Caballero B. (ed.) Encyclopaedia of Food Science, Food Technology and Nutrition. Ten-Volume Set
Подождите немного. Документ загружается.


combination of methanol (or acetonitrile) and water,
and in some cases may be entirely nonaqueous.
0027 Many foods, including poorly characterized or un-
known samples, will usually be assayed subsequent to
saponification, which offers the option of employing
either chromatographic mode, and permits analytical
simplification through conversion of multiple esters
to a single retinol form. Milks, dairy products, and
fortified cereals, may, alternatively, be tested by direct
injection of a total lipid extract. Figure 3 illustrates a
typical chromatogram obtained for a milk-based
infant formula by this procedure. While retinyl acet-
ate is the additive, the long-chain endogenous esters
are visible as a composite peak, which are resolvable
by independent nonaqueous, reversed-phase chroma-
tography, as illustrated in the insert.
0028In fats and oil products, retinyl esters can
be assayed by direct dissolution in hydrocarbon,
followed by clarification and injection into the
chromatographic system. Analytical success with
these strategies relies on the selective nature of spec-
tral detection modes, the narrow elution window of
the various esters, and their extinction equivalence.
Retinyl
acetate
cis-isomers
Trans-
retinyl
acetate
Long-chain esters
S
O
10 min
P
05
Time (min)
10 15
cis-isomers
fig0003 Figure 3 The lipids from 2 g of milk-based infant formula
(22% fat) were extracted without saponification into 5 ml of
organic solvent: 100 ml was injected directly into high-perform-
ance liquid chromatography (HPLC) equipped with a silica
column and a fluorescence detector (325 nm excitation, 470 nm
emission). Mobile-phase composition was hexane:2-propanol
(99.92:0.08 v/v). Insert: The endogenous esters were collected
and separated by nonaqueous reversed-phase HPLC on a C
18
column with an isocratic mobile phase of acetonitrile:dichloro-
methane (80:20). The major identified long-chain esters are
retinyl oleate (O), retinyl palmitate (P), and retinyl stearate (S).
Time (min)
0
24
3⬘
3
1⬘
1
12
fig0004Figure 4 Retinyl acetate was extracted from a multivitamin
formulation without saponification, using dimethyl sulfoxide
(DMSO). High-performance liquid chromatographic (HPLC)
analysis was achieved on a Waters C
18
Radial-PAK column with
a methanol (100%) mobile phase flowing at 2 ml min
1
. Detection
was achieved by ultraviolet at 280 nm, 0.3 aufs. Vitamins D, E, and
K can be assayed concurrently. Peak identification: 1, retinyl
acetate (vitamin A); 2, cholecalciferol (vitamin D
3
); 3, tocopheryl
acetate (vitamin E); 4, phylloquinone (vitamin K
1
). The elution
positions of retinol (1
0
) and tocopherols (3
0
) are also indicated.
4956 RETINOL/Properties and Determination

Limitations of these approaches include the require-
ment for normal-phase conditions in order to avoid
triglyceride column fouling, and anhydrous or iso-
hydric eluents to obtain reproducible retention. In
addition, any nonesterified retinol present in the
sample will not be concurrently viewed owing to its
tenacious retention on silica relative to the esters.
0029 High-potency pharmaceutical preparations and
vitamin premix formulations can also be successfully
assayed without the need for saponification. Thus,
providing the carrier materials are free from signifi-
cant fat, the retinyl esters can be extracted into a
suitable solvent (e.g., dimethylsulfoxide), insuring
release from encapsulation, diluted into ethanol,
clarified, and injected on to a reversed-phase column.
Such products usually contain vitamins D, E, and K,
all of which can be concurrently assayed using both
wavelength and sensitivity attenuation. However,
under these conditions, it is difficult to analyze for
retinyl acetate and palmitate concurrently, as the
latter is highly retained by nonpolar columns. This
problem can be circumvented by performing the
analyses subsequent to saponification and thereby
recovering total vitamin A as retinol. Figure 4 illus-
trates a typical chromatogram of retinyl acetate in a
pharmaceutical product. The elution position of
retinol is also indicated.
See also: Antioxidants: Natural Antioxidants;
Carotenoids: Occurrence, Properties, and
Determination; Physiology; Chromatography: Thin-layer
Chromatography; High-performance Liquid
Chromatography; Gas Chromatography; Emulsifiers:
Uses in Processed Foods; Food Fortification;
Spectroscopy: Fluorescence
Further Reading
Ball GFM (1988) Fat-Soluble Vitamin Assays in Food
Analysis. A Comprehensive Review. London: Elsevier
Applied Science.
Ball GFM (1998) Bioavailability and Analysis of Vitamins
in Food, pp. 115–161. London: Chapman and Hall.
Bates CJ (2000) Vitamins: fat and water soluble: analysis.
In: Meyers RA (ed.) Encyclopedia of Analytical
Chemistry, pp. 1–35. Chichester: John Wiley.
De Leenheer AP, Lambert WE and Boxdaer R (2000) Vita-
min A: retinol, carotenoids and related compounds.
Modern Chromatographic Analysis of the Vitamins,
Chromatographic Series, vol. 84, pp. 1–74. New York:
Marcel Dekker.
Eitenmillar RR and Larder WO (1999) Vitamin Analysis
for the Helath and Food Sciences, pp. 3–75. Boca Raton,
USA: CRC Press.
Friedrich W (1988) Vitamins, pp. 63–140. Berlin: de
Gruyter.
Klaui HM, Hausheer W and Huske G (1970) Technological
aspects of the use of fat-soluble vitamins and carotenoids
and of the development of stabilised marketable forms.
In: Morton RA (ed.) Fat Soluble Vitamins; International
Encyclopaedia of Food and Nutrition, vol. 9, pp. 113–
159. Oxford: Pergamon Press.
McLaren DS and Frigg M (2001) Sight and Life. Guide-
book on Vitamin A in Health and Disease. Basel, Switz-
erland: Roche.
Noll GN and Becker C (2000) HPLC of non-polar retinoid
isomers. Journal of Chromatography A 881: 183–188.
Olson JA (1991) Vitamin A. In: Machlin LJ (ed.) Handbook
of Vitamins, pp. 1–57. New York: Marcel Dekker.
Pitt GAJ (1985) Vitamin A. In: Diplock AT (ed.) Fat-Soluble
Vitamins. Their Biochemistry and Applications, pp.
1–75. London: Heinemann.
Thiex N, Smallidge R and Beine R (1996) Sources of error
in vitamin A analysis. Journal of the Association of
Official Analytical Chemists 79: 1269–1275.
Thompson JN (1986) Review: official methods for meas-
urement of vitamin A. Journal of the Association of
Official Analytical Chemists 69(5): 727–738.
United States Department of Agriculture (2002) USDA
National Nutrient Database for Standard Reference,
Release 15. Beltsville, MD, USA: Agricultural Research
Services.
Wagner AF and Folkers K (1975) Vitamins and Coenzymes,
pp. 280–307. New York: Robert E. Krieger.
Wyss R (1990) Chromatography of retinoids. Journal of
Chromatography 531: 481–505.
Physiology
H C Furr, Craft Technologies, Wilson, NC, USA
M M McGrane, University of Connecticut, Storrs,
CT, USA
Copyright 2003, Elsevier Science Ltd. All Rights Reserved.
Absorption, Bioavailability, Transport, and
Distribution
0001‘Vitamin A’ is the collective term for compounds that
show the biological properties of retinol, including
maintenance of epithelial tissue and visual function.
This classification includes retinol, retinyl esters, and
retinal. Retinoic acid is included, even though it does
not sustain visual function, because it has been found
to be the active metabolite that carries out many of
the functions of vitamin A. These are isoprenoid
compounds, having in common an 11-carbon poly-
ene chain attached to a trimethyl substituted cyclo-
hexenyl ring (Figure 1). The term ‘retinoids’ refers to
all compounds, natural or synthetic, that show some
biological activity typical of vitamin A, such as pro-
moting differentation of cells in culture; not all reti-
noids can support all the functions of vitamin A, such
RETINOL/Physiology 4957

as vision. Vitamin A compounds are not found as
such in plant tissues, but rather are characteristic of
the animal kingdom; the notable exception is 13-cis
retinal, which serves as a chromophore in the purple
membrane of certain halobacteria.
0002 Dietary vitamin A is provided by two sources: pre-
formed vitamin A and provitamin A carotenoids.
Preformed vitamin A (mostly as esters of retinol
with long-chain fatty acids) comes from animal prod-
ucts or from dietary supplements; retinyl esters, e.g.,
retinyl acetate and retinyl palmitate, are more stable
chemically than is free retinol. Provitamin A carote-
noids arise mostly from plant products: b-carotene is
the most active and is widely distributed in plants,
but other carotenoids (such as a-carotene and
b-cryptoxanthin and b-apo-carotenals) can be im-
portant sources of vitamin A from particular foods
(See Carotenoids: Physiology). The relative import-
ance of these sources of vitamin A is very dependent
on diet. Other carotenoids, such as lycopene and
xanthophyls (including lutein and zeaxanthin), are
major carotenoids in some foods and may have
other important physiological functions, but they
have no provitamin A activity in higher animals.
0003Typical estimates of dietary vitamin A absorption
efficiency are 70–85%. Estimates of carotenoid ab-
sorption are usually much lower, but are confounded
by slow intestinal absorption and rapid metabolism;
there is considerable species variability in absorption
efficiency and in metabolism of carotenoids. Animal-
feeding studies show that the biological matrix of
food carotenoids has profound effects on their bio-
availability.
0004Since both vitamin A and carotenoids are lipids,
intestinal micelle formation with bile acids is essential
for their absorption. Human subjects with impaired
bile acid formation or flow (e.g., with biliary atresia)
may require intramuscular supplementation with
vitamin A and the other fat-soluble vitamins. Within
the intestinal lumen, vitamin A esters (retinyl esters)
CH
2
OH
all-trans Retinol
(vitamin A alcohol)
CH
2
OOC
16
H
31
all-trans Retinyl Palmitate
CH
O
all-trans Retinal
COH
O
all-trans Retinoic acid
(Tretinoin)
13-cis Retinoic acid
(Isotretinoin, Accutane)
C
O
O
COH
O
O
COOH
O
Retinoyl β-glucuronide
4-oxo Retinoic acid
CH
2
OH
3,4-didehydroretinol (vitamin A
2
alcohol)
OH
C
COH
O
11-cis Retinal
CH
O
N
H
4-hydroxyphenylretinamide (4-HPR, Fenretinide)
O
COC
2
H
5
Acitretin (Etretinate, Ro 10-9359)
TTNPB (an arotinoid)
β-Carotene
COOH
O
OH
OH
HO
fig0001 Figure 1 Chemical structures of some representative retinoids.
4958 RETINOL/Physiology
are hydrolyzed to free retinol and are absorbed as
such; this free retinol is promptly reesterified within
the intestinal cells. (Figure 2). Provitamin A carote-
noids, such as b-carotene, are often cleaved within
intestinal cells; this metabolism is predominantly by
central cleavage (mediated by the enzyme carotene
15,15
0
-dioxygenase), although asymmetric cleavage
followed by chain shortening may play a role. Retinal
(also called retinaldehyde or vitamin A aldehyde), the
final product of carotenoid cleavage, is enzymatically
reduced to retinol and is then esterified. The physio-
logical ligand for this esterification process seems to
be retinol bound to an intracellular retinol-binding
protein (CRBP II, molecular weight (MW) approxi-
mately 14 600 Da, one of several small intracellular
retinoid-binding proteins); the primary retinyl ester
synthesizing activity transfers fatty acid from phos-
phatidyl choline (lecithin:retinol acyltransferase,
LRAT), although an acyl-coenzyme A-dependent
esterifying activity (acylCoA:retinol acyltransferase,
ARAT) is also present.
0005 Regardless of their dietary source, the retinyl esters
are incorporated in the core of chylomicra and trans-
ported in the lymph. After removal of triacylglycerols
by lipoprotein lipase as the lipoprotein particle
circulates through peripheral tissues, the chylomicron
remnants are rapidly taken up by the liver, and the
vitamin A esters are hydrolyzed by retinyl ester
hydrolase there. The resulting retinol is then either
reesterified (primarily by LRAT, although ARAT ac-
tivity may be important at high retinol concentra-
tions) and stored in the liver, or released into the
plasma as a complex with plasma retinol-binding
protein (RBP). The two cell types involved in liver
vitamin A metabolism are the hepatocytes (parenchy-
mal cells) and the lipocytes (also called stellate cells,
Ito cells, or fat-storing cells). The hepatocytes are the
major site of RBP synthesis and retinol-RBP release;
some retinyl esters are also found here. The lipocytes
store retinyl esters in cytoplasmic lipid droplets,
which also contain triacylglycerols and some choles-
teryl esters within a phospholipid coat. It has been
suggested that RBP transfers vitamin A, as retinol,
between lipocytes and hepatocytes.
0006Because most forms of vitamin A are hydrophobic,
the transport, metabolism, and function of vitamin A
are dependent on a series of binding proteins, each
specific for its ligand and tissue. Plasma RBP typically
has molecular weight of approximately 21 000 Da
in mammalian species; the complete amino acid com-
position has been determined for several species, and
the gene from some species has been cloned. RBP
binds retinol with 1:1 stoichiometry. The hydropho-
bic retinol molecule fits into a ‘barrel’ within the
protein, shielded from interactions with the aqueous
environment. Holo-RBP, in turn, binds transthyretin
in plasma (TTR; formerly called prealbumin); a TTR
tetramer can bind up to four molecules of RBP. Usual
concentrations of human plasma RBP are 1.9–2.4 mM
(40–50 mgml
1
); typically, total circulating RBP is
80–90% saturated with retinol ligand. Although ret-
inol, retinal, and retinoic acid bind to RBP with simi-
lar affinity, retinol is present in highest concentrations
and is the predominant ligand. Retinyl esters have
much less affinity for RBP and are transported by
serum lipoproteins instead.
0007The release of holo-RBP from liver hepatocytes is
carefully controlled to maintain levels of circulating
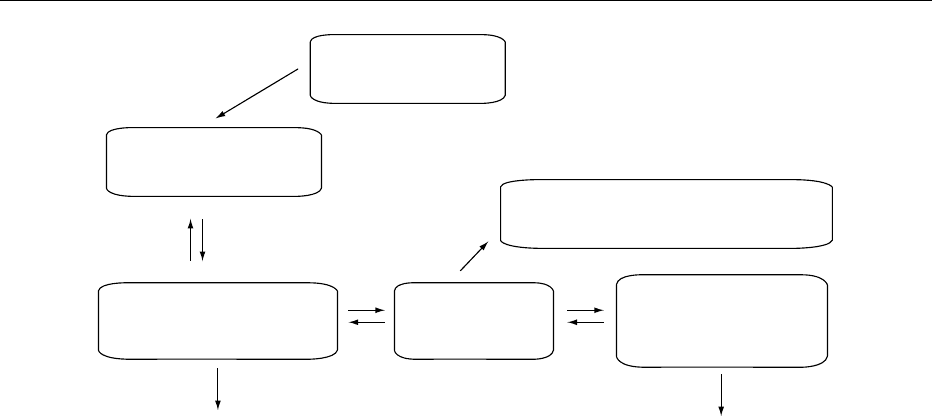
Dietary Vitamin A,
Carotenoids
Eye (visual cycle)
(11-cis Retinal <==> all-trans Retinal)
Intestines
(carotenoid cleavage)
Chylomicrons
(retinyl esters)
Liver
(retinol <===> retinyl esters)
Metabolism, excretion
(including retinoid glucuronides)
Serum
(retinol-RBP-TTR)
Extrahepatic tissues
(retinol, retinyl esters,
retinoic acid)
Metabolism,
excretion
fig0002 Figure 2 Overview of vitamin A metabolism.
RETINOL/Physiology 4959

retinol within narrow limits, but the mechanism of
this control is not yet clear. In the absence of adequate
vitamin A, apo-RBP accumulates in the liver, ready to
be released as soon as vitamin A is available (this is
the basis of the Relative Dose Response assay for
vitamin A deficiency, as discussed below). Clearly,
this mechanism has developed because of the dichot-
omy of vitamin A action: vitamin A is essential, in
small amounts, for proper differentiation and main-
tenance of epithelial cells, but excesses of vitamin A
are toxic and must be avoided by the organism.
Metabolism, Storage, and Excretion
0008 Vitamin A in excess of immediate requirements is
stored in the liver as esters of long-chain fatty acids
(Figure 2); the ratio of liver retinol to retinyl esters
decreases as total liver vitamin A increases, but typic-
ally, 95% of total liver vitamin A is presentas the esters.
Retinyl palmitate is the primary ester in the liver of the
human, rat, cow, sheep, rabbit, cat, frog, trout and
polar bear, althoughsignificant amounts ofother esters
are found (especially oleate and stearate), and the liver
retinyl ester composition can be affected by dietary
fatty acid composition. (Remarkably, the predominant
vitamin A ester in rat adrenal cortex is retinyl stearate.)
Liver frequently contains as much as 90% of total body
vitamin A, although other organs, such as kidney,
testes, and adrenal glands, also contain detectable
retinyl esters. The efficiency of liver storage of vita-
min A is particularly noteworthy in the polar bear,
where concentrations as much as 36 mmol (10 380 mg)
per gram of liver have been reported. Typical values
for human liver vitamin A (autopsy specimens) are
0.44–0.74 mmol g
1
(126–211 mgg
1
) in the USA.
0009 Kinetic studies carried out in rats have shown that
there is extensive cycling of liver vitamin A stores, and
extensive recycling of vitamin A between liver and
other tissues; this assures an ample supply of retinoid
to tissues in response to immediate needs. It is certain
that retinol-RBP is the major form of transport from
liver to peripheral tissues; although the transport of
vitamin A from other tissues back to the liver may be
via retinyl esters transported by lipoproteins (normally
present at about 5–10% the serum concentration of
retinol-RBP), transport via retinol on RBP synthesized
in the outlying tissues may be an important pathway
(mRNA for plasma RBP has been detected in a number
of other tissues in addition to liver). In vitamin A-
deficient rats, the recycling is even more extensive,
and catabolism of vitamin A is markedly diminished.
0010 The rat has served as a valuable model for vitamin A
metabolism in the human, particularly for aspects that
can not be studied readily in humans. In the rat, catab-
olism of vitamin A occurs by oxidation of the cyclo-
hexenyl ring (particularly at the 4-position), epoxide
formation at the 5,6-positions, hydroxylation of the
ring methyl groups, and chain shortening. The
resulting metabolites are generally inactive, although
some may have a little biological activity. These more
polar retinoids are excreted in the urine and in the bile.
0011Retinol can be reversibly oxidized to retinal (vita-
min A aldehyde); retinal can be oxidized to retinoic
acid, but retinoic acid cannot be converted back to
the other forms. Thus, retinol, retinyl esters, and
retinal have equal biological activity because they
are freely interconverted; retinoic acid fills some
(but not all) of the functions of retinol, but it and
its derivatives are not stored. Typical human serum
concentrations of retinoic acid are 5–10 nM (1.5–
3ngml
1
), compared with typical retinol concentra-
tions of 1–2 mM. Retinoyl b-glucuronide and retinyl
b-glucuronide (formed in the liver from retinoic acid
and retinol, respectively) are secreted into the bile but
can be hydrolyzed and reabsorbed in the intestine
(enterohepatic circulation); both these compounds
have vitamin A activity in a variety of tests, are
found in human blood, and may have regulatory
roles in vitamin A function.
Roles in the Body
Vision
0012The major roles of vitamin A are in vision and in
control of gene expression (including differentiation
of epithelial tissues, and in the immune system, as well
as a host of other systems). Metabolism of vitamin A
in the retina of the eye is unique, in keeping with the
unusual role of vitamin A in that tissue. Vitamin A is
stored in the retinal pigment epithelium as retinyl
esters. All-trans retinyl esters are simultaneously
hydrolyzed and isomerized to 11-cis retinol by an
isomerohydrolase enzyme that uses the free energy of
hydrolysis of the retinyl esters to drive the isomeriza-
tion reaction. 11-cis Retinol, a compound unique to
the eye, is then oxidized to 11-cis retinal by an alcohol
dehydrogenase. 11-cis Retinal is transferred from the
pigment epithelium cells to the rod cells by interstitial
retinoid binding protein (IRBP), a distinct binding
protein (MW 140 000). In the rod cells, 11-cis retinal
binds (as a Schiff base with the e-amino group of a
specific lysine residue) to the protein opsin to form the
visual pigment, rhodopsin. When a photon of light is
absorbed by a rhodopsin complex, the 11-cis retinal is
isomerized to all-trans retinal and released from the
protein complex; the resulting conformation change
of the protein initiates a cascade of reactions, resulting
in a neural signal to the brain. The protein opsin is
then available to bind another molecule of 11-cis
4960 RETINOL/Physiology
retinal for another round of the visual cycle. The all-
trans retinal that was released from the protein com-
plex is transferred back to the pigment epithelium via
IRBP, enzymatically reduced to all-trans retinol, and
esterified again. In contrast to the high turnover rates
of vitamin A in other tissues, vitamin A in the eye is
highly conserved, with little leakage back to the liver.
Prolonged vitamin A deficiency, however, leads to re-
duced sensitivity to light, usually first noted as
impaired vision at night (night blindness). These
effects of vitamin A deficiency are generally reversible
by subsequent vitamin A supplementation.
0013 In a very different role of vitamin A in the eye, the
cornea depends on vitamin A for proper cell differen-
tiation and for secretion of protective glycoproteins.
In vitamin A deficiency, these tissues are susceptible
to attack by opportunistic bacteria; such attack may
result in permanent scarring and permanent vision
loss. These effects of vitamin A deficiency, unlike
those in the retina, may not be reversible by subse-
quent vitamin A supplementation. Such vitamin A-
dependent corneal degeneration (given the general
name ‘xerophthalmia’) accounts for an estimated
500 000 new cases of blindness in preschool children
in the world each year.
Control of Gene Expression
0014 Overview of control of gene expression Vitamin A
and its metabolites are required for embryonic
development, cellular differentiation, growth, repro-
duction, adult tissue homeostasis, vision, and main-
tenance of the immune system. Over the past several
years, it has become clear that retinoids mediate
many of their pleiotropic effects by regulating the
activation or inhibition of gene transcription. It is
well documented that retinoids regulate the expres-
sion of many genes involved in their own transport
and metabolism. The genes for different retinoid re-
ceptor subtypes, the cellular retinol binding proteins
(CRBP) I and II, and the cellular retinoic acid binding
protein II (CRABP II) are regulated by either all-trans
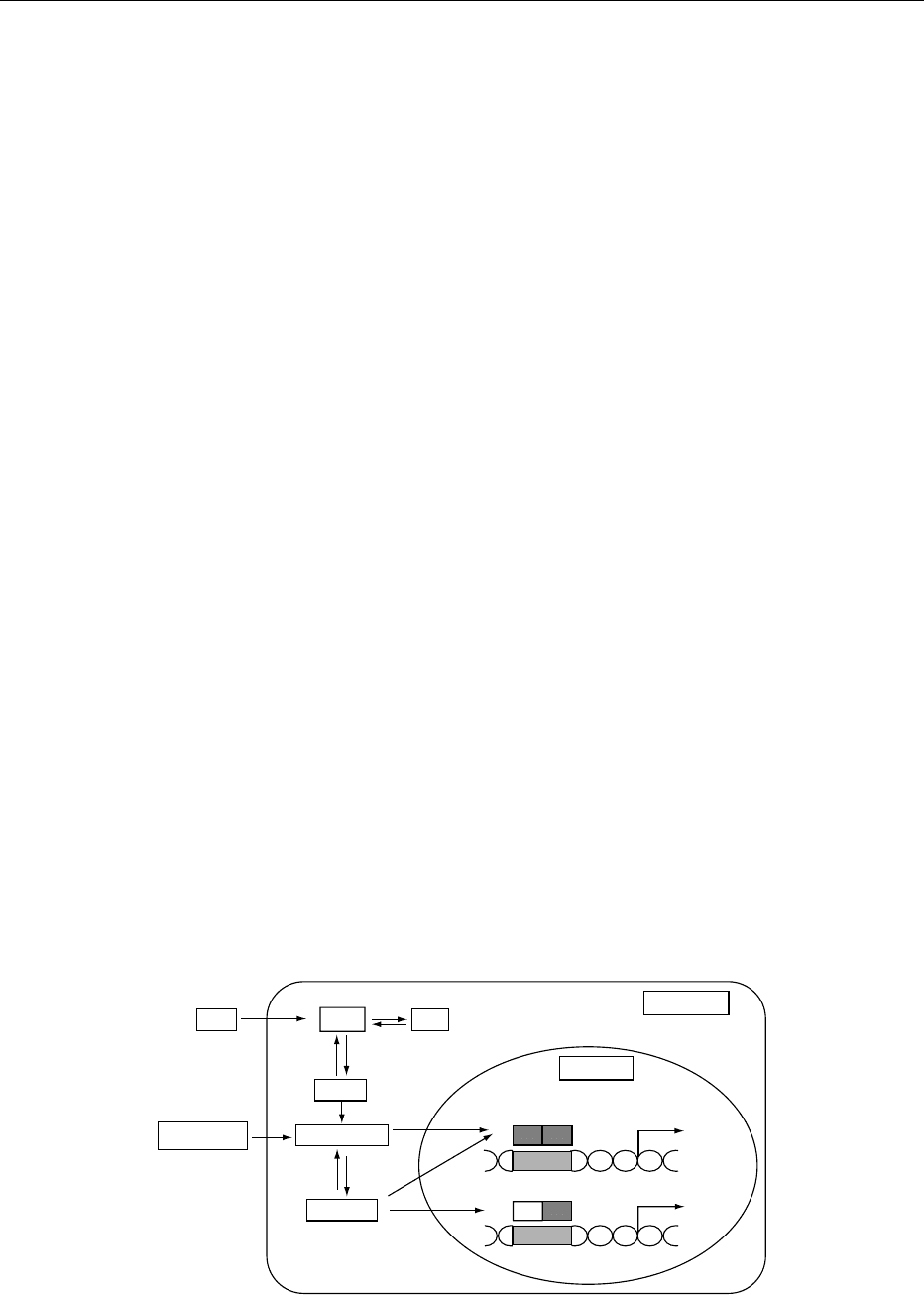
retinoic acid (RA) or 9-cis RA (Figure 3). These vita-
min A metabolites exhibit hormonal action by bind-
ing to their nuclear receptors that interact with DNA
responseelementswithintargetgenestoaffectgenetran-
scription. The retinoid receptors are members of the
steroid/thyroid/retinoid superfamily of nuclear recep-
tors. There are two distinct families of nuclear
retinoid receptors: the retinoic acid receptors
(RARs) and the retinoid X receptors (RXRs). It has
been suggested that the two classes of receptors have
a distinct functional activity, the RARs being import-
ant in cell development and differentiation, and the
RXRs being important in metabolic regulation. Nu-
clear receptors of the steroid/thyroid/retinoid super-
family can act as either inhibitors or activators of
transcription. The mechanism whereby these regula-
tory properties are propagated is via the recruitment
of accessory proteins to the nuclear receptor. In the
absence of ligand, such as all-trans RA, corepressor
proteins are bound to retinoid receptors and cause
inhibition of transcription of the target gene.
Conversely, binding of ligand causes a conforma-
tional change in the receptor such that corepressor
proteins are released, and coactivator proteins are
recruited to the retinoid receptor. The association of
coactivators with the nuclear retinoid receptor causes
induction of transcription of the target gene, initiated
by retinoic acid binding to its specific nuclear receptor.
0015Nuclear retinoid receptors As indicated previously,
there are two distinct classes of nuclear retinoid re-
ceptors, the RARs and RXRs. Both RARs and RXRs
bind the naturally occurring metabolites of vitamin A
with a similar order of affinity (all-trans RA > reti-
nal > retinyl acetate > retinol), but RXRs require from
10- to 40-fold more ligand for comparable tran-
scriptional activation. A downstream metabolite of
ROH
RCHO
ROH RE
all-trans RA
all-trans RA
9-cis RA
RAR
RXR RXR
RXRE
Nucleus
Cytoplasm
RXR
RARE
fig0003 Figure 3 Retinoid action in the nucleus.
RETINOL/Physiology 4961

all-trans RA, 9-cis RA, has been shown to be the
naturally occurring, high-affinity, ligand for the
RXR. The binding of ligand to nuclear retinoid recep-
tors induces dimer formation and initiates transcrip-
tional activation of a target gene via the DNA
response element(s) in the promoter/regulatory region
of the gene. Retinoid receptors act in a dimeric con-
formation and bind to specific RA response elements
(RARE) within the 5
0
promoter/regulatory regions of
target genes. The DNA response elements are usually
a direct repeat (DR) of six nucleotides in a specific
sequence separated by a defined number of nucleotides
that vary depending on the type of DNA response
element. RAR/RXR heterodimers can activate or re-
press gene transcription, depending on the type of
RARE to which they bind in the promoter region of
a target gene. If the RAR/RXR heterodimer binds to a
DR-5 RARE, the RAR/RXR complex activates tran-
scription in response to the RAR ligand all-trans RA.
In contrast, RAR/RXR heterodimers bound to a
DR-1 RARE exhibit no response to RAR-specific
ligands and repress the response to RXR-specific
ligands such as 9-cis RA. Therefore, the RAR/RXR
heterodimer can be stimulatory, neutral, or inhibi-
tory, depending on ligand, all-trans RA or 9-cis RA,
and the DNA RARE to which it is bound. This is
partially explained by the fact that there is a specific
polarity to the binding of RAR and RXR hetero-
dimeric partners, determined by the DNA itself.
RAR/RXR heterodimers bind with the RXR occupy-
ing the upstream half-site of a DR-5 element, but this
orientation is reversed on a DR-1 element. Further
investigation of this polarity of RAR/RXR heterodi-
mer binding has shown that transactivation by RAR
bound to a DR-5 RARE involves the ligand-depend-
ent dissociation of a bound corepressor (discussed
below), but RAR bound to a DR-1 RARE does not
respond to ligand by dissociation of a bound core-
pressor. Therefore, transcription is not stimulated in
the later response. In this regard, the DNA itself
is considered an allosteric regulator of receptor–
accessory protein interaction.
0016 All of the RAR and RXR subtypes, a, b,andg, are
encoded by separate genes at distinct chromosomal
loci. However, the protein products encoded by these
genes have certain common modular structures, par-
ticularly in the DNA-binding and ligand-binding
domains, which define, in part, their membership
within the steroid/thyroid/retinoid superfamily. The
amino acid sequence of the RARs can be divided into
six regions (A–F). The amino acid sequence of
the RXR genes has a similar domain structure
to the RARs. Among the retinoid receptors, the
DNA-binding domain (C) is a highly conserved
region of approximately 70 amino acids that contains
two zinc fingers; the zinc-finger configuration is re-
sponsible for specific DNA response element recogni-
tion. According to Evans and colleagues, a sequence
of three amino acids within the first zinc finger (the P
box) determines recognition of the half-site sequence
of a DNA response element, and five amino acids in
the second zinc finger (the D box) are critical
for recognition of the spacing between half-sites.
Nuclear magnetic resonance spectroscopy of the
DNA-binding domain indicates a secondary structure
comprising three a-helices; the first helix is postulated
to fit across the major groove of the DNA response
element. In this orientation, the third helix interacts
with the minor groove of the DNA, such that Lys and
Arg residues make contact with the phosphate back-
bone and stabilize protein–DNA interactions. This
third helix in the DNA-binding domain is also
thought to mediate protein–protein interaction re-
quired for dimer formation. The ligand-binding
domain (E) is also highly conserved among the RAR
and RXR genes and mediates the ligand-dependent
transcriptional activation function. The amino-
terminal region (A) is less well conserved and is sub-
type-specific. Both regions A and B are responsible for
ligand-independent transcriptional activation. The
amino-terminal part of domain D may function as a
nuclear localization site.
0017In addition to subtype differences in RARs and
RXRs, there are also different mRNA isoforms for
each gene. These isoforms are generated by different
transcription start sites and alternative splicing,
which affects the 5
0
untranslated region (UTR) and
the A domain such that either a deleted or altered A
region is combined with a common B–F region.
0018The biological activity of vitamin A metabolites is
further extended by the interaction of the retinoid
receptors with other members of the receptor super-
family. The RXRs serve as heterodimeric partners
with RARs, thyroid hormone receptors (TR), and
the vitamin D receptors (VDR) and have been
shown to increase the affinity of RAR, VDR, and
TR, for their respective response elements (favored
DR motifs DR-2/DR-5, DR-3, and DR-4, respect-
ively), and to activate transcription in the presence
of the appropriate ligand. This indicates a central role
for the RXR as a heterodimeric partner with the other
nuclear receptors of this superfamily. Interestingly,
only one ligand is required to stimulate the hetero-
dimeric complex and affect transcriptional acti-
vation. However, the RXR is unique in that it can
function as a homodimer. In this capacity, it can
stimulate gene transcription favoring the DR-1 hor-
mone response element and is dependent upon the
9-cis RA ligand. The RXR also forms a heterodimeric
complex with the peroxisome proliferator-activated
4962 RETINOL/Physiology

receptor (PPAR) and, for example, mediated by a DR-
1 motif in the promoter of the acyl-CoA oxidase gene,
stimulates transcription in response to both 9-cis RA
or clofibric acid (a peroxisomal proliferator). Re-
cently, it has been shown that dietary polyunsaturated
fatty acids (PUFA) activate PPAR, and via PPAR–
RXR heterodimer formation, dietary PUFA regulate
both lipid and glucose metabolism. Clarke refers to
dietary PUFA as ‘fuel partitioners’ that act, via PPAR
activation, to increase glycogen storage and fatty acid
oxidation while decreasing triglyceride synthesis.
0019 The multiplicity of nuclear retinoid receptors re-
flects developmental and tissue-specific differences in
their expression. In the mouse embryo, RARa is ubi-
quitously expressed, whereas RARb and RARg are
more restricted, and nonoverlapping, in their expres-
sion. RXRb genes are widely expressed in the mouse
embryo, whereas RXRa is found in liver, kidney,
intestine, and skin, and RXRg is more restricted
still. In the adult, both RARa and RXRb are found
in most tissues, whereas RARb,RARg, RXRa, and
RXRg are more limited. RXRa is highly expressed in
liver, kidney, spleen, and visceral tissues, indicating
a possible role for this RXR in retinoid, lipid,
and carbohydrate metabolism. The demonstrated
RXR responsiveness of numerous genes including
the CRBP II, apolipoprotein (apoAl), acyl-CoA oxi-
dase and the phosphoenolpyruvate carboxykinase
(PEPCK) genes further supports this hypothesis.
0020 Nuclear coactivators and corepressors associated
with nuclear retinoid receptors Recently, several
coactivators and corepressors that associate with nu-
clear retinoid receptors and affect their ability to
regulate target genes have been cloned and character-
ized. It has been established that retinoid binding
induces conformational changes in nuclear retinoid
receptors and promotes their association with a
diverse group of nuclear proteins that function as
coactivators of transcription. Alternatively, unbound
receptors recruit nuclear proteins that repress tran-
scription of target genes. It has been demonstrated
that all-trans RA binding to RAR, with subsequent
activation of transcription of a target gene, requires
dissociation of corepressor proteins and recruitment
of coactivator proteins. It is thought that corepressors
contribute to the formation of condensed chromatin
structure, prohibiting activation of target genes. Con-
versely, most coactivators cause a loosening of chro-
matin conformation, allowing nuclear receptors to
activate transcription of target genes.
0021Regulation of the PEPCK gene by retinoids The
multifaceted action of vitamin A is indicated by its
role in regulation of the gene encoding PEPCK, the
rate-determining enzyme of gluconeogenesis in liver
(Figure 4). All-trans RA and 9-cis RA, in equal con-
centrations, induce the PEPCK promoter by approxi-
mately sevenfold in hepatoma cells in culture. Studies
in mice in vivo have shown that vitamin A deficiency
causes a decrease in PEPCK gene expression. Specif-
ically, with food deprivation, hepatic PEPCK mRNA
levels are not induced in vitamin A-deficient mice.
Induction of the PEPCK gene can be reestablished
with retinoid treatment of vitamin A deficiency. Reti-
noid regulation of the PEPCK gene has been tested
in vivo in two lines of transgenic mice. Results from
these experiments indicate that expression driven by
both the 460/þ73 and 355/þ73 PEPCK DNA
sequences is decreased with vitamin A deprivation.
Subsequent treatment of vitamin A deficiency with
all-trans RA increases expression driven by the
460/þ73 promoter or the minimal 355/þ73 pro-
moter. The second retinoid isomer, 9-cis RA, however,
9-cis RA
DR-1 RARE:
Required for 9-cis RA induction
Inhibitory for all-trans RA
DR-5 RARE:
Mediates all-trans RA induction
No response to 9-cis RA
DR-1
RARE
TGACCT
TTGGCCG
−451 −439
DR-5
RARE TATA
AGCCCT
GTCCTTGACCC
−337 −321
RXR RXR
all-trans RA
RXR RXR
fig0004 Figure 4 Retinoic acid response elements involved in regulation of the PEPCK gene.
RETINOL/Physiology 4963

increases expression driven by the 460/þ73 pro-
moter, but not the 355/þ73 promoter. Therefore,
the two active retinoid isomers in liver have different
molecular signaling mechanisms and activate the
PEPCK gene at different DNA response elements.
0022 Additionally, it has been shown that appropriate
developmental regulation of the PEPCK gene during
fetal development and the perinatal period is depend-
ent on adequate vitamin A nutriture. Vitamin A defi-
ciency causes a decrease in hepatic PEPCK mRNA
during late gestation, a stage in development when
the PEPCK gene is first expressed in liver. At birth,
with adequate vitamin A nutriture, the PEPCK gene
is rapidly induced. This is consistent with the need of
the newborn to conduct gluconeogenesis because of
removal from the maternal glucose supply. The rapid
induction of PEPCK gene expression at birth is sig-
nificantly decreased with vitamin A depletion in the
newborn. Overall, adequate vitamin A is required
for the appropriate developmental expression of the
PEPCK gene in late gestation and at birth. The fact
that vitamin A is an active regulator of expression of
the PEPCK gene indicates a significant role for this
nutrient in the regulation of carbohydrate metabol-
ism overall. This is a new area of investigation of
vitamin A regulation of gene expression as it expands
the known role of vitamin A beyond the regulation of
genes involved in vitamin A metabolism itself.
Human Health
Infection and Disease
0023 Early in the study of vitamin A requirements, it was
appreciated that vitamin A-deficient animals are
more susceptible to disease. Attempts to extend
these observations to human health long gave incon-
sistent results, however. Recently, it has been shown
that supplementation of children with vitamin A re-
duces the severity of measles and diarrhea (although
vitamin A supplementation may not reduce the inci-
dence and duration of diarrhea). Although vitamin A
prophylaxis may not reduce measles severity, sup-
plementation of measles patients with vitamin A
(200 000 international units (IU), i.e., 60 000 mgof
RE, 209 mmol) during infection does reduce hospital
stay and mortality rate and is now recommended by
WHO in communities where vitamin A deficiency is
prevalent. Currently, it is not believed that vitamin A
supplementation can provide any benefit for other
respiratory diseases (nonmeasles pneumonia). The
possible role of vitamin A status in tuberculosis, mal-
arial infection, and HIV infection is not clear. How-
ever, vitamin A public health programs can have a
tremendous impact on child morbidity and mortality
in developing countries.
0024The mechanism(s) by which retinoids affect the
immune system have not yet been delineated, but it
seems safe to assume that control of gene expression is
involved. In at least some experiments, the function of
vitamin A can be filled by all-trans or 13-cis retinoic
acid, consistent with current knowledge of retinoids
and gene expression. Both antibody-mediated and
cell-mediated immune responses are reduced in vita-
min A deficiency. Interpretation of human studies of
the relationship between vitamin A status and immune
response is complicated by the fact that plasma retinol
concentrations (often used as a convenient measure of
vitamin A status) fall during infection even in vitamin
A-replete subjects, and chronic infection reduces vita-
min A absorption and mobilization.
0025Dermatology One of the first pharmaceutical appli-
cations of retinoids was the use of 13-cis retinoic acid
(isotretinoin) to combat cystic acne, ichthyosis, and
follicular keratosis; 13-cis RA inhibits sebaceous
gland size and activity by promoting CRABP II
mRNA expression. The process of keratinization
(differentiation and maturation) of skin cells is
affected both by deficiency and by excess of retinoids.
Dehydroretinol and its esters (vitamin A
2
) are found
in human skin (more so in epidermis than in dermis)
and may have specific roles there. Retinoids are now
used clinically to treat psoriasis, ichthyosis, Darier’s
disease, and other disorders of keratinization, acne,
and photoaged skin. Synthetic retinoids, such as etre-
tinate and the ethyl ester of the arotinoid TTNPB
(Figure 1), as well as the naturally occurring retinoyl
b-glucuronide, have been developed in attempts to
reduce the toxicity of the naturally occurring retinoic
acids without losing efficacy.
0026Cancer prevention and treatment Retinoids can in-
hibit cell transformation and tumor development in a
variety of cell and animal models, but the effects are
not uniform with all malignancies. In general, it
seems that retinoids may not prevent initiation of
tumors, but more likely interfere with the promotion
process. They exert these effects by modulating
growth, differentiation, and apoptosis of cells; most,
if not all, of these effects may be explained by the role
of retinoids in control of gene expression.
0027Retinoids have been tested against a variety of
cancers in human and animal studies. N-(4-Hydroxy-
phenyl)-retinamide (4-HPR, Fenretinide) has been
tested clinically for prevention of recurrence of
human breast cancer; although results are promising,
some toxicity (owing to interference with normal
retinol metabolism) has been observed. Incidence of
skin tumors associated with xeroderma pigmentosum
is reduced by treatment with 13-cis retinoic acid.
4964 RETINOL/Physiology

Perhaps the most dramatic application is the use of
retinoic acid (tretinoin) against promyelocytic leuke-
mia; this disease results from a chromosomal trans-
location producing the fusion of the RARa gene with
the PML gene to produce an oncogene and responds
well to high doses of all-trans retinoic acid.
Requirements and Recommended Intakes
0028 Because of confusion among the various units for
presenting vitamin A values, the concept retinol
equivalent (RE) has been proposed. One retinol
equivalent equals 1 mg of all-trans retinol, either free
or as the retinyl component of a retinyl ester; 1 RE ¼
3.33 IU of vitamin A, or 3.5 nmol of retinol or retinyl
ester. Although the exact vitamin A value of caro-
tenoids depends on several factors (See Carotenoids:
Occurrence, Properties, and Determination), the ret-
inol equivalent was defined as 6 mg of all-trans b-
carotene or 12 mg of other provitamin A carotenoids
(10 IU of provitamin A carotenoid). To account for
updated findings on provitamin A value of carote-
noids, the retinol activity equivalent (RAE) has been
defined more recently as 1 mg of all-trans retinol,
12 mg of all-trans b-carotene, or 24 mg of other pro-
vitamin A carotenoids.
0029 Liver concentrations provide the best appraisal of
vitamin A status. Human liver specimens can be diffi-
cult to obtain, but analysis of autopsy specimens can
be useful in evaluating the vitamin A status of a
population. As indicated above, serum retinol levels
are maintained nearly constant and are not generally
useful in assessing vitamin A status, except when liver
vitamin A reserves fall well below 0.07 mmol g
1
(20 mg per gram of liver). Serum (or plasma) retinol
levels are normally 1–2 mM (290–570 ng ml
1
) across
a wide range of mammalian species.
0030 Because liver concentrations of vitamin A are not
readily measured, and serum retinol values are not an
adequate indicator of an individual’s vitamin A
status, several indirect methods have been developed.
Conjunctival impression cytology (CIC) evaluates the
development of squamous metaplasia (enlarged epi-
thelial cells) and loss of goblet cells from the conjunc-
tiva of the eye by histological examination of cells
transferred from the cornea to filter paper. The rela-
tive dose response (RDR) depends on the release of
retinol-RBP from liver into serum after a large oral
dose of vitamin A (typically, 450 mg of retinyl acetate
in human studies); as described above, apo-RBP ac-
cumulates in the liver in vitamin A depletion. The
RDR is calculated as:
RDR ¼
A
5
A
0
A
5
100%,
where A
5
represents the serum retinol concentration
at 5 h after the oral dose, and A
0
represents the fasting
serum retinol concentration at the time of the dose.
Studies in both humans and rats indicate that an RDR
value greater than 20% indicates liver vitamin A
reserves less than 0.07 mmol g
1
(< 20 mg per gram of
liver), i.e., inadequate vitamin A status. The RDR
assay requires, of course, that the oral dose be nor-
mally absorbed (an intramuscular injection has been
used in human subjects with biliary atresia), and that
protein metabolism is normal: protein deficiency or
zinc deficiency or liver cirrhosis impairs the dose
response. An alternate approach, the modified rela-
tive dose response (MRDR), uses the vitamin A
analog 3,4-didehydroretinol (vitamin A
2
, a form
found in some freshwater fish and found in very low
levels in human skin). The ratio of vitamin A
2
to
vitamin A
1
in serum retinol at 5 h after an oral dose
is used as a measure of vitamin A status, with high
values (ratio > 0.03 after an oral dose of 100 mg per
kilogram of body weight) indicating poor vitamin A
status. Chlorinated vitamin A analogs have been
used in a similar fashion. The RDR and MRDR, as
well as CIC, have been used successfully in human
population studies. Isotope dilution of tracer-labeled
vitamin A (radioactive or stable-isotope labeling) has
been used to estimate human vitamin A status, but
technical difficulties have so far prevented general use
of the technique.
0031Nutritional requirements for vitamin A have not
been well defined because of the diversity of vitamin
A functions. In animal studies, intakes of 3–8 RE per
day per kilogram of body weight (10–28 nmol per day
per kilogram of body weight) cure deficiency symp-
toms and intakes of 30–60 RE per day per kilogram of
body weight (100–210 nmol per day per kilogram
of body weight) produce optimal growth. Kinetic
studies of vitamin A metabolism in rats show that
irreversible loss of vitamin A is decreased on low
vitamin A intakes. Studies on vitamin A requirements
have not yet addressed functional criteria such as
immune function and possible anticancer effects.
0032To provide an adequate and safe human intake,
current WHO/FAO dietary recommendations (1988)
and suggested recommended dietary intakes (RDIs)
for adults are based on intakes of 9.3 RE per day per
kilogram of body weight (33 nmol of vitamin per day
per kilogram of body weight). The European Union
Scientific Committee for Food has recommended
population reference intakes of 700 RE per day
(men) and 600 RE per day (women), with estimated
average requirements of 500 and 400 RE, respect-
ively, and lowest threshold intakes of 300 and
250 RE, respectively. In contrast, the recommended
dietary allowances (RDAs) of the US National
RETINOL/Physiology 4965
