Jean-Laurent Mallet. Geomodeling
Подождите немного. Документ загружается.

18
CHAPTER
1.
DISCRETE MODELING
FOR
NATURAL OBJECTS
Generalization
of the
DSI
method
In
practical applications,
the
function
(p
of the
discrete model
Ai
n
(0,
TV,
<^,
C]
may
have several components interacting through "cross" constraints
(see
page
9).
Thus
the
simplified
version
of the DSI
method presented
in the
previous section
has to be
extended
somewhat
to
take
into
account
the
mul-
ticomponent case where:
The
multicomponent problem
can be
solved with generalized local
DSI
equa-
tions identical
in
form
to the one
obtained
in the
case where
(p
has
only
one
component.
As
will
be
seen
later
on, a
slight change
in the
notations
used
in
the
definition
of the
local roughness criterion
R((f>\a]
together with
the
degree
of
violation
of the
constraints
p(f\c)
suffices:
In
the
above
definition
of the
roughness
R(y>\a),
r
v
is a
given
positive weight-
ing
coefficient
allowing
the
contribution
of the
function
<£>"(•)
to the
local
roughness
R((p\ot)
to be
modulated.
If the
functions
{^{-)}
have approxi-
mately
the
same magnitude,
then
it is
possible,
for
example,
to
choose
all the
coefficients
{r"}
equal
to 1.
The
equation allowing
the
value
(p
v
(oi)
in the
iterative
DSI
algorithm
to
be
updated remains almost unchanged
in its
form
(see
equation
(4.46))
and
becomes:
The
only noticeable
difference
in the
monocomponent case
is a
slight
modifi-
cation
of the
term
T
v
(a\(f>)
in the
case
of
cross constraints involving several
components
of
(p
(see page
163).
When there
is no
such cross constraint
at-
tached
to the
node
a,
then
the
above
formulas
are
absolutely identical
to the
ones
in the
monocomponent case.
Implementation
of the DSI
method
In
spite
of its
apparent complexity,
the
local
DSI
equation
is
very simple
to
program
and the
very heart
of the
iterative algorithm introduced
on
page
17
can be
implemented
in
about
30
lines
of
source code.
The
real programming
difficulty
comes
from
the
discrete model
.A/l
n
(O,
N,
<p,C),
which
has to be
implemented
to
•
optimize
the
access
to the
neighborhoods
N(a)
and the
values
<p"(oi)
used
by
the DSI
equation,
and
•
take
into
account
its
"self-consistency"
when modifications
occur
in
Q,
AT(-),
</?,
or C.

1.4.
EXAMPLES
OF
APPLICATIONS
19
Figure
1.10
Each
point
Pi is
assumed
to
hold
data
4>(Pi)
which
are
projected
onto
the
triangulated
surface
in
direction
A(Pi)
specific
to Pi.
Data
4>(Pi)
are
transformed
into
constraints
affecting
the
nodes
corresponding
to the
vertices
of
the
triangle
hit by
A(Pi).
If
the
direction
A(Pi)
is
changed,
then
a
different
triangle
is
hit and the
constraints
have
to be
updated.
Implementing
a
totally
general algorithm
that
takes
into account
these
two
items
may
result
in a
large complex software with more
than
100
thousand
lines
of
source code playing
the
role
of a
server
for the 30
lines corresponding
to the
DSI
algorithm. Such heavy machinery
is not
always necessary
and its
presentation
is
beyond
the
scope
of
this book.
A
look
at
figure
(1.10) gives
an
idea
of the
nature
of the
problem: this
figure
represents
a
triangulated surface
and a set of
data
points
{Pi,
P
where
each point
Pi is
assumed
to
hold
data
0(P;)
that
are
projected onto
the
triangulated
surface
in a
direction
A(P^)
specific
to Pi. The
data
tf>(Pi) are
transformed
into constraints
affecting
the
nodes corresponding
to the
vertices
of
the
triangle
hit by
A(Pj).
As a
result
of
this
situation:
• If the
direction
A(Pi)
is
changed,
then
a
different
triangle
is hit and the
associated
constraint
c
(E
C has to be
updated.
• If the
density
of the
nodes
in a
region
of the
model
is
changed
to
obtain
a
better
numerical
approximation,
then
the set fi and the
neighborhoods
{N(a)
:
a G
Q}
are
changed
and it is
necessary
to
update
all the
constraints
c
G
C in
order
to
take into account these global
modifications.
The
implementation
may be
even more complex
if, at
each
DSI
iteration,
the
direction
of
projection
A(Pj)
is
optimized automatically
to hit the
closest
point
of the
surface.
1.4
Examples
of
applications
Applications
of
discrete models
and
Discrete Smooth Interpolation introduced
in
this chapter
are
numerous,
and the
choice
of a
specific
example
is
difficult.
20
CHAPTER
1.
DISCRETE MODELING
FOR
NATURAL OBJECTS
Figure
1.11
Modeling salt dome
with
DSI:
(A)
initial model
and
data.
(B) final
model. (Data courtesy
of
T-Surf)
Figure
1.12
Modeling seismic velocities
in a
parallelepipedic volume
of the
subsurface.
The
volume
is filled by a
rectilinear
3D
grid
cut by
geological horizons
and
faults. Variations
of
the
velocities stored
in the
grid
are
represented
by
varying
degrees
of
shading (EAEG-overthrust model
[6]).
In
this section,
we
present some examples
of
particularly interesting applica-
tions
in the
realm
of
subsurface
and
natural object modeling.
Interpolation problems
The
most straightforward
and
frequent
use of DSI is in
interpolating problems
related
to the
geometry
and
properties
of
natural objects.
As an
example
of
this kind
of
application,
figures
(1.11)
and
(1.12) show
two
discrete models
corresponding
to the
modeling
of
geological structures:

1.4.
EXAMPLES
OF
APPLICATIONS
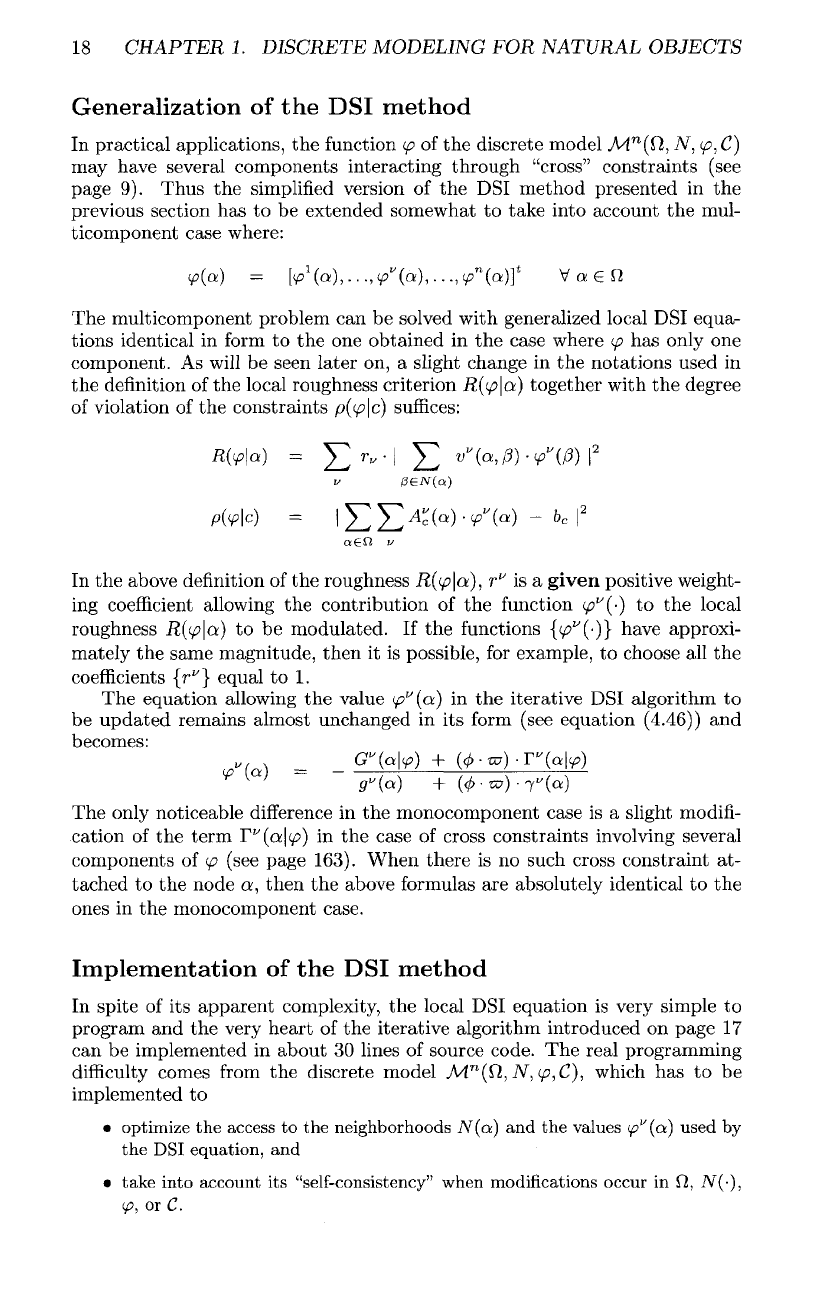
The first
example presented
in figure
(1.11)
is
related
to the
modeling
of
the
geometry
of a
salt
dome.
In figure
(l.ll)-A,
there
are
about
100
data
points
Pi,
which
are
assumed
to be
located
on the
surface
to be
constructed.
As
suggested
in figure
(1.10), each
of
these
data
points
is
projected onto
the
surface according
to a
specific
direction
A(P;),
hitting
a
triangle
Ti(ctQ,
&i,
a
2
]
at a
point
P*.
It is
possible
to
define
a
linear constraint
c —
c(i)
on the
nodes
(ao,
&i,
&%),
specifying
that
Pi
should
be
coincident with
P*.
The
modeling process
is
split into three
steps:
1.
First,
an
initial
discrete
model
.A/f
n
(f2,
JV,
<£,
C),
consisting
of a
rectan-
gular
triangulated
surface,
is
created
in the
neighborhood
of the
data
points
{Pi}.
2.
Each
data
point
Pi is
projected
onto
the
closest point
of
this initial
surface
and is
transformed into
a
constraint
c —
c(i)
G C
applied
to the
components
(<p
x
,
ip
y
,
<f>
z
)
that
corresponds
to the
location
of the
nodes
of
the
surface.
3.
The
DSI
algorithm
is
applied.
After
convergence
if
necessary,
to
give
more flexibility
to the
surface,
the
triangles with
the
distance
d(Pi,P*)
bigger
than
a
given threshold
can be
split
before returning
to
item
(2)
above. Moreover,
as
suggested
in figure
(1.10), between
two
iterations
of
the DSI
algorithm,
it is
also possible
to
optimize
the
directions
of
projection
A(Pj).
After
some iterations
of
this modeling process,
we
obtain
the
model
represented
in figure
(l.ll)-B.
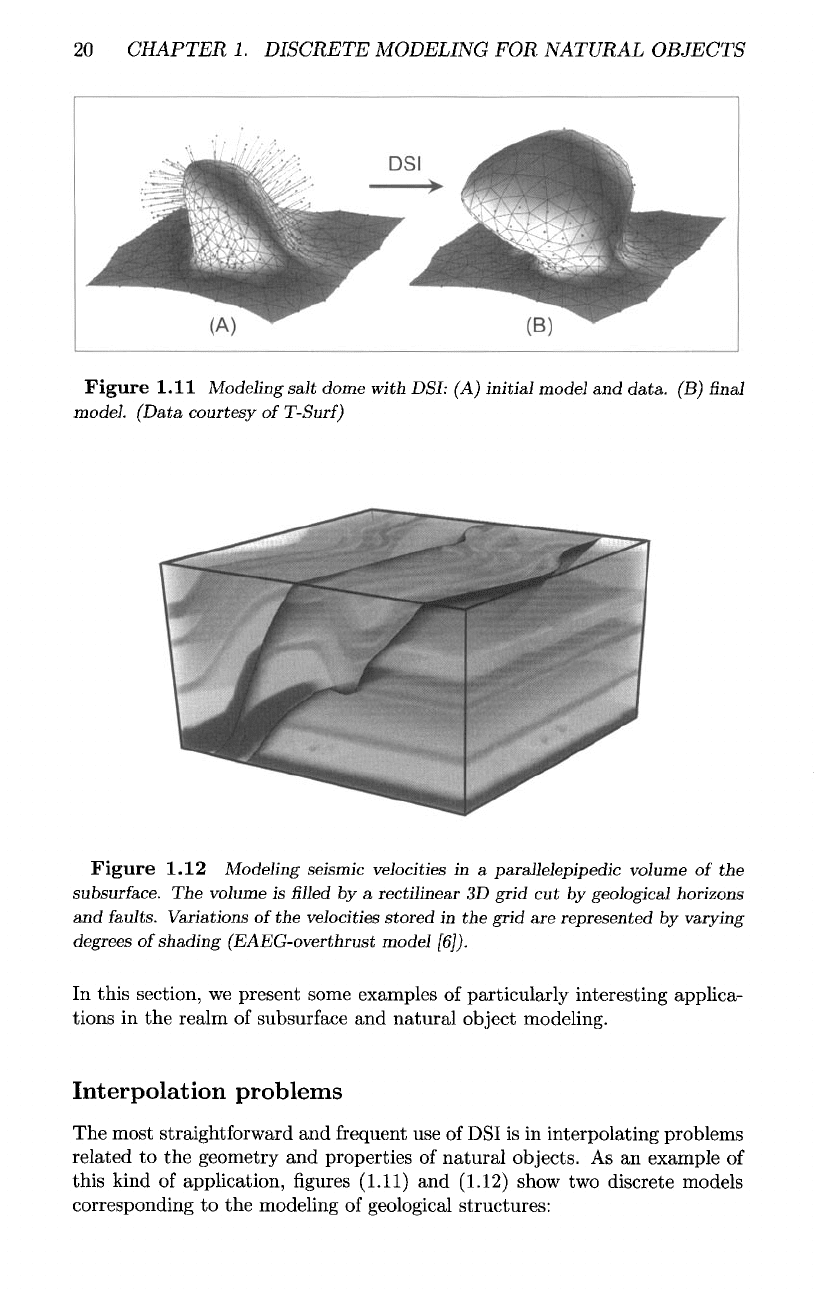
The
second example presented
in figure
(1.12)
corresponds
to the
mod-
eling
of a
seismic velocity
in a
parallelepipedic
volume
of the
subsurface.
This volume
is filled by a
regular rectilinear
3D
grid having
200
steps
in
the
^-direction,
200
steps
in the
y-direction
and 100
steps
in the z-
direction. This grid
is
assumed
to be cut by
some horizons
and
listric
faults
and the
velocities
(j)
v
(Pi}
are
known
at
about
150
points
{Pi}
irregularly distributed
in the
volume. Each
data
point
{Pi,(j)
v
(Pi}}
is
transformed
into
a
constraint
specifying
that
the
velocity
(f>
v
(a,o)
of the
closest node
ao
should
be
equal
to the
given value
0
v
(Pj).
The
varia-
tions
of the
velocities, restricted
to one
layer,
are
shaded
on the
faces
of
the
parallelepipedic volume shown
in figure
(1.12).
As you can
see,
all
the
discontinuities corresponding
to the
intersections
of the
grid with
the
horizons
and
faults
have been strictly honored.
However,
it
should
be
noted
that,
if
necessary, partial continuity
can be
maintained across
these surfaces
by
keeping
a few
connections
at
random between nodes
located
on
both sides;
in
this case,
the
density
of the
connections
to be
retained could
be
modeled
as a
property
of the
surfaces.
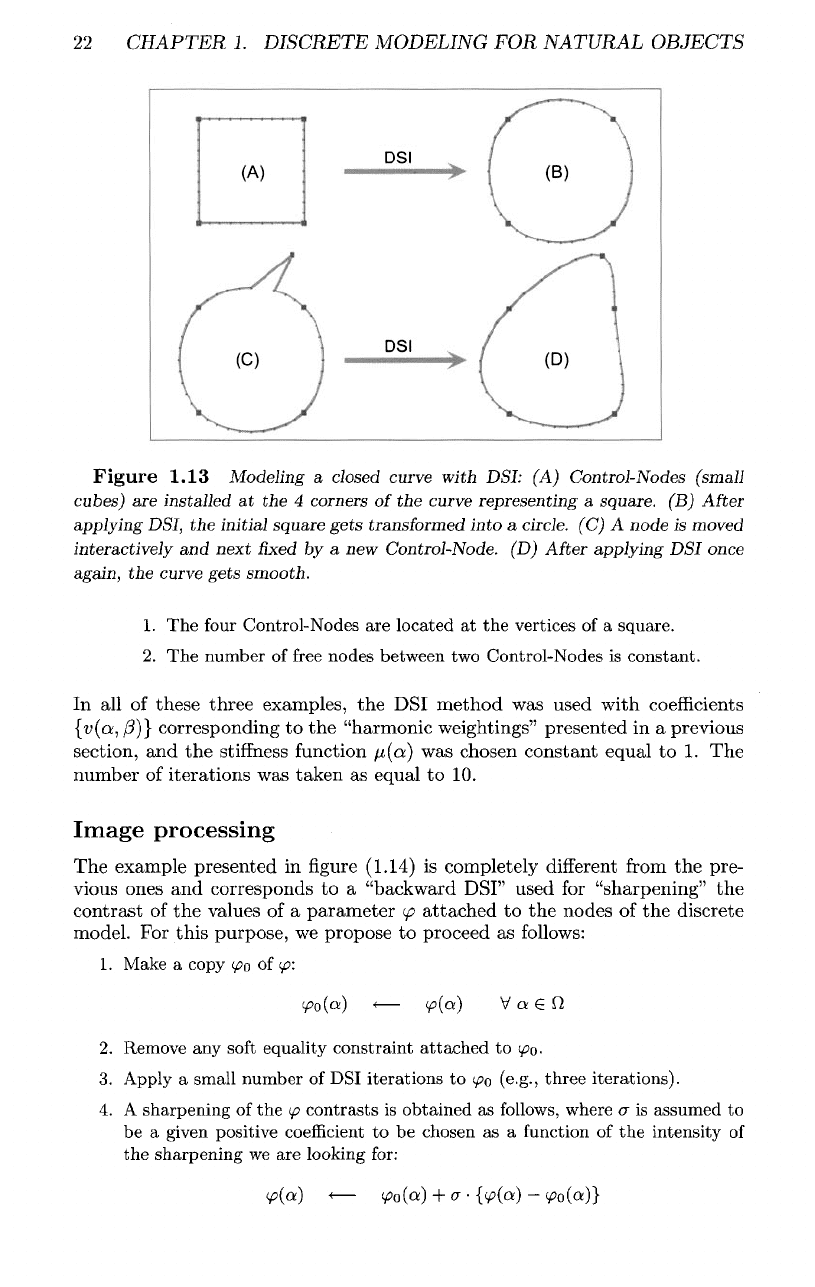
The
third example presented
in figure
(1.13)
is a
very simple tutorial
example showing
the
behavior
of the DSI
interpolation when modeling
a
closed polygonal curve controlled
by
four
Control-Nodes displayed
as
small cubes.
It can be
shown
that
the
resulting discrete curve
fits a
perfect
circle
if the two
following
conditions
are
respected:
21
22
CHAPTER
1.
DISCRETE MODELING
FOR
NATURAL
OBJECTS
Figure
1.13
Modeling
a
closed
curve
with
DSL (A)
Control-Nodes
(small
cubes)
are
installed
at the 4
corners
of the
curve representing
a
square.
(B)
After
applying
DSI,
the
initial square gets
transformed
into
a
circle.
(C) A
node
is
moved
interactively
and
next
fixed by a new
Control-Node.
(D)
After
applying
DSI
once
again,
the
curve gets
smooth.
1.
The
four
Control-Nodes
are
located
at the
vertices
of a
square.
2.
The
number
of
free
nodes between
two
Control-Nodes
is
constant.
In all of
these three examples,
the DSI
method
was
used with
coefficients
{v(a,
j3}}
corresponding
to the
"harmonic weightings" presented
in a
previous
section,
and the
stiffness function
//(a)
was
chosen constant equal
to 1. The
number
of
iterations
was
taken
as
equal
to 10.
Image processing
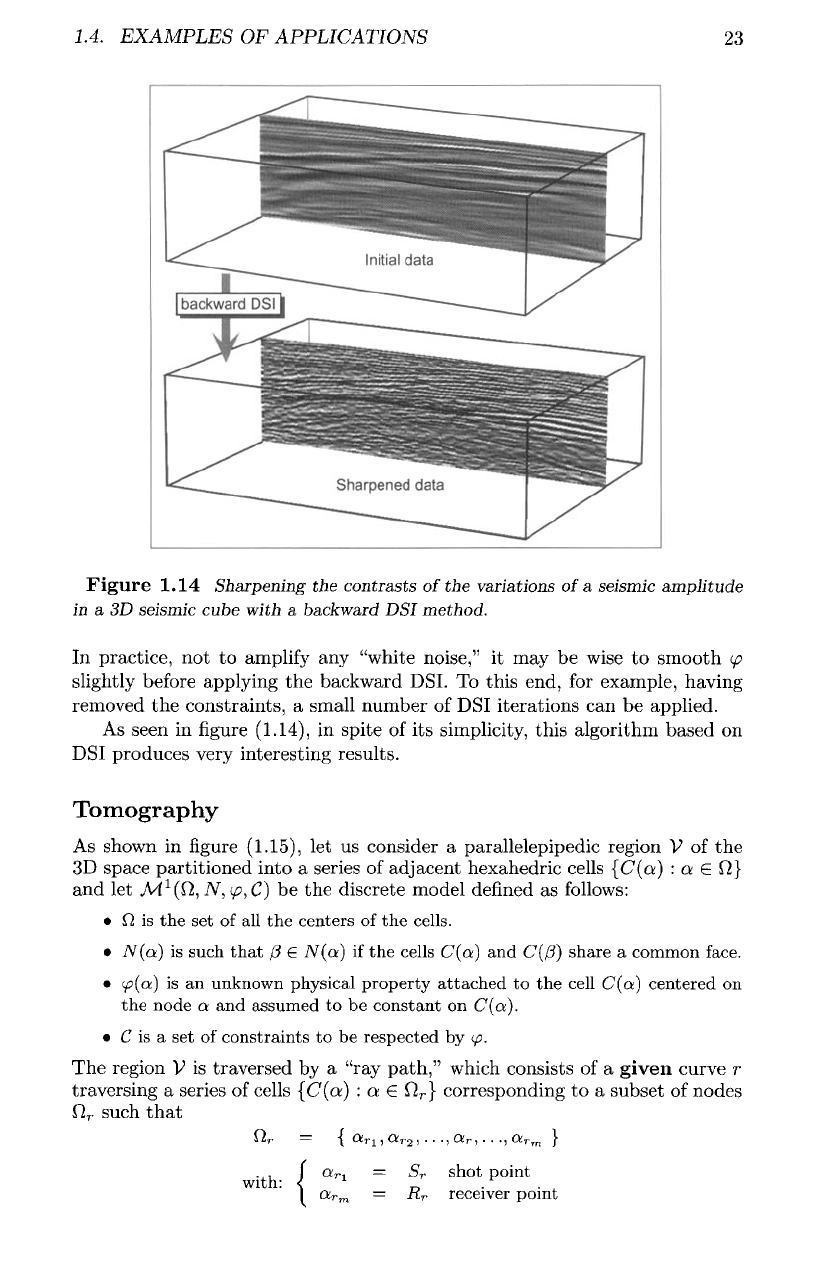
The
example presented
in
figure
(1.14)
is
completely
different from
the
pre-
vious ones
and
corresponds
to a
"backward DSI" used
for
"sharpening"
the
contrast
of the
values
of a
parameter
<p
attached
to the
nodes
of the
discrete
model.
For
this
purpose,
we
propose
to
proceed
as
follows:
2.
Remove
any
soft
equality constraint attached
to
(/?o-
3.
Apply
a
small number
of DSI
iterations
to
tpo
(e.g.,
three iterations).
4. A
sharpening
of the
(p
contrasts
is
obtained
as
follows,
where
a is
assumed
to
be a
given positive
coefficient
to be
chosen
as a
function
of the
intensity
of
the
sharpening
we are
looking
for:
1.
Make
a
copy
</?o
of
(p:
1.4.
EXAMPLES
OF
APPLICATIONS
23
Figure
1.14
Sharpening
the
contrasts
of
the
variations
of
a
seismic
amplitude
in
a 3D
seismic
cube
with
a
backward
DSI
method.
In
practice,
not to
amplify
any
"white noise,"
it may be
wise
to
smooth
(f>
slightly
before
applying
the
backward DSI.
To
this end,
for
example, having
removed
the
constraints,
a
small number
of DSI
iterations
can be
applied.
As
seen
in figure
(1.14),
in
spite
of its
simplicity, this algorithm based
on
DSI
produces very interesting results.
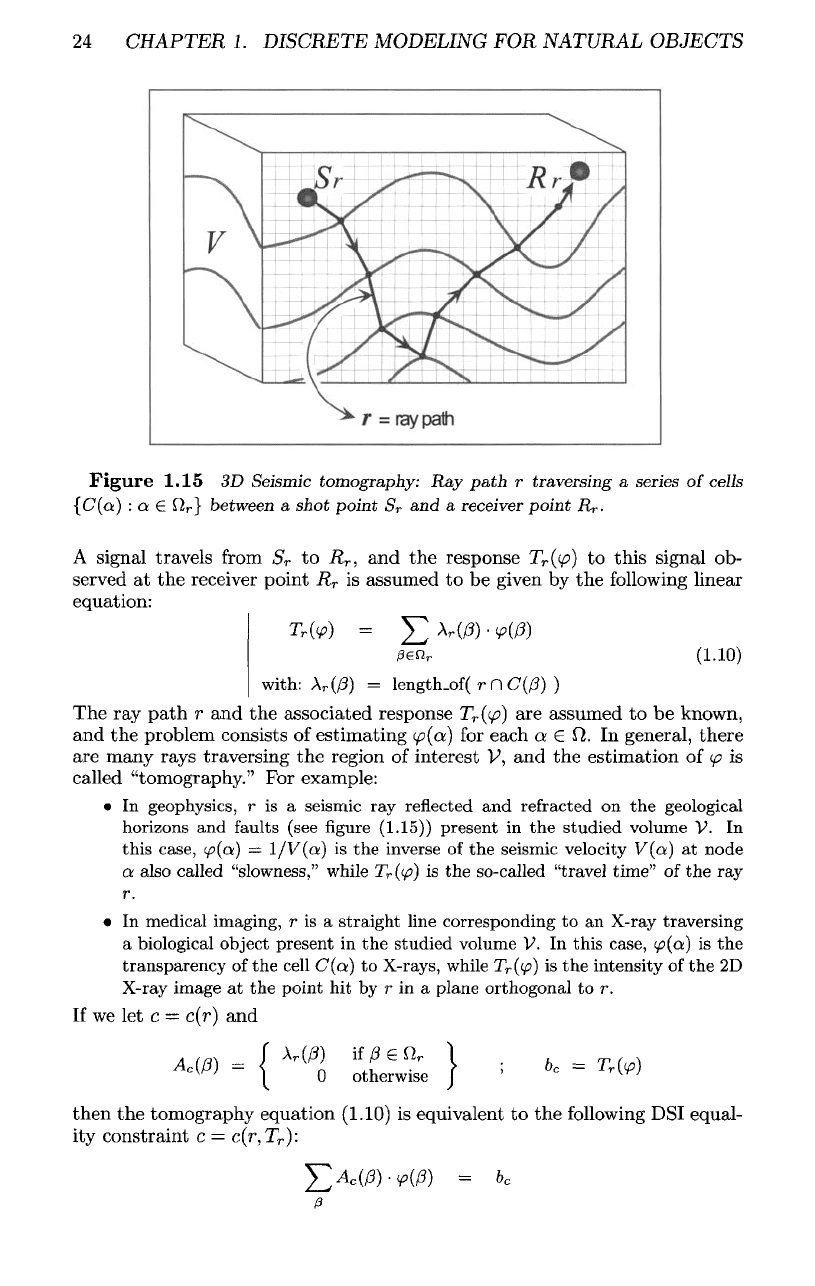
Tomography
As
shown
in figure
(1.15),
let us
consider
a
parallelepipedic region
V of the
3D
space partitioned into
a
series
of
adjacent hexahedric cells
{C(a}
:
a 6
il}
and let
A
/
1
1
(S7,7V,
</?,C)
be the
discrete model
defined
as
follows:
•
Q
is the set of all the
centers
of the
cells.
•
N(a)
is
such
that
(3
G
N(a)
if the
cells
C(a)
and
C(/3)
share
a
common
face.
•
(p(oi)
is an
unknown
physical
property
attached
to the
cell
C(a)
centered
on
the
node
a and
assumed
to be
constant
on
C(a).
• C is a set of
constraints
to be
respected
by
(p.
The
region
V is
traversed
by a
"ray path,"
which
consists
of a
given
curve
r
traversing
a
series
of
cells
{C(a}
:
a €
Q
r
}
corresponding
to a
subset
of
nodes
O
r
such
that
24
CHAPTER
1.
DISCRETE
MODELING
FOR
NATURAL OBJECTS
Figure
1.15
3D
Seismic tomography:
Ray
path
r
traversing
a
series
of
cells
{C(a)
:
a G
O
r
}
between
a
shot point
S
r
and a
receiver
point
R
r
.
A
signal
travels
from
S
r
to
R
r
,
and the
response
T
r
((p)
to
this
signal
ob-
served
at the
receiver
point
R
r
is
assumed
to be
given
by the
following
linear
equation:
The ray
path
r and the
associated
response
T
r
((p)
are
assumed
to be
known,
and the
problem
consists
of
estimating
</?(a)
for
each
a E
£7.
In
general,
there
are
many
rays
traversing
the
region
of
interest
V, and the
estimation
of
(p
is
called
"tomography."
For
example:
• In
geophysics,
r is a
seismic
ray
reflected
and
refracted
on the
geological
horizons
and
faults (see
figure
(1.15))
present
in the
studied volume
V. In
this case,
(p(oi)
—
l/V(a)
is the
inverse
of the
seismic velocity
V(a)
at
node
a
also
called "slowness," while
T
r
(tp)
is the
so-called
"travel time"
of the ray
r.
• In
medical imaging,
r is a
straight line corresponding
to an
X-ray traversing
a
biological object present
in the
studied volume
V. In
this case,
<p(a)
is the
transparency
of the
cell
C(ce)
to
X-rays, while
T
r
(if>)
is the
intensity
of the 2D
X-ray image
at the
point
hit by r in a
plane
orthogonal
to
r.
If
we let c —
c(r]
and
then
the
tomography
equation
(1.10)
is
equivalent
to the
following
DSI
equal-
ity
constraint
c =
c(r,
T
r
):
1.4.
EXAMPLES
OF
APPLICATIONS
25
Figure
1.16
3D
Medical tomography
with
DSI:
Imaging some vessels
of
a
human
brain from
six 2D
X-ray views corresponding
to six
different
directions
of
X-rays
in
the 3D
space (contrary
to
seismic rays, X-ray
paths
are
rectilinear).
Figure (1.16) gives
an
example
of the 3D
variations
of
(p
obtained with
DSI
in
the
case
of the
imaging
of the
vessels
of a
human brain.
In
this example,
each
cell
was
traversed
by six
rays corresponding
to 2D
X-ray images taken
in
six
different
directions. Experimental results have shown
that
this approach
is,
at
least,
as
efficient
as
classical methods (see [124,
125]).
Similar results
could
be
obtained
in
geological applications.
Deconvolution
Let
A
/
(
1
(O,
TV,
y?,
C]
be a
discrete
model
defined
as follows:
•
SI
is the set of the
nodes
of a
regular
ra-dimensional
grid.
•
N(a)
is
such
that
/3
€
N(a)
if a and
(3
are two
adjacent
nodes
of the
grid.
•
<p(°i)
i
s
an
unknown physical property
attached
to the
node
a.
• C is a set of
constraints
to be
respected
by
<p.
There
are
many problems
in
physics where
one can
obtain some information
on the
unknown
function
(p
through
a
convolution equation with
the
following
form
where:
(a 0 ft.) is the
node
of the
grid,
if
any, which
can be
reached
from
a
when
taking
a
step
equal
to ft.
H is a
given
set of
steps
on the
grid.
QH
is the
subset
of
SI
such
that

26
CHAPTER
1.
DISCRETE MODELING
FOR
NATURAL OBJECTS
w
a
(h)
is a
given
function
denned
on
H.
It
should
be
noted
that,
if
necessary,
w
a
(h)
can
also
depend
on a
G
fi.
tl>(a)
is a
given
function
defined
on
Q#.
For
each node
a
e
Q#,
we
would
like
to
define
a
DSI
constraint
c =
c(a,
w,
H,
T/J)
on
(p
specifying
that
the
convolution
equation
(1.11)
should
be
respected.
For
this purpose,
let us
define
the
coefficients
A
c
(j3)
and
b
c
as
follows:
It is
easy
to
check
that,
for
each node
a E fi#, we
have:
This
is a
classic linear equality constraint, which
can
easily
be
taken into
account
by
DSI.
Among
the
most traditional applications
of
deconvolution
in
seismic pro-
cessing,
we can
mention
the
reconstitution
of the
seismic
impedance
5
<p(oi)
as
a
function
of the
seismic amplitude
i^(ot)
observed
at the
nodes
of a
regular
3D
grid similar
to the one
shown
in figure
(1.14).
In
this case,
the set H
corresponds
to a
vertical window, while
the
function
w
a
(h)
is a
"wavelet"
corresponding
to the
seismic signal.
1.5
Conclusions
The few
examples presented above represent only
a
very small
part
of the
wide
scope
of
applications
that
can be
addressed using
the DSI
method
in
association with
the
notion
of a
discrete model.
A
large part
of
this book
is
dedicated
to an
extensive presentation
of the
mathematics
and the
appli-
cations
of
DSI.
Before
diving into elaborate mathematical concepts,
the few
examples above have
already
provided
an
understanding
of the
crux
of
this
method.
To sum up, DSI
uses
two
ingredients:
DSI
applies
the old
"divide
and
conquer"
principle:
any
global
complex
problem
is
split
into
a
series
of
small
localized
linear
problems
that
are in
turn
transformed
into
DSI
constraints.
DSI
uses
a
"minimum energy"
principle
to
link,
as
smoothly
as
possible,
the
small
localized
problems:
this
corresponds
to the
minimization
of the DSI
roughness,
whose
only
role
is to
ensure
the
existence
and
uniqueness
of the
solution
(see
section
(4.3)).
The
real secret
of the
efficiency
and the
simplicity
of DSI is to be
found
by
combining
these
two
principles.
5
In
seismic
processing,
"seismic impedance"
is
defined
as the
product
<p(a)
of the
seismic
velocity
V(a)
by the
density
p(a)
of the
subsurface:
Chapter
2
Cellular
Partitions
The
notion
of
discrete model introduced
in the
previous chapter relies strongly
on a
decomposition
of
objects
into
a finite set of
connected nodes. However,
as
pointed
out in figure
(1.4),
such nodes connectivities
are not
sufficient
to
describe
without
ambiguity
the
topology
of
these objects.
In
this
chapter,
we
will
see how
cellular
partitions
can
achieve this goal
in a
completely general
way, whatever the object (manifold/non-manifold) or the dimension of the
embedding space
(ID,
2D,
...,
nD).
2.1
Introduction
It is
well known
that
complex biological objects, also called organs, consist
of
sets
of
adjacent "biological
3D
cells," which
are
structured
to
respect
the
external
and
internal boundaries
of
these objects.
It can be
observed
that
•
from
a
mathematical
point
of
view,
each
3D
cell
is in
fact
an
element
of
volume
which
can be
transformed
into
a 3D
ball,
continuously,
and
•
each
3D
cell
holds
attributes
consisting
of
information
attached,
for
example,
to its
genes.
Similarly,
as
shown
in
figures
(2.1),
(2.35),
(3.17),
and
(3.23),
the
subsur-
face
can be
decomposed into "geological
3D
cells" having attributes (physical
properties)
and
bounded
by
geological horizons
and
faults.
As
suggested
in
figure
(2.2),
the
same approach
can be
used
to
decompose
a
geological cross
section
or a
geological
map
into
2D
cells (which
can be
transformed into
a 2D
ball,
continuously).
Over
the
past
decade,
this
cellular paradigm
has
been used
by
several
computers scientists
as a
template
for
modeling
the
topology
of
complex
ob-
jects (e.g.,
see
[35,
37, 66, 67, 68,
134,
181]).
A
complete presentation
of the
27
