Hermanson G. Bioconjugate Techniques, Second Edition
Подождите немного. Документ загружается.

separations in one or two dimensions can be done to fractionate the peptides in complex sam-
ples by charge, size, or hydrophobicity before being injected into the mass spec.
Another major technique to simplify the analysis of protein samples is to use mass tags. Mass
tags are modifi cation reagents that contain a reactive group for coupling to biomolecules and
another component of known mass, which behaves predictably upon MS separation. The mass
tag also may contain a functional group for capture and separation on an affi nity support,
which permits further fractionation of the proteome. MS analysis of mass tagged peptides can
be done by focusing only on those peptides that contain an additional mass component rep-
resenting the tag ’s known mass contribution. Thus, all other peaks on the MS spectrum can
be ignored, which greatly reduces the complexity of the sample. Mass tag reagents have been
developed with reactive groups to modify specifi cally only certain low frequency amino acids
within proteins. For instance, a thiol-reactive iodoacetyl group on a mass tag can be used to
modify only those peptides having cysteine residues, thus removing from the analysis window
all other peptides not containing cysteine.
The design of mass tags also can be combined with stable isotope labels to create more than
one mass unit for each tag type (Schneider and Hall, 2005). For example, certain hydrogen
atoms on one mass tag can be replaced with deuterium atoms on another derivative. Everything
else on the tag is identical except for the isotope substitutions. Thus, the two mass tag analogs
will differ in molecular weight by exactly the mass difference represented by the isotopic substi-
tutions. Such tags can be used to modify a test sample with the stable isotope tag versus a con-
trol sample modifi ed with the normal tag. If the two samples then are combined and analyzed
by mass spec, their signal peaks generated from the tagged peptides will differ in mass units by
the isotopic mass differences in the two tags. Identifi cation of the peptides from both samples
is done by looking for peptide peak pairs differing by the characteristic mass amount, therefore
greatly reducing the complexity of sample analysis, and allowing simultaneous investigation of
two samples. In this way, a test sample ’s protein expression levels can be compared to a control
sample by measuring the different areas of the paired peptide peaks. The ability to analyze pro-
tein expression in two samples is vitally important to drug discovery and life science research
applications studying the proteome.
Mass tags also can be broad spectrum in their modifi cation properties to derivatize all pep-
tides as they are formed upon proteolysis. For instance, one of the simplest mass tagging sys-
tems is to use the oxygen isotope
18
O in the water used during the enzymatic digestion of a
protein sample (Miyagi and Rao, 2007). Upon hydrolysis by trypsin, the resultant C-terminal
carboxylates that are formed each incorporate two
18
O atoms. Thus, peptides formed from
18
O digestion will be four mass units heavier than peptides formed by proteolysis using normal
water. Mass spec analysis of this difference can identify the peptide pairs resulting from a con-
trol sample and a test sample run simultaneously.
Other broad-spectrum mass tag modifi cation agents are designed to modify all amine groups
and yield tags on every peptide at their N-terminal amines. For instance, small molecule tags
using deuterium labeled forms and regular hydrogen labeled ones, such as the use of isotopi-
cally labeled propionic anhydride (Zappacosta and Annan, 2004), provide differentiation in
the mass spec signals of peptides from test samples and controls. To eliminate interference,
side chain lysine amines are blocked by guanidination with O-methylisourea hemisulfate and
cysteine thiols are blocked with iodoacetamide (Leitner and Lindner, 2004). Some mass tag
reagents of this type are able to differentiate peptides from 6 to 10 samples analyzed at the
same time (see section on isobaric tags, this chapter).
650 16. Mass Tags and Isotope Tags
The following sections describe some of the major mass tag types and discuss the general
protocols for their use.
1. ICAT Reagents
Isotope coded affi nity tags (ICAT) are bifunctional mass tagging agents containing a reactive
group on one end of the molecule and an affi nity capture group on the other end (Gygi et al .,
1999; Aebersold, 2003) ( Figure 16.1 ). In addition, a portion of the tag can contain stable iso-
tope substitutions, usually designed to be in the cross-bridge between the reactive group and
the biotin handle. The original ICAT reagent contained eight deuterium atom substitutions on
the outer ends of an ethylene oxide spacer. The isotope tagged version thus differs from its nor-
mal atom analog by exactly eight mass units.
Most ICAT style compounds contain a thiol-reactive iodoacetyl group on one end and a
biotin handle on the other end of a spacer arm ( Figure 16.2 ). Reagents of this type are highly
specifi c for reacting with cysteine thiols in proteins to result in stable thioether modifi cations
containing a terminal biotin group ( Figure 16.3 ). After enzymatic digestion, modifi ed peptides
then can be isolated using immobilized (strept)avidin, which specifi cally binds only to those
peptides containing the biotin tag and allows the other peptides to be discarded. Thus, the sam-
ple complexity can be reduced to analyze only peptides that contain a cysteine residue, which
in the human proteome represents about 26.6 percent of the total tryptic peptides in a sample.
This translates into the ability to cover 96.1 percent of all the proteins in the human proteome
by targeting only cysteine-containing peptides (Zhang et al ., 2004; Yan and Chen, 2005).
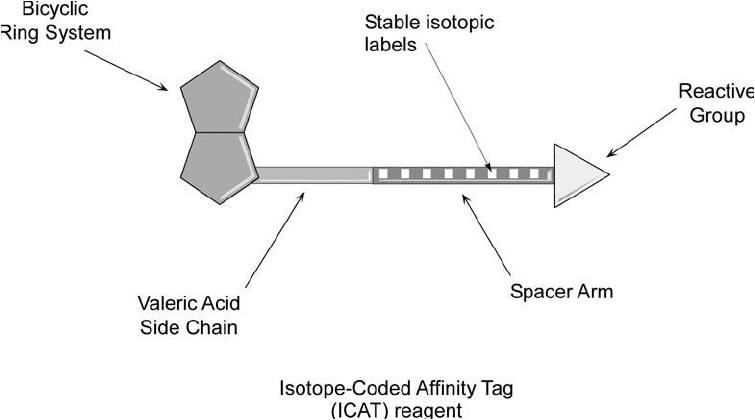
Figure 16.1 The general design of an ICAT reagent consists of a biotinylation compound with a spacer
arm containing stable isotope substitutions. The reactive group is used to label proteins or peptides at par-
ticular functional groups and the biotin affi nity tag is used to isolate labeled molecules using immobilized
(strept)avidin.
1. ICAT Reagents 651
652 16. Mass Tags and Isotope Tags
ICAT reagents can be used to compare two different samples by mass spec analysis. For
instance, one cell population can be treated with a drug candidate, while another one remains
untreated and acts as a control. Alternatively, one cell population can represent a disease
state and the control population is the normal cell line. After cell lysis, the proteins in each
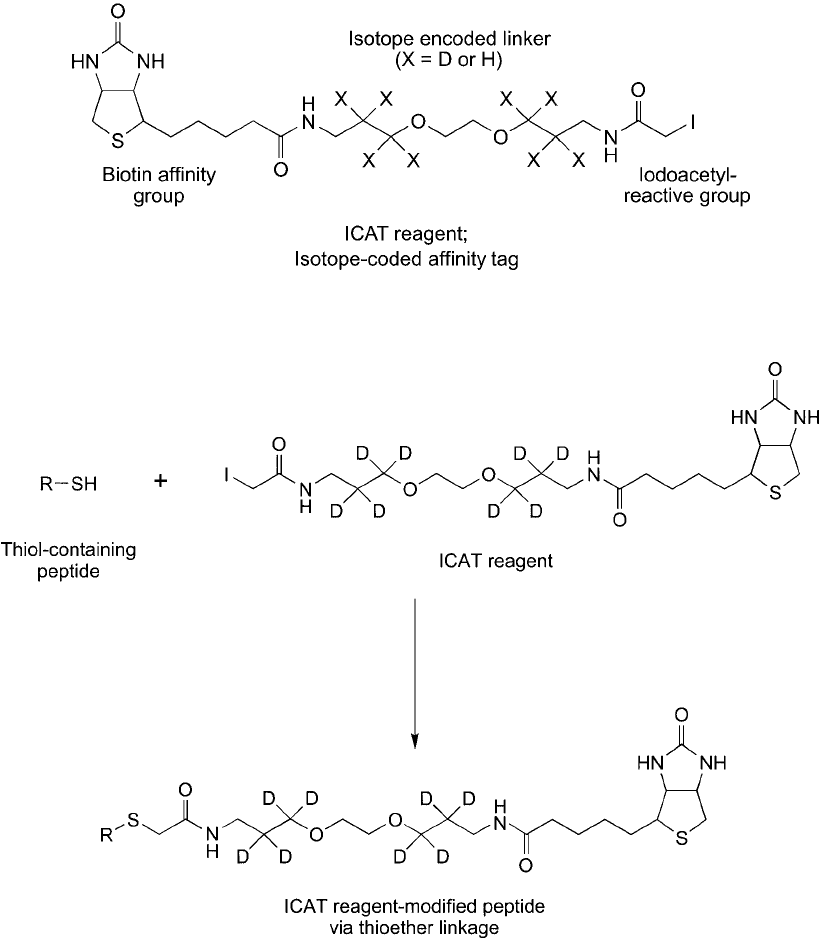
Figure 16.3 The ICAT reagent reacts with cysteine-containing peptides to form a thioether bond.
Figure 16.2 The original design of the ICAT compound. The iodoacetyl group provides reactivity with thiol
groups. The isotopically labeled spacer arm typically is substituted with eight deuterium atoms.
sample are denatured and reduced to make available all of the cysteine thiols for modifi ca-
tion. One sample then is reacted with the heavy atom ICAT reagent, while the other sample
is reacted with the normal isotope compound. The two samples next are combined and enzy-
matically digested with trypsin to generate peptide fragments, some of which will contain ICAT
labeled cysteine groups. This combined peptide sample is affi nity separated on an immobilized
(strept)avidin column (or monomeric avidin column), which binds biotin labeled peptides from
both sample populations equally. After removal of the non-biotinylated peptides by washing
the column followed by elution of the ICAT labeled peptides, the sample is subjected to capil-
lary reverse phase chromatography leading into ESI or MALDI mass spec analysis. The fi nal
HPLC separation again reduces the complexity of the sample set by further fractionating the
peptides based on comparative hydrophobicity. In the MS spectrum, the relative peptide con-
centrations are determined by comparing all peaks separated by exactly the mass unit differen-
tial between the heavy atom mass tag and the normal atom mass tag. Each peptide sequence
then is identifi ed by fragmentation of the peptides into amino acid ions in a second dimen-
sion MS separation (MS/MS). Comparison of the amino acid sequence of each peptide peak
to known sequence databases can identify the protein from which it came. Thus, the resultant
peptide peak ratios are directly proportional to the relative amounts of the corresponding pro-
teins present in the cell population.
The original ICAT design was found to have a number of defi ciencies that often prevent the
reagent from providing acceptable MS results. First, the deuterium isotope-labeled compound
has a tendency to behave differently than the normal hydrogen isotope during reverse phase
separation (Regnier et al., 2002). If the labeled peptides that are identical except for the pres-
ence or absence of a D
8
ICAT modifi cation don ’t elute at precisely the same point in an HPLC
separation, then the MS analysis won ’t provide the peak pairs necessary for quantifi cation. To
solve this problem, a second-generation ICAT compound was designed containing
13
C isotopes
instead of deuterium atoms. This type of reagent facilitates precise chromatographic separation
of the labeled peptides and thus gives far superior performance upon MS analysis.
A second problem in the original ICAT design relates to the presence of the biotin tag. The
biotinylated peptides often give undesirable fragmentation patterns during MS/MS analy-
sis, which interferes with the smooth identifi cation of peaks. Removing the biotin tag before
mass spec analysis therefore would be benefi cial to interpreting the MS results. Another issue
with using a biotin tag is the elution step from the immobilized (strept)avidin column. Only
under severely denaturing conditions is the interaction between biotin and (strept)avidin dis-
rupted. However, even when using such conditions, the bound peptides do not always get
released reproducibly from the column. The result is ineffi cient recovery of labeled peptides,
which directly translates into a lack of precision in the MS data. Even using an immobilized
monomeric avidin column does not completely solve this problem, because this affi nity sup-
port sometimes has higher affi nity binding sites or binds non-biotinylated peptides nonspe-
cifi cally. To solve these issues, new cleavable ICAT designs were created that contain a bond
within the cross-bridge that can be chemically broken (Li et al., 2003). After binding to the
(strept)avidin column, elution can be accomplished by cleaving the biotin arm, not by breaking
the (strept)avidin–biotin interaction. The cleavage site can consist of a disulfi de group within the
cross-bridge (Turecek, 2002) that can be reduced for elution from the (strept)avidin column or
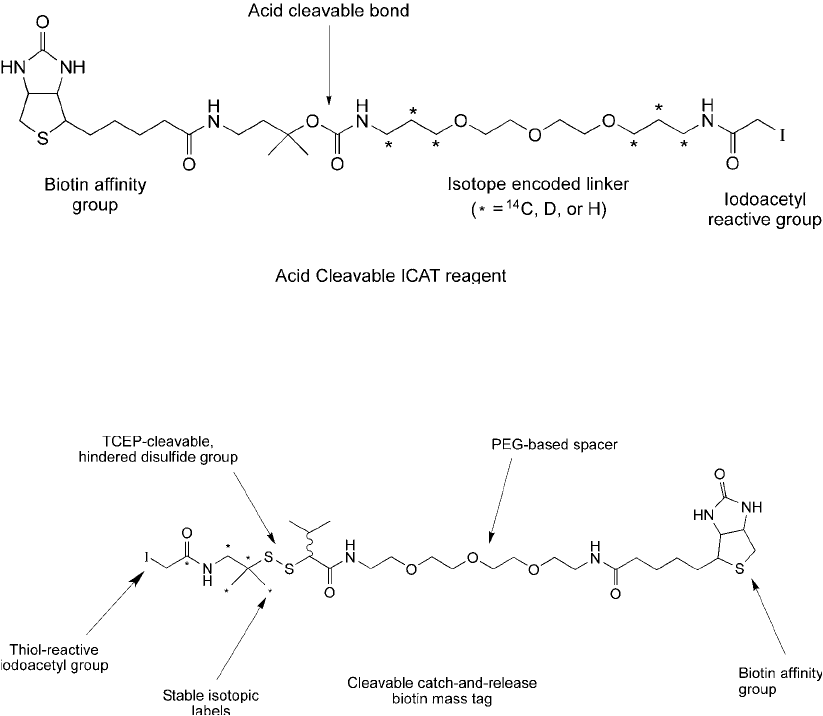
it can consist of an acid cleavable linker arm (e.g., a carbamate bond) within the ICAT structure
(Fauq et al., 2006) ( Figure 16.4 ). Either method dramatically improves the recovery of labeled
peptides from the affi nity column and thus provides increased precision in the samples leading
1. ICAT Reagents 653
654 16. Mass Tags and Isotope Tags
into the LC–MS analysis. The
13
C labeled, acid cleavable ICAT reagent has been used to identify
successfully low-level protein expression in highly complex samples (Hansen et al., 2003).
Another new ICAT design, termed a “catch-and-release” tag, contains a constrained, steri-
cally hindered disulfi de linkage with bulky alkyl groups on both sides. The hindered nature
of the disulfi de makes it stable to standard protein reduction procedures, but it can be specifi -
cally reduced upon the addition of tris(2-carboxyethyl)phosphine(TCEP) (Gartner et al., 2007)
(Figure 16.5 ). This allows proteins to be labeled with the catch-and-release ICAT compound
that have undergone reduction using dithiothreitol (DTT) to cleave protein disulfi des but not
affect the disulfi de group in the reagent cross-bridge. Only after capture of labeled peptides on
Figure 16.5 A catch-and-release ICAT design incorporates a gem-methyl group and an isopropyl group on
either side of a disulfi de bond within its spacer arm. The hindered disulfi de permits the use of standard reducing
gel electrophoresis conditions using DTT without reduction. After purifi cation on a (strept)avidin affi nity col-
umn, however, the disulfi de group can be cleaved with TCEP, which provides recovery of the labeled peptides
prior to mass spec separation.
Figure 16.4 A more advanced ICAT design uses an acid-cleavable spacer arm to facilitate elution of labeled
peptides from a (strept)avidin affi nity column. The use of
14
C isotopes instead of deuterium labels permits
precise reverse phase separations prior to mass spec that show no elution peak time differences between
isotope-labeled and normal atom-labeled peptides.
a (strept)avidin column is the cross-bridge cleaved by the addition of TCEP and the labeled
peptide recovered.
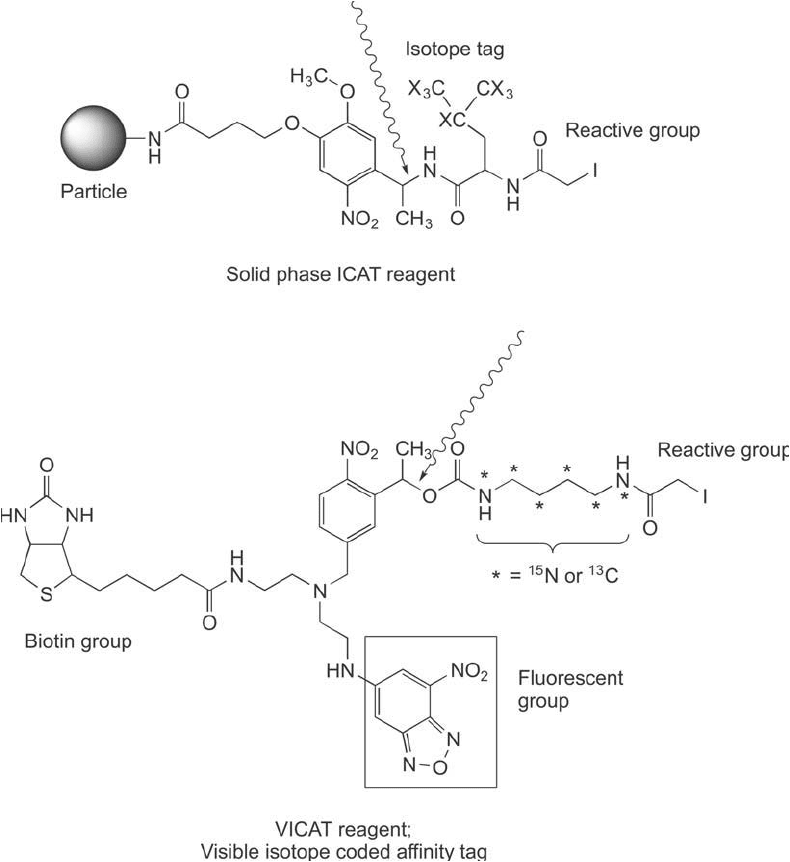
A variation on the ICAT mass tag concept was made by immobilizing the label on a solid
phase (Zhou et al., 2002). Using this design, cysteine-containing peptides are modifi ed directly
on a beaded insoluble support. After washing away non-cysteine peptides, the linked peptides
can be cleaved from the matrix by use of a photo-cleavable group and eluting off the peptides
with an isotope tag modifi cation. This approach results in cleaner and more effi cient isolation
of tagged peptides and simplifi es the ICAT labeling process ( Figure 16.6 ).
Another novel mass tag design involves a spectrally visible ICAT variant developed to
include a fl uorescent group for detection purposes (Lu et al., 2004). Like the original ICAT
reagent, the VICAT compound includes a thiol-reactive iodoacetyl group, a cleavable cross-
bridge, an isotopically labeled portion, and a biotin handle. It also has another arm, however,
that contains the chromogenic label, which is detectable by absorption at 493 nm and emission
at 503 nm.This group allows detection of peptides in samples separated by chromatographic
or electrophoretic means. Quantifi cation of the fl uorescent tag in isolated peptides can provide
absolute information regarding the level of proteins present in a cell.
ICAT type compounds are designed to enrich for peptides containing one particular amino
acid residue, usually cysteine. The affi nity capture step removes other non-cysteine peptides
and thus reduces the complexity of the MS data set. ICAT reagents also can be designed with
a different reactive group that is able to covalently couple to other amino acid groups (or even
sites of post-translation modifi cation) to change the selectivity of the peptide population being
analyzed. However, it is best to target amino acids or functional groups present in limited
amounts within proteins, otherwise the tag may capture more peptides than could be conve-
niently measured by mass spec. Han et al. (2007) developed a hydrazide-ICAT compound to
identify proteins modifi ed by 2-alkenals derived from lipid peroxidation (LPO). This type of
mass tag should be useful for the study of other oxidative changes on proteins, such as those
resulting in aldehyde or ketone modifi cations (see Chapter 1, Section 1).
The following protocol describes the use of an acid-cleavable ICAT reagent, currently avail-
able from Applied Biosystems. This is not meant to be a detailed method describing every
aspect concerning the use of mass spectrometry, but only to describe the modifi cation reaction
of the ICAT compound with proteins.
Protocol
1. Grow cells to 70–80 percent confl uence and harvest by scraping the cells into 5 ml PBS,
5 mM EDTA, pH 7.4. Aliquot cell counts of approximately 2.5–5 10
6
for processing.
Lyse cells using a detergent lysis buffer (e.g., Poppers, Thermo Fisher) or by mechan-
ical means. Centrifuge and discard the cellular debris. Measure the total protein con-
centration using the BCA assay (Thermo Fisher) and adjust the protein concentration to
1.5 mg/ml using 50 mM Tris, 0.1 percent SDS, pH 8.0. Separately process a test sample
and a control sample made up of different cell populations.
2. Reduce disulfi des in the two protein samples by the addition of 2 l of 50 mM TCEP
(Thermo Fisher) to each 100 l aliquot of protein solution. Cover and boil the samples
for 10 minutes in a water bath to completely denature and reduce the proteins. Avoid the
use of thiol-containing reductants, such as DTT, as these will react with the iodoacetyl
group on the ICAT compound.
1. ICAT Reagents 655
656 16. Mass Tags and Isotope Tags
3. Dissolve one vial of heavy isotope, cleavable ICAT reagent (Applied Biosystems) in 20 l
acetonitrile (use a fume hood for handling solvents). Dissolve a second vial contain-
ing the normal isotope ICAT compound in 20 l acetonitrile. Vortex mix each vial to
dissolve.
Figure 16.6 The solid phase ICAT reagent provides a thiol-reactive iodoacetyl group to capture cysteine pep-
tides, a spacer containing stable isotopic labels, and a photo-cleavable group that can release the captured
peptides for mass spec analysis. The VICAT mass tag is a solution phase labeling agent that also has a photo-
cleavable site to release isolated peptides from a (strept)avidin affi nity resin. This compound adds a fl uorescent
group to better detect labeled peptides as they are being isolated from a sample.
2. ECAT Reagents 657
4. Add 100 g of the control protein solution to one vial of dissolved normal isotope ICAT
reagent. Mix to dissolve. Add 100 g of the test protein solution to one vial of dissolved
heavy isotope ICAT reagent. Mix to dissolve.
5. React both solutions for 2 hours at 37°C.
6. Combine the test sample with the control sample in a single vial. Mix well.
7. Prepare a solution of TCPK-trypsin in 50 mM ammonium bicarbonate, pH 8.0, at a con-
centration of 100 ng/ l. Add 10 l of the trypsin solution to every 10 g of combined,
labeled protein solution from Step 6. Incubate at 37°C for 12–16 hours or overnight
with mixing.
8. Centrifuge the digested peptide mixture to remove any insoluble material. The bioti-
nylated peptides then are purifi ed on 20 l immobilized monomeric avidin column. The
column fi rst is primed with elution buffer (0.4 percent TFA in 30 percent acetonitrile)
and then washed with binding buffer (100 mM ammonium bicarbonate, pH 8.0). The
sample is applied and washed through with binding buffer until all not-bound peptides
are completely removed. The acetonitrile/TFA elution is subsequently used to cleave off
the biotin group from the eluted peptides. Note: Additional fractionation may be done to
reduce further the complexity of the sample, such as the use of ion-exchange chromatog-
raphy. The eluted, labeled peptides fi nally are analyzed by LC–MS/MS.
2. ECAT Reagents
Element-coded affi nity tags represent a new type of isotope tag for mass spec analysis
(Corneillie et al., 2003, 2004; Whetstone et al., 2003; Meares et al., 2007). This system uses
a bifunctional metal chelate group that securely coordinates a lanthanide metal ion. A reactive
group also is present for the modifi cation of certain amino acids in proteins, which typically
consists of a bromoacetyl group. This group reacts similarly to an iodoacetyl group and forms
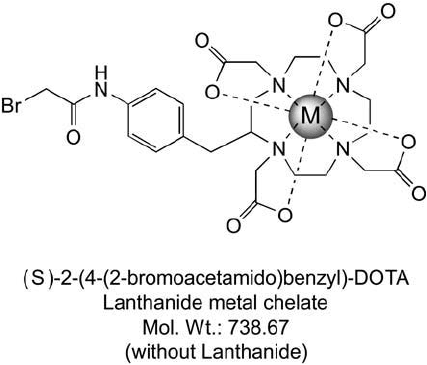
thioether linkages with cysteine thiols. The ECAT design includes a DOTA chelating group
(Chapter 10) containing four nitrogen atoms and four carboxylates to complex with any of
the lanthanide series metal ions via eight coordination bonds. Simply by using different lantha-
nide elements within the complex the result will be a unique set of isotope tags having differ-
ent mass signatures by MS. At least in theory, up to 15 different ECAT mass tag compounds
could be created by using all of the different lanthanide metals representing elements 57–71. In
addition, the lanthanides are naturally mono-isotopic in that they occur mainly in nature with
only a single isotope. Only cerium contains a high percentage of another isotope in nature (88
percent
140
Ce and 11 percent
142
Ce), all the other lanthanides are 97 percent a single isotope.
This is important for mass spec separations, as the resultant peaks won ’t contain extraneous
mass signatures due to multiple isotopes ( Figure 16.7 ).
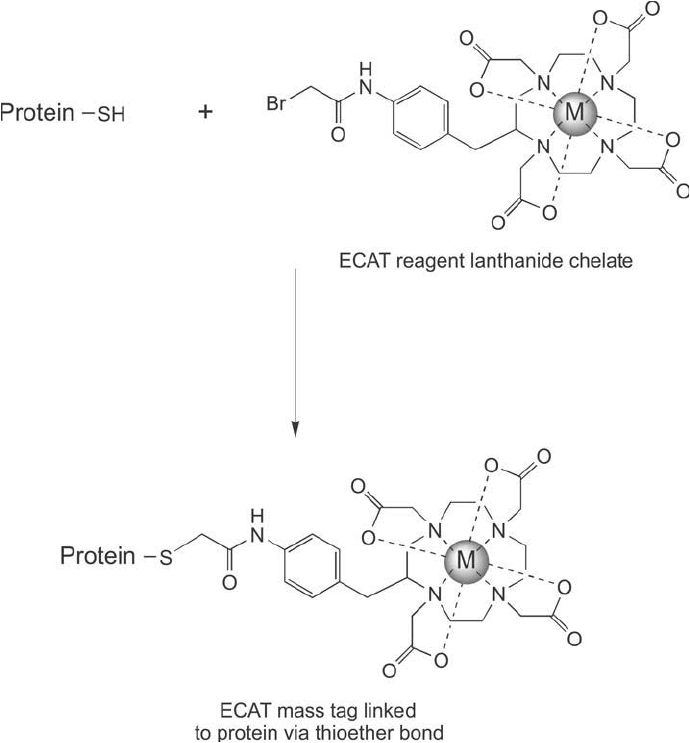
The ECAT reagent bromoacetamidobenzyl-DOTA (BAD) can be used like an ICAT tag to
label only those peptides containing cysteine residues ( Figure 16.8 ), thus reducing the total
number of peptides having to be analyzed in a mass spec separation. Unlike the ICAT reagent
design, the ECAT compound does not contain a biotin handle for affi nity separation. Instead,
a monoclonal antibody has been developed with specifi city for the DOTA chelate containing
a bound lanthanide metal. The antibody will recognize any lanthanide element bound in the
chelate and thus function as an affi nity ligand for separating ECAT-labeled peptides. The affi n-
ity of the monoclonal for ECAT chelate structure allows highly stringent washes to be done
658 16. Mass Tags and Isotope Tags
prior to elution of the labeled peptides. The affi nity column typically is washed with low pH,
high salt, high pH, and 30 percent acetonitrile before elution is done. This removes all traces of
nonspecifi cally bound peptides, so they can ’t interfere with the mass spec analysis. The ECAT
labeled peptides then are eluted with 20 percent acetonitrile containing 0.1 percent TFA.
The ECAT system has an advantage over ICAT reagents in being available with more mass
signatures than is achievable using
13
C or
2
H labeled tags, which are diffi cult to synthesize. In
addition, it is not hampered by the binding idiosyncrasies of a biotin group interacting with
(strept)avidin or the fragmentation problems a biotin tag gives on MS analysis. The ECAT
chelate does not generate fragmentation products during the mass spec analysis. Also, the mass
defect characteristic of lanthanide metals results in a mass signature upon MS separation that
occurs in a relatively unoccupied region of the m/z spectrum (Schneider and Hall, 2005). Thus,
identifi cation of ECAT labeled peptides potentially is simpler than using ICAT reagents.
Another variant of ECAT reagent technology has been developed to analyze the products of
protein oxidation (Lee et al., 2006). Called “oxidation-dependent, carbonyl-specifi c element
coded affi nity tag ” (O-ECAT), the compound contains the same DOTA lanthanide chelating
group, but instead of a thiol-reactive bromoacetyl group, it has an aldehyde- or ketone-reactive
aminoxy group ( Figure 16.9 ). The aminoxy functional group can covalently link to aldehydes
or ketones to give an oxime bond, which is stable under aqueous conditions ( Figure 16.10 ).
The O-ECAT reagent is a superior alternative to the use of 2,4-dinitrophenylhydrazine
(DNPH; Chapter 1, Section 1.1) in the study of protein oxidation. DNPH modifi cation pro-
duces detectable complexes, but it does not provide information as to what amino acids are
involved. O-ECAT modifi es carbonyl end products of protein oxidation and in addition, it can
provide exact information as to the amino acids that were oxidized. Mass spec analysis of mod-
ifi ed proteins performed after proteolysis gives the exact amino acid sequences including the
sites of O-ECAT reagent modifi cation. The same antibody that is specifi c for the metal chelate
portion of the standard ECAT reagent also can be used to capture and detect the O-ECAT
Figure 16.7 The ECAT mass tag consists of a DOTA metal chelate group that can coordinate a lanthanide
metal ion and a bromoacetyl group for coupling to cysteine-containing proteins.
labeled proteins or peptides. Thus, O-ECAT-modifi ed proteins can be detected in Western blots
or the sites of oxidation quantifi ed using ELISA-based assays.
3. Isobaric Tags
The use of mass tagging reagents to analyze proteomic data has greatly improved the ability
to compare samples for protein expression differences. However, a major limitation of the
ICAT procedure (Section 1, this chapter) is that it can only compare two samples simultane-
ously, usually a test and a control. Even with the ECAT design (Section 2) using multiple lan-
thanide metals to make a series of different mass tag signatures, it is diffi cult to extend the
3. Isobaric Tags 659
Figure 16.8 Reaction of the ECAT reagent with a cysteine-containing protein results in the formation of a
stable thioether bond.
