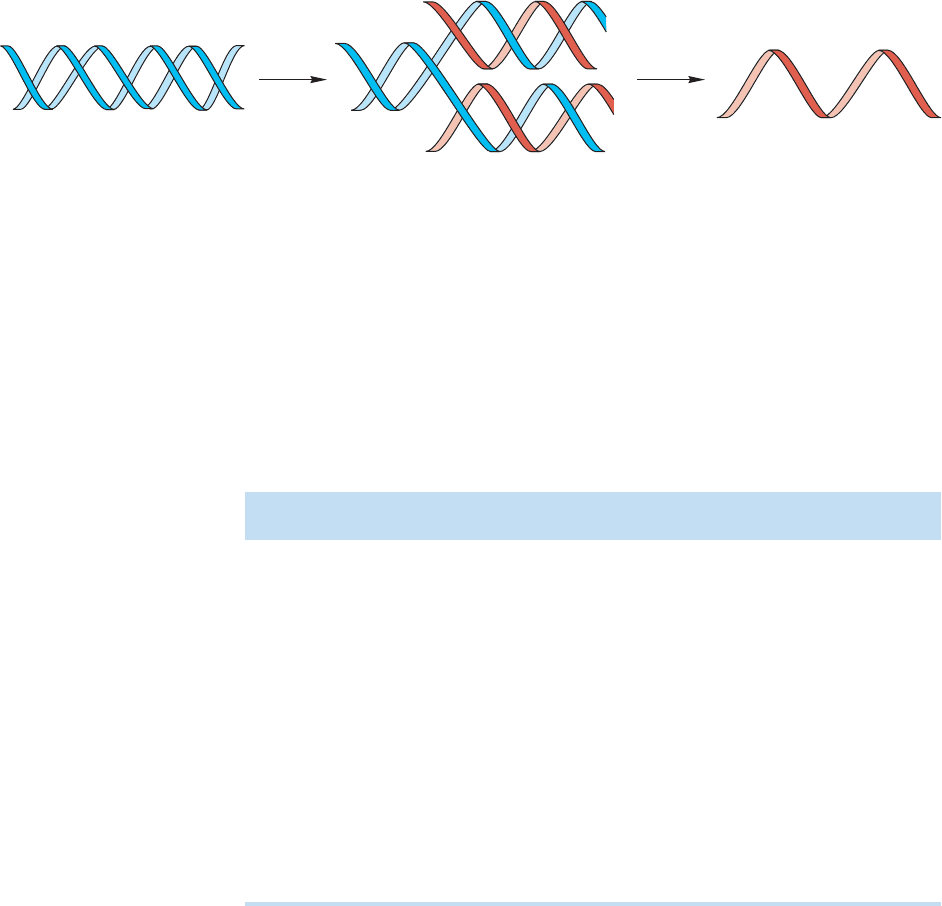
1214 CHAPTER 23 Amino Acids and Polyamino Acids (Peptides and Proteins)
Protein synthesis starts with a similar, but not quite identical process that
involves RNA. A DNA strand can also act as a template for the production of
a strand of RNA. In the RNA construction the base uracil is used instead of
thymine. The RNA produced is called messenger RNA (mRNA) (Fig. 23.61).
mRNA molecules
DNA
Now replication uses a ribose, not deoxyribose, based
nucleotide. In RNA, the bases are A, C, G, and U not T
FIGURE 23.61 Synthesis of mRNA.
TABLE 23.2 Codons for Amino Acids and Simple Commands
First Second Base Third
Base in Sequence U C A G Base in Sequence
UPheSerTyrCysU
Phe Ser Tyr Cys C
Leu Ser Stop Stop A
Leu Ser Stop Trp G
CLeuProHisArgU
Leu Pro His Arg C
Leu Pro Gln Arg A
Leu Pro Gln Arg G
A Ile Thr Asn Ser U
Ile Thr Asn Ser C
Ile Thr Lys Arg A
Met
a
Thr Lys Arg G
G Val Ala Asp Gly U
Val Ala Asp Gly C
Val Ala Glu Gly A
Val Ala Glu Gly G
a
The AUG codon translates to Met when in the middle of a message, but it translates as the Start message when it
is at the beginning of a sequence.
This new RNA molecule directs protein synthesis in the following spectacularly
simple way: Synthesis of each of the 20 common amino acids in proteins is directed
by one or more sequences of three bases, called codons. How many codons can be
constructed from the available four RNA bases? The answer is 4
3
64, more than
enough to code for the 20 common amino acids.Indeed, some amino acids are coded
by more than one codon. Some extra codons are needed to deal with the problems
of when to start amino acid production and when to stop.Table 23.2 gives the three-
base codons for the 20 common amino acids and the frequently used commands.
As an example of how this process works, let’s look at a strand of DNA with the
sequence T A C C G A A G C A C G A T T. Base pairing must produce the mRNA
fragment A U G G C U U C G U G C U A A (remember that in RNA the base U
replaces T). A look at Table 23.2 shows that the first codon, A U G, is interpreted as the
Start command (find the first letter of the codon in the left column, the middle letter of
the codon from the corresponding column, and the final letter from the right column).
The second codon of this mRNA fragment is G C U which corresponds to Ala, the
next codon is U C G which corresponds to Ser, then codon U G C which corresponds
to Cys, and the last codon U A A which instructs the peptide synthesis to Stop. The
fragment translates into the following message: “Start-add Ala-add Ser-add Cys-Stop.”

23.6 Summary 1215
23.6 Summary
New Concepts
Proteins are α-amino acids assembled into long polymeric chains.
The secondary structure of these peptides or proteins involves the
regions of α-helical or β-pleated sheet arrangements, which are
separated from each other by disordered sections of the chains
called random coils. Disulfide bonds, electrostatic forces, van der
Waals forces, and hydrogen bonding twist these molecules into
shapes characteristic of individual proteins, the tertiary structure.
Secondary and tertiary structure can be destroyed, sometimes only
temporarily, by any of a number of denaturing processes. Finally,
intermolecular forces can hold a number of these protein chains
together to form supermolecules, the quaternary structure.
These higher ordered structures are affected by the identities
of the R groups on the constituent amino acids. It is the electronic
and steric properties of the R groups that generate the particular
secondary, tertiary, and quaternary structures of proteins.
These new structures require new analytical and synthetic
techniques. Methods for determining the sequence of peptides
and proteins involve physical techniques such as X-ray diffraction,
as well as chemical techniques for revealing the terminal amino
acids of the chains, and even for sequencing entire polymers.
Other biopolymers, the nucleic acids, are constructed not of
amino acids, but of nucleotides, which are sugars bearing het-
erocyclic base groups and joined through phosphoric acid link-
ages. These nucleic acids also have higher order structures, the
most famous being the double-helical arrangement of DNA.
Most remarkable of all is the ability of these molecules to carry
the genetic code, to uncoil and direct the assembly not only of
replicas of themselves, but also, through the mediation of RNA,
of the polyamino acids called proteins. In the nucleic acids, the
information is not carried in a diverse supply of R groups as it
is in peptides and proteins, but in a small number of base
groups. Hydrogen bonding in base pairs allows for the replica-
tion reactions, and three-base units called codons direct the
assembly of amino acids into proteins.
WORKED PROBLEM 23.24 Why would 20 amino acids be coded for by 64 three-
base sequences? Why is there so much overlap? Consider the alternative method
of having two-base sequences coding for amino acid production. How many pos-
sible two-base sequences are there for four bases?
ANSWER There are only 2
4
possible combinations of four objects taken two at a
time. So there are only 16 things that could be coded by two-base sequences of
four different bases, which is not sufficient to code for the 20 common amino
acids that make up proteins.Three-base codons turn out to be the most econom-
ical way to code for a system of 20 amino acids.
One further problem remains. How does mRNA direct amino acid synthesis?
The answer is that another type of RNA is used,called transfer RNA (tRNA),which
is relatively small and designed to interact with an enzyme, aminoacyl tRNA syn-
thetase, to acquire one specific amino acid and carry it to the mRNA where it is
added to the growing chain at the position of the correct codon.
Key Terms
α-amino acids (p. 1175)
amino terminus (p. 1189)
base pair (p. 1212)
binding site (p. 1193)
tBoc (p. 1205)
carboxy terminus (p. 1189)
Cbz (p. 1205)
codon (p. 1214)
cyanogen bromide (BrCN) (p. 1200)
denaturing (p. 1193)
deoxyribonucleic acid (DNA) (p. 1211)
dicyclohexylcarbodiimide (DCC)
(p. 1205)
disulfide bridges (p. 1190)
Edman degradation (p. 1198)
electrophoresis (p. 1180)
essential amino acid (p. 1177)
Gabriel synthesis (p. 1182)
gel-filtration chromatography (p. 1195)
α-helix (p. 1191)
ion-exchange chromatography (p. 1195)
isoelectric point (pI ) (p. 1179)
kinetic resolution (p. 1186)
messenger RNA (mRNA) (p. 1214)
ninhydrin (p. 1187)
nucleic acid (p. 1211)
nucleoside (p. 1210)
nucleotide (p. 1211)
peptide bond (p. 1175)
β-pleated sheet (p. 1191)
primary structure (p. 1190)
quaternary structure (p. 1194)
random coil (p. 1191)
ribonucleic acid (RNA) (p. 1211)
Sanger degradation (p. 1196)
secondary structure (p. 1191)
side chain (p. 1175)
Strecker synthesis (p. 1184)
tertiary structure (p. 1192)
thiourea (p. 1198)
zwitterion (p. 1179)
1216 CHAPTER 23 Amino Acids and Polyamino Acids (Peptides and Proteins)
Syntheses
Reactions, Mechanisms, and Tools
Reactions important in determining the amino acid sequence of
a protein include several techniques for end-group analysis.
There are enzymes (proteins themselves) that cleave the amino
acid at the carboxy terminus of the polymer. The amino termi-
nus can be found through the Sanger reaction.
In principle, an entire sequence can be unraveled using the
Edman degradation in which phenylisothiocyanate is used in a
procedure that cleaves the amino acid at the amino terminus
and labels it as a phenylthiohydantoin.
Other agents are available for inducing specific cleavages
in the amino acid chain that yield smaller fragments for
sequencing through the Edman procedure. These agents
include various enzymes and small chemical reagents such
as cyanogen bromide.
Synthesis of these polymeric molecules requires nothing
more than the sequential formation of a large number of amide
bonds. The DCC coupling procedure is effective, but it requires
that the amino acid monomers be supplied in a form in which
the ends to be attached are activated and other reactive points
blocked, or protected. Those protecting and activating groups
must be added in appropriate places and removed after the
reaction is over.
If we need to synthesize the amino acids themselves, the
best known routes are the Gabriel and Strecker procedures.
Naturally occurring amino acids are optically active (except
for glycine, which is achiral) and occur in the
L form.
Resolution can be achieved through the classical technique of
diastereomer formation followed by crystallization, or through
other methods. The most clever of these involves a kinetic reso-
lution in which advantage is taken of the ability of enzymes to
attack only the naturally occurring (S) enantiomers, while the
(R) isomer is left alone.
1. Amino Acids
Strecker synthesis
O
O
–
R
+
NH
3
–
2. R
—
X, base
1.
3. H
3
O/H
2
O/⌬
+
Gabriel synthesis. The R
—
X must be S
N
2 active
O
O
–
R
+
NH
3
1. KCN/NH
3
2. H
2
O/H
3
O
+
ROOC
ROOC
Br
CHOR
C
O
N
C
O
O
R
OH
Simple alkylation of ammonia using an α-bromo
acid; overalkylation is a problem
Br
O
O
–
R
1. NH
3
2. neutralize
+
NH
3
2. Disulfide Bridges
thiols or
air
(oxygen)
O
O
NH
NH
SH
O
R
O
SH
NH
O
O
NH
NH
S
O
R
O
R'
S
NH
NH
Oxidation can convert the SH groups of cysteines into disulfide bonds between or within chains
NH
R'

23.6 Summary 1217
3. Optically Active Amino Acids
1. optically
active
alkaloid
2. separate
diastereomers
3. regenerate
amino acids
O
Racemic mixture
of (R) and (S)
O
–
R
+
NH
3
(R)
(S)
O
–
R
+
NH
3
O
Classical resolution
O
–
R
O
+
NH
3
+
1. Ac
2
O/HOAc
2. hog-kidney
acylase
O
(R,S)
O
–
R
+
NH
3
(R)
(S)
O
–
R
+
NH
3
O
The free (S) amino acid is produced in this kinetic
resolution, along with acylated (R) amino acid
OH
R
O
NHAc
+
glutamate
dehydro-
genase
NH
3
,
NADH
(S)
A typical enzymatic synthesis of only the (S) enantiomer of
an amino acid through reductive amination
O
O
HOOC
OH
NH
3
+
HOOC
COO
–
4. Protected Amino Acids
O
O
–
R
O
H
N
R
OH
O
A tBoc-protected
amino acid
(CH
3
)
3
CO
(CH
3
)
3
CO
H
3
N
O
O
OC(CH
3
)
3
O
+
O
O
–
R
A Cbz-protected amino acid
PhCH
2
O
H
3
N
Cl
O
+
O
H
N
R
OH
O
PhCH
2
O
1. DCC
2. removal of
protecting
groups
O
H
2
N
H
3
N
OR
R
O
COO
–
O
H
N
RR
OH
+
R
N
H
+
P
P = tBoc or Cbz
DCC-Mediated coupling; notice that the starting amino acids are prepared
for reaction with protecting groups that are removed in later steps
1218 CHAPTER 23 Amino Acids and Polyamino Acids (Peptides and Proteins)
23.7 Additional Problems
RNH
2
CNO
NHR
Addition of an amine to an isocyanate
C
O
H
2
O
CN
R
R
N
NHR
RHN
Hydration of a carbodiimide
C
O
R
RHN
7. Urea s
(a)
O
N
H
OH
COO
–
H
3
N
+
(b)
O
OH
N
H
COO
–
H
3
N
H
2
N
+
PROBLEM 23.28 Draw the 3-D perspective of (S)-serine and
(S)-proline using solid and dashed wedges.
PROBLEM 23.29 Write Fischer projections for L-serine and
L-proline.
–
+
N
1
O
O
PROBLEM 23.30 Give the structures for the amino acids His,
Arg, and Phe at pH 3 and 12.
PROBLEM 23.31 Use the pI data in Table 23.1 to determine
which electrode (anode or cathode) the amino acids Asp and
Lys will migrate toward at pH 7.
PROBLEM 23.32 All of the amino acids listed in Table 23.1,
except the achiral glycine, have the
L configuration. Of the
19 amino acids with the
L configuration, 18 are (S) amino
acids. Which amino acid has an (R) configuration? Explain
why the configuration of this
L-amino acid is (R) rather than (S).
PROBLEM 23.33 In Section 23.3c, we saw that all the amino
acids in Table 23.1, except proline, react with ninhydrin to form
the same purple compound. In contrast, proline reacts with
ninhydrin to give a yellow compound thought to have structure
1. Propose a mechanism for the formation of 1.
PROBLEM 23.25 (a) Amides protonate on oxygen rather than
nitrogen. Why? (b) Amides (pK
b
~14) are weaker bases than
amines (pK
b
~5). Why?
PROBLEM 23.26 Write structures for the following tripeptides
and name them:
(a) (b) (c)
PROBLEM 23.27 Name the following dipeptides:
Val
.
Asp
.
HisMet
.
Phe
.
ProAla
.
Ser
.
Cys
PROBLEM 23.34 (a) A peptide of unknown structure is treat-
ed with the Sanger reagent, 2,4-dinitrofluorobenzene, followed
by acid hydrolysis.The result is formation of a compound con-
taining signals integrating for eight aromatic hydrogens in its
1
H NMR spectrum. What information can you derive about
the structure of this peptide?
5. Thiohydantoin
Ph
NCS
R
O
O
O
O
–
O
–
NH
3
+
OS
N
N
H
Ph
R
The Edman degradation
H
3
N
+
+
NH
R
R
RNH
2
CNS
NHR
Addition of an amine to an isothiocyanate
C
S
R
RHN
6. Thioureas
