Hermanson G. Bioconjugate Techniques, Second Edition
Подождите немного. Документ загружается.

680 17. Chemoselective Ligation: Bioorthogonal Reagents
added to the reaction mixture to prepare the fi nal concentration desired. Mix well to dis-
solve before immediately exposing the solution to the slide surface.
3. React for at least 1 hour at room temperature with gentle mixing.
4. Wash the slide with 10 volumes of water.
5. Prepare a 1 M solution of hydroxylamine in coupling buffer suffi cient to again treat the
slide. The pH of the coupling buffer should be adjusted to pH 10 after dissolving the
hydroxylamine into it. Expose the slide to the hydroxylamine solution in the same man-
ner as the crosslinker treatment. The hydroxylamine will react with the methyl ester
groups on the salicylic acids and form hydroxamate functionalities suitable for conjuga-
tion with the P(D)BA-modifi ed protein from above.
6. React the slide with the hydroxylamine solution for 16–24 hours at room temperature.
7. Thoroughly wash the slide with coupling buffer and at least 10 volumes of water.
Coupling PDBA-Modifi ed Protein to SHA-Modifi ed Slide
Proteins modifi ed with boronic acid groups may be covalently linked to SHA-modifi ed slides
simply by spotting the PDBA-protein onto the slide surface in 0.1 M sodium bicarbonate buffer,
pH 8.0. The arraying technique may use pin spotters, piezoelectric contactless printers, or even
pipette dispensing into masked wells on the surface. The quantity of PDBA-protein placed on
the surface should at least be in 2-fold molar excess to the theoretical density of SHA groups
present. Large quantity protein spotting may obviate a covalent attachment strategy by build-
ing up a “mountain” of dried protein, as typically is done using simple surface adsorption of
protein to form array spots.
4. Click Chemistry: Cu(I)-Promoted Azide–Alkyne [3 ⴙ 2] Cycloaddition
The 1,3-dipolar cycloaddition reactions to unsaturated carbon–carbon bonds have been known
for quite some time and have become an important part of strategies for organic synthesis of many
compounds (Smith and March, 2007). The 1,3-dipolar compounds that participate in this reac-
tion include many of those that can be drawn having charged resonance hybrid structures, such
as azides, diazoalkanes, nitriles, azomethine ylides, and aziridines, among others. The heterocyclic
ring structures formed as the result of this reaction typically are triazoline, triazole, or pyrrolidine
derivatives. In all cases, the product is a 5-membered heterocycle that contains components of
both reactants and occurs with a reduction in the total bond unsaturation. In addition, this type of
cycloaddition reaction can be done using carbon–carbon double bonds or triple bonds (alkynes).
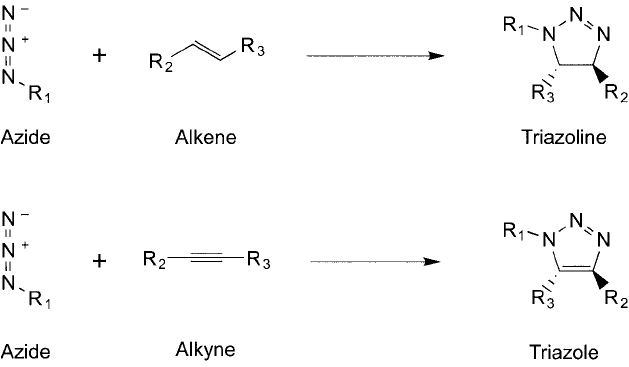
The reaction between an azide and an alkyne has been referred to as the Huisgen cycload-
dition reaction, after the name of its originator (Huisgen et al., 1964). This type of reaction
is shown in Figure 17.9 for both alkenes and alkynes, which results in similar heterocycles
only differing in a single carbon–carbon double bond within the ring. The Sharpless group has
coined the term “click chemistry ” to describe these reactions, because of the seemingly “spring-
loaded” nature of the electrophiles that participate in it (for review, see Kolb et al., 2001). It
also was noticed that such reactions appear to be accelerated in aqueous solution compared to
the same reactions done in organic solvent.
Click chemistry reactions historically are done at elevated temperatures, and sometimes ele-
vated pressures, to increase the rate of reaction and make the yield of heterocycle formation
acceptable. However, it was discovered that in the presence of Cu(I), the reaction kinetics are
dramatically accelerated to provide high yields even at room temperature and ambient pres-
sures (Rostovtsev et al., 2002; Tornøe et al., 2002; Sharpless et al., 2005). There was an early
indication that Cu(I) could catalyze this process (L ’abbé, 1984), but it was not pursued further
at that time, especially as a potentially useful bioconjugation method. In fact, in aqueous solu-
tion, the presence of only a small catalytic amount of Cu(I) in a click chemistry reaction can
increase the rate of cycloaddition by about a million-fold, making the reaction biocompatible.
The copper-catalyzed azide–alkyne cycloaddition process has resulted in a proliferation of
applications in organic synthesis and bioconjugation. There are hundreds of references to the
use of this conjugation reaction for small molecule synthesis, protein conjugation, activity-based
protein profi ling (ABPP), nucleic acid conjugation, surface modifi cation, detection schemes, and
in vivo targeting of molecules on cells. One of the primary reasons for the increasing popularity
of the click chemistry reaction is the bioorthogonal nature of the two reacting groups.
Azides in particular are convenient electrophilic participants in the click chemistry reaction
due to their ease of formation and stability. Alkyl azides undergo nearly no side reactions and
are extremely stable in aqueous solution, even in the presence of complex biological material.
Note that the aryl azides, which often are used as photoreactive crosslinking agents, are highly
unstable to UV light or reducing agents and probably should be avoided for this purpose
(although they may be perfectly good substrates for the click reaction).
Alkyne groups also are remarkably stable in biological solutions, provided they don ’t have an
activating group adjacent to them, such as a carbonyl, which would make them predisposed to
Michael-type addition reactions, especially with thiols. Adding an alkyne group to a modifi ca-
tion reagent or a crosslinker can be as simple as coupling an activated carbonyl group with prop-
argylamine, which forms the propargylamide linkage and creates a terminal acetylene group for
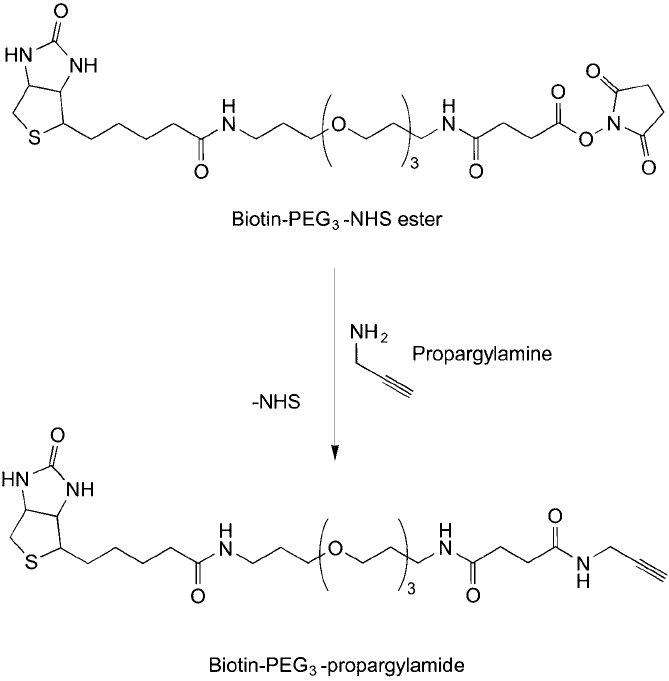
conjugation. Link et al. (2004) synthesized a biotin–PEG-alkyne modifi cation reagent using this
strategy, which then could be used to modify proteins containing azide amino acids ( Figure 17.10 ).
The functional groups used for click chemistry conjugations are completely unreactive toward
biological molecules and virtually free of side reactions, which otherwise would cause reagent
Figure 17.9 A general Huisgen reaction involves the cycloaddition of an azide with an alkene or an azide with
an alkyne. The products of these reactions are a triazoline ring or a triazole ring, respectively.
4. Click Chemistry: CU(I)-Promoted Azide–Alkyne [32] Cycloaddition 681
682 17. Chemoselective Ligation: Bioorthogonal Reagents
instability in aqueous environments. This means that a molecule modifi ed to contain an azide
functionality would be able to react specifi cally with another molecule containing an alkyne group,
even in the presence of biological fl uids, cells, or cell lysates. In addition, without the presence of
Cu(I), the azido-molecule and the alkyne-molecule would not react to an appreciable extent at
room temperature even when placed together in solution. Only upon the addition of Cu(I) in suf-
fi cient concentration would the cycloaddition reaction take place and a triazole linkage be formed.
The Cu(I) source used to drive the click reaction can be generated in several ways. The use
of CuBr can be done to add Cu(I) directly to the reaction; however, Cu(I) salts are relatively
impure and in solution they are labile and may degrade by oxidation to a signifi cant extent
over the time course of the reaction. The use of Cu(I) salts also has been found to result in
some degree of side reaction products as well as needing an organic base and organic co-solvent
during the reaction to effi ciently drive the cycloaddition process. For these reasons, the source
for Cu(I) typically is generated in situ using Cu(II) in the presence of a reducing agent.
Figure 17.10 Propargylamine can be used to add an alkyne group to amine-reactive reagents, such as the NHS
ester group on the biotin–PEG
3
compound.
The Cu(II) salt, CuSO
4
, is particularly convenient, as it is readily available and easily converted
to Cu(I) with a reducing agent, such as sodium ascorbate or TCEP. In solution, Cu(II) is reduced
to Cu(I) by ascorbate with concomitant oxidation of ascorbate to dehydroascorbate. For reactions
between pure click chemistry components in solution, the amount of catalyst addition only has
to be from about 0.25 mole percent to 2 mole percent relative to the amount of reactants present,
with a 5-fold molar excess of ascorbate over the amount of Cu(II). Therefore the reaction is initi-
ated by production of only a small amount of Cu(I), which catalytically gets oxidized and then
regenerated by reduction during the cycloaddition process. The proposed mechanism for the click
chemistry reaction has been illustrated as a catalytic cycle by both the Meldal group (Tornøe et al.,
2002) and the Sharpless group (Rostovtsev et al., 2002), giving a cyclic intermediate azide-Cu(I)-
alkyne complex, which then goes on to form the 5-membered triazole ring.
Another source for Cu(I) in the click reaction is to use elemental copper metal fi lings, which
generate Cu(I) ion in solution slowly by oxidation. This last option, however, is considerably
slower in generating the necessary Cu(I) than the other methods and will result in reactions
needing to be done for at least 24 hours.
For click reactions done in complex solutions, such as in the presence of biological molecules,
the amount of Cu(II) and ascorbate addition typically is at a concentration of at least 0.1 mM
CuSO
4
and 0.2 mM ascorbate. In this type of environment, the labeling reaction usually is done
on azide or alkyne targets at very low concentration levels and for extended times. At this con-
centration of metal salt and ascorbate, cells may not remain viable for long periods and may die.
In some cases, click chemistry ligation reactions may not be appropriate for labeling within
cells if continued cell viability is important. Live cell labeling requires that the conjugation
chemistry not adversely affect cell viability or dramatically alter protein expression or pathway
activation. Due to this limitation, the click chemistry reaction has been said to be undesirable
for performing conjugations within a living cell, and only useful for labeling targets on live cell
surfaces (Link and Tirrell, 2003; Prescher and Bertozzi, 2005).
However, some groups have worked around these issues and developed strategies for live cell
labeling wherein the fi rst step occurs in vivo, but then subsequent steps use in vitro cycload-
dition for detection. Speers and Cravatt (2004a, b) used a click chemistry reactant to label
enzymes in vivo at their active sites with an azide-substrate analog. ABPP typically involves
using a binding probe along with a reactive group and a detectable tag, which is able to tar-
get specifi cally the binding site of an enzyme. The reactive group covalently links the affi nity
molecule to the active site, while the tag is used to image the enzyme in vitro. Using the click
chemistry strategy, the active site binder in ABPP doesn ’t contain the detectable tag, but only
possesses an azide group. The azide functionality is extremely stable in vivo, so the affi nity
reagent can be used in living cells or whole organisms. After incubation with the azide affi n-
ity component, the probe specifi cally interacts with the enzymes being targeted. Subsequently,
the tissue or cells can be lysed (or fi xed) and probed for bound enzyme using an alkyne-labeled
reagent. This can be a fl uorescent probe or an affi nity handle, such as biotin, for purifi cation.
In fact, most cell-based assays are done using fi xed cells, not live cells, which makes click
chemistry reactions imminently practicable. In this approach, a test population and a con-
trol population of cells is grown and after treating the test population of cells with a potential
drug candidate or another modulator of cellular processes, they are compared relative to the
expression of a biological component or the activity of a biomolecule. Most high content
screening assays are done on cells after a formaldehyde fi xation step followed by a permea-
bilisation process to allow passage of molecular probes into the cells (refer to Thermo Fisher
4. Click Chemistry: CU(I)-Promoted Azide–Alkyne [32] Cycloaddition 683
684 17. Chemoselective Ligation: Bioorthogonal Reagents
Scientifi c, Cellomics). For these applications, the use of azide/alkyne reagents in a click chem-
istry strategy is entirely appropriate and may be the best choice of all conjugation reactions,
because of its exquisite chemoselectivity, bioorthogonality, and excellent reaction kinetics.
The triazole ring generated by the reaction of an azide and an alkyne is a very stable linkage
and not likely to undergo hydrolysis or any other breakdown reaction to cleave the linkage.
Even under relatively extreme conditions used in some biological operations involving the addi-
tion of denaturants, detergents, chaotropic agents, organic solvents, or acidic or basic condi-
tions, the triazole ring will survive and remain intact. Click chemistry reactions are thus highly
chemoselective and result in strong conjugation bonds for use in any application.
Another important advantage to the use of click chemistry for cell-based targeting is the abil-
ity to create bio-monomer analogs containing either azido or alkyne functionalities, which then
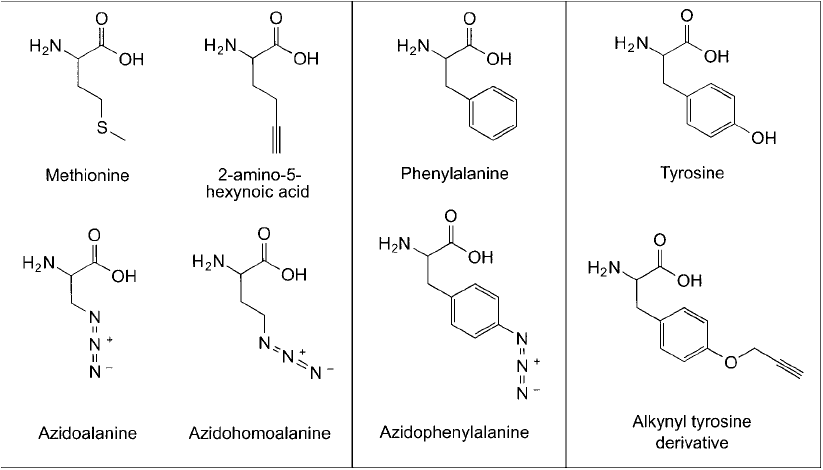
can be incorporated into biopolymers using a cell ’s native enzymatic machinery. Methionine
and phenylalanine analogs containing side chain alkyne or azide groups have been synthesized
and proven able to be introduced into proteins at normal methionine sites in a sequence-specifi c
manner (Link et al., 2004; Prescher and Bertozzi, 2005). Kiick et al. (2002) also demonstrated
that both azido and alkynyl amino acid derivatives could be used as methionine surrogates and
get integrated into proteins with nearly the same effi ciency as normal methionine. In addition,
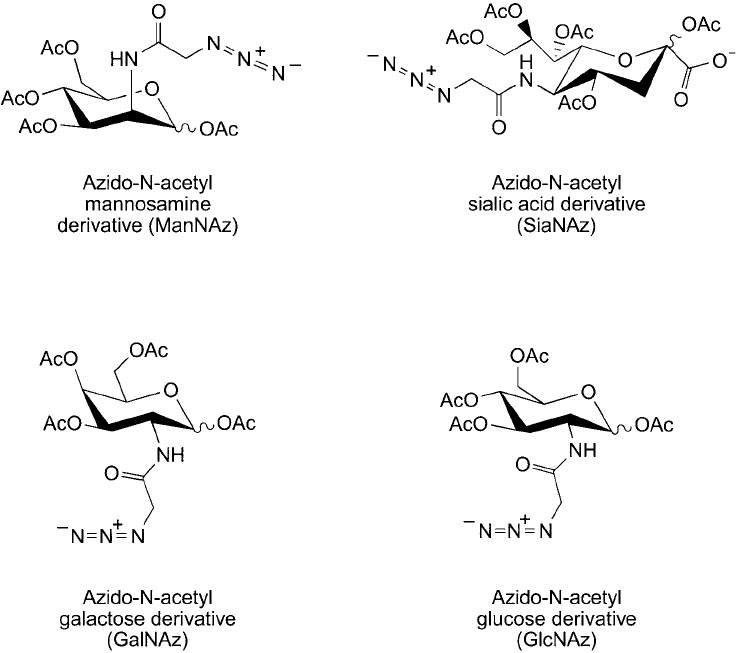
azide-sugar derivatives have been prepared that are capable of being incorporated into glycans
and glycoconjugates using normal enzymatic biosynthetic pathways in cells (Saxon and Bertozzi,
2000). Thus, proteins and carbohydrates can be specifi cally tagged to contain non-canonical
amino acids or sugars for subsequent bioconjugation using reagents containing the opposite
click chemistry reactant ( Figures 17.11 and 17.12 ). Cells grown in the presence of azido or
Figure 17.11 Amino acid analogs containing either azido or alkyne modifi cations can be fed to cells and these
monomers incorporated into expressed proteins.
alkyne monomer analogs will utilize these as building blocks for creating biopolymers. Proteins
and glycans within the cells and on cell surfaces afterward display discrete bioconjugation tar-
gets for subsequent detection, crosslinking, or capture using click chemistry applications.
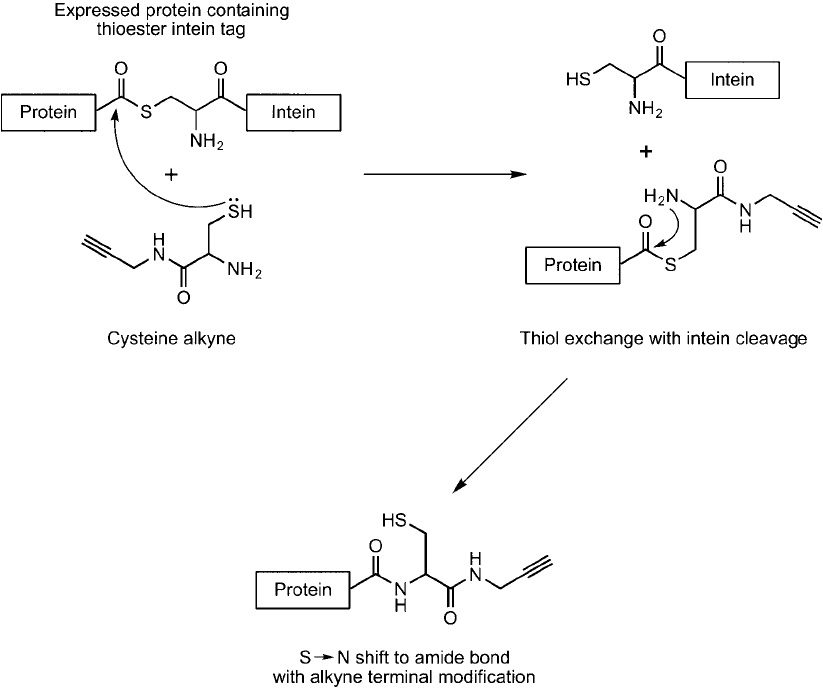
Lin et al. (2006) used click chemistry combined with site-specifi c labeling of recombinant
protein using expressed protein ligation (EPL) (Muir, 2003) to couple proteins to array sur-
faces. A propargylamido-cysteine reagent was used to modify an expressed protein containing a
thioester intein at its C-terminal. Reaction of the free thiol on the propargylamido-cysteine with
the thioester linkage on the protein resulted in transthioesterifi cation followed by an immediate
S;N shift to give an amide bond between the alkyne compound and the protein ( Figure 17.13 ).
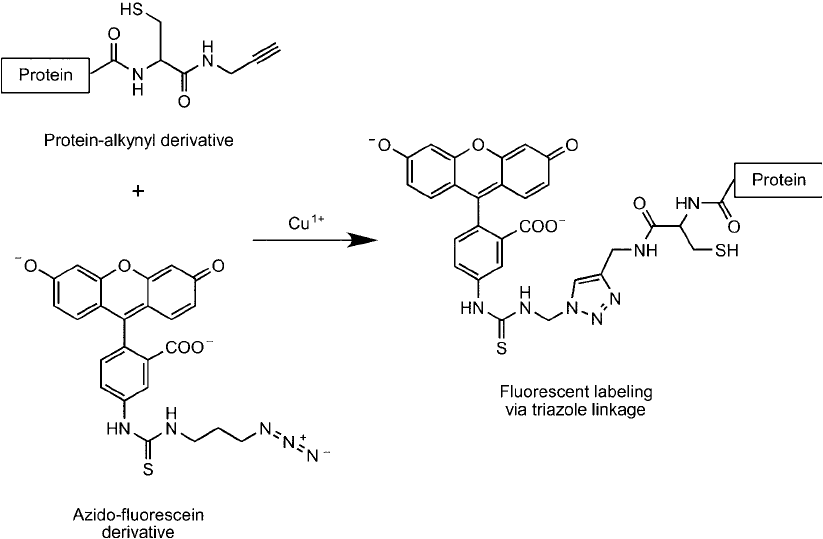
Once the protein is modifi ed to contain an alkynyl group at its C-terminal it can be used
to covalently link to its click chemistry reactant partner, an azide on the surface of an array.
Other azido molecules also can be conjugated with an alkyne-protein to facilitate the detection
or capture of the protein using affi nity techniques. For instance, an azido-fl uorescein reagent
can be used to detect fl uorescently the expressed protein in complex samples or an azido-biotin
Figure 17.12 Azido derivatives of sugars can be used as monomers for glycan and carbohydrate synthesis by
cells. Such modifi cations can be probed using click chemistry or Staudinger ligation reactions.
4. Click Chemistry: CU(I)-Promoted Azide–Alkyne [32] Cycloaddition 685
686 17. Chemoselective Ligation: Bioorthogonal Reagents
derivative can be used to biotinylate the protein at its C-terminal for subsequent purifi cation or
detection ( Figure 17.14 ).
In another application of coupling proteins to surfaces using click chemistry, Duckworth
et al. (2006) carried out prenylation of a protein using a farnesyl azide derivative and the
enzyme farnesyl transferase for subsequent chemoselective ligation to alkyne-functionalized
agarose beads. The result is a highly discrete, site-specifi c attachment of the protein to the solid
phase at a single location.
Bonnet et al. (2006) used the click chemistry reaction to synthesize receptor ligands con-
taining fl uorescent probes or biotin groups to specifi cally tag and detect or isolate receptor
proteins. The effi ciency of the click cycloaddition reaction catalyzed by Cu(I) presented many
benefi ts over doing such synthesis by other routes. If receptor ligands can be modifi ed to con-
tain an alkyne group or an azide, then the opposite click chemistry reactant can be used on any
number of probes to conjugate with the ligand for subsequent receptor probing applications.
Figure 17.13 Expressed proteins containing a thioester intein tag can be specifi cally modifi ed using a cysteine-
alkyne derivative by transthioesterifi cation followed by an internal S ; N shift.
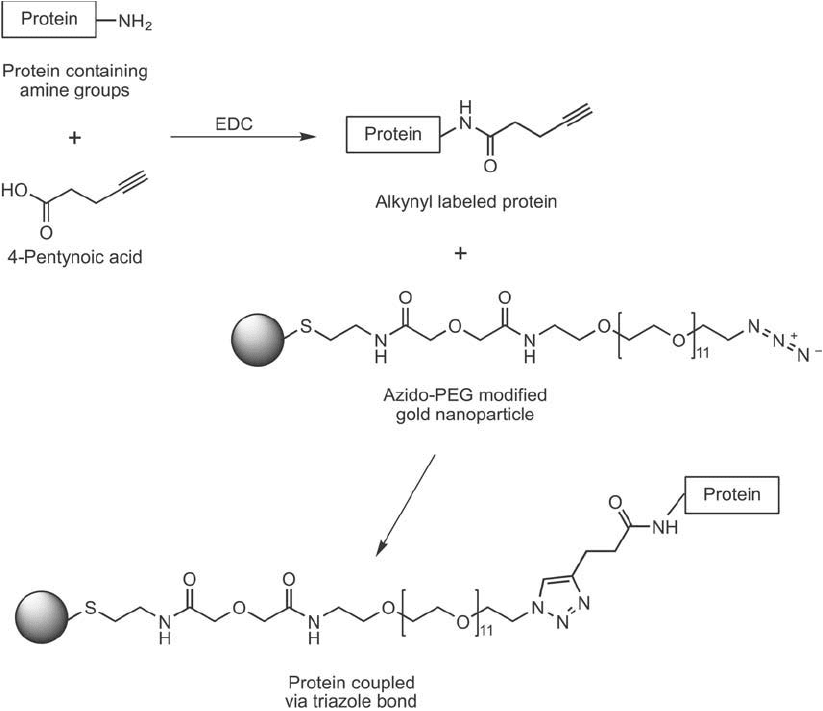
Nanoparticles also can be modifi ed with one of the click chemistry reactants to facilitate pro-
tein or ligand coupling to them in high yield. Brennan et al. (2006) modifi ed gold nanoparticles
with a thiol-azido spacer to produce azide functional groups on the particles for subsequent
coupling of alkynyl-lipases. The thiol end formed a dative bond with the gold surface, which
produced a long, hydrophilic, PEG-containing spacer terminating in an azide ( Figure 17.15 ).
Interaction of the alkynyl-lipase with the modifi ed particles in the presence of Cu(I) resulted
in triazole linkages, which effi ciently immobilized the enzyme on the surface. The resultant
enzyme-gold particles retained high enzymatic activity with an average of 7 protein molecules
conjugated per nanoparticle.
The following protocol adapted from Brennan et al. (2006) describes the coupling of alkyne-
modifi ed protein to 14 nm gold nanoparticles. The lipase enzyme was engineered to con-
tain a single lysine residue that was accessible to the aqueous environment for labeling with
4-pentynoic acid using carbodiimide coupling. This procedure also is applicable to other proteins
containing lysine residues to add alkynyl groups for subsequent click chemistry conjugation
procedures.
An alternative method of modifying proteins to contain alkynyl groups is to use the propargyl–
PEG
1
–NHS ester compound described in Chapter 18, Section 2. This reagent will react sponta-
neously with available amine groups in proteins to form an amide bond without the need to use
EDC, as in the following protocol.
Figure 17.14 An expressed protein containing a thioester intein tag that was subsequently modifi ed by native
chemical ligation to contain an alkyne group then can be labeled using an azido-fl uorescein probe by the click
chemistry reaction in the presence of Cu
1
.
4. Click Chemistry: CU(I)-Promoted Azide–Alkyne [32] Cycloaddition 687
688 17. Chemoselective Ligation: Bioorthogonal Reagents
Protocol
Modifi cation of Protein with Alkynyl Groups
1. Dissolve a protein to be modifi ed with an alkynyl group in 1.2 ml of 20 mM sodium
phosphate, pH 7.0 (PBS), at a concentration of at least 20 M.
2. Prepare a stock solution of 4-pentynoic acid by dissolving it at a concentration of 100 mM
in 50 percent THF/PBS. Add 6.7 l of this solution to the protein solution with mixing.
3. Prepare a stock solution of EDC just prior to use by dissolving it in PBS at a concentration
of 50 mM. Note: EDC is unstable in aqueous environments and should be used immedi-
ately after the solution is made. Quickly add 13 l of the EDC solution to the protein solu-
tion with mixing.
Figure 17.15 The small carboxylate-alkyne compound 4-pentynoic acid can be used to modify proteins at their
amine groups with EDC to provide alkyne sites for click chemistry-mediated conjugation. The subsequent reac-
tion of an azido-PEG-modifi ed gold nanoparticle with the alkynyl-protein in the presence of Cu
1
yields the
triazole-coupled protein.
4. Add to the reaction solution with mixing 150 l of THF and 130 l of PBS.
5. React with mixing in a fume hood overnight at room temperature.
Note: this reaction may be complete within 2–4 hours, as carbodiimide conjugations usu-
ally proceed to completion within this time frame.
6. Purify the alkynyl-protein by dialysis or gel fi ltration using PBS to remove excess reac-
tants and solvent.
Modifi cation of Gold Nanoparticles with Thiol-PEG-Azide Linker
1. Gold nanoparticles (or other metallic or semiconductor particles) are functionalized
with azide groups by suspending a 2.8 nM concentration of particles in 20 ml of water
and adding a 20 mole amount of a thiol-PEG-azide spacer ligand to the suspension with
stirring.
2. Mix for 18 hours at room temperature.
3. Purify the modifi ed particles from excess linker by repeated centrifugation (at least 3
times at 15,000 g) and resuspending each time with water.
Coupling Alkyne-Protein to Azide-Nanoparticles
1. Suspend the azide-nanoparticles in water at a concentration of 13 nM.
2. With mixing, add a quantity of alkyne-modifi ed protein to the particle suspension to pro-
vide at least a 10-fold molar excess over the quantity of azide groups present on the par-
ticles. The high molar excess is important to prevent particle aggregation if the modifi ed
protein has more than one alkyne group, which could crosslink more than one particle
with a single protein, if the protein concentration is too low. However, if the modifi ed
protein only has a single alkyne modifi cation, reacting it at a lower molar ratio is okay.
For example, Brennan et al. (2006) added 36 l of alkyne-lipase (69 M) in PBS buffer to
192 l of azide-nanoparticles (13 nM).
3. Add 2.5 l of 10 mM CuSO
4
5H
2
O, 50 mM ascorbic acid dissolved in water per ml of
the protein and particle mixture. Mix to dissolve.
4. React at room temperature for up to 3 days (or at 4 °C, if the protein is not stable at
ambient temperature). The optimal time of reaction is dependent on the protein being
coupled and the number of alkyne reactive groups available. An alkynyl-protein added
to the nanoparticles at a high molar ratio probably would reach maximal coupling yield
in a matter of hours.
5. Purify the protein-particles by repeated centrifugation (at least 3 times @ 15,000 g) and
resuspending each time with water.
In another application to couple ligands to surfaces, Sun et al. (2006) used two chemoselec-
tive ligation reactions, a Diels–Alder cycloaddition followed by an azide–alkyne (click chem-
istry) reaction to immobilize protein, biotin, or carbohydrate ligands on glass slides. In this
strategy, glass slides were prepared containing maleimidocaproyl groups, which were used
in the Diels–Alder reaction to couple a PEG
4
spacer containing an alkyne on one end and a
cyclopentadiene at the other end. The Diels–Alder reaction was used to link the cyclopentadi-
ene to the maleimide groups, while the alkyne groups at the other end were used in the click
chemistry reaction to attach the azide-containing ligands (see previous Figure 17.3 ).
Click chemistry reactant pairs used for surface immobilization have the advantage of being sta-
ble to aqueous conditions and long-term storage. Unlike many of the other coupling chemistries
4. Click Chemistry: CU(I)-Promoted Azide–Alkyne [32] Cycloaddition 689
