Zelditch M.L. (и др.) Geometric Morphometrics for Biologists: a primer
Подождите немного. Документ загружается.


chap-14 4/6/2004 17: 29 page 368
368 GEOMETRIC MORPHOMETRICS FOR BIOLOGISTS
this issue must be addressed before the first one is even relevant; coding becomes a moot
issue if there are no characters to code, and if there are no characters, we cannot test the
hypothesis that they are especially homoplastic.
It is clear that partial warps should not be used as characters (for the reasons discussed
below), but it is not clear what ought to be used instead. It is not even clear that the
problem has a solution. The major objective of the first part of this section is to define the
problem we had hoped to solve using partial warps, then to explain why our approach was
flawed. In the next section we discuss two alternatives, both of which rely on conventional
multivariate methods, but neither is precisely tailored to the problem.
Defining the problem
The general problem we face is to find features that differ among taxa and are shared by a
subset of them. The differences indicate evolutionary novelties and the similarities indicate
common ancestry, although we will not be able to determine which are novelties until we
have completed the phylogenetic analysis. We would not expect that an entire shape is a
character because species rarely have exactly the same shape (whether we are comparing
whole bodies or parts of them). If we think of the problem from the perspective of whole
landmark configurations, we will not make any progress. On the other hand, if we do
not think of the problem in terms of whole landmark configurations, we may be led to
theoretically invalid solutions. Therefore the problem is to analyze entire configurations of
landmarks, and find features that differ among taxa and are similar among a subset of taxa.
Additionally, to say that we have a character we must be able to say where it is, and over
how large a spatial expanse it extends. A primary objection to traditional morphometric
variables is that they are lines, having no spatial extent as individual variables. As soon
as we try to determine their spatial location and extent, even by multivariate analyses,
we face one of the most severe limitations of traditional morphometric data – their poor
ability to localize morphological differences.
When looking for these similarities and differences, we are not concerned with the
magnitude of the difference, nor its degree of localization. Small differences (so long as
they are large enough to be considered a difference at all) count as much as large ones, and
spatially large-scale differences count as much as localized ones. Consequently, neither the
Procrustes distance between taxa nor the bending energy of the transformation has any
relevance to the problem. This is one of the reasons why it is so difficult to solve – neither
of the metrics used in geometric morphometrics is germane to the problem, and if there is
a relevant metric, it has yet to be defined.
When we first approached this problem, we focused on one major limitation of conven-
tional (qualitative) approaches: that organisms are often dissected arbitrarily, along lines
of convention. Conventional anatomical subdivisions are often not biologically meaningful
except in the context of a particular problem. For example, if we are interested in locomo-
tion and foraging, we can subdivide an organism into parts that are used in locomotion and
parts that are used in foraging. Alternatively, if we are interested in development, we can
subdivide the organism into parts that have a common germ-layer origin, or that develop
from the same type of bone, or that undergo the same kinds of epigenetic interactions,
etc. These subdivisions have long been regarded as arbitrary, except to the extent that
they are useful in a particular investigation. These subunits are not suitable for dissecting

chap-14 4/6/2004 17: 29 page 369
MORPHOMETRICS AND SYSTEMATICS 369
an organism in systematic studies when a single character crosses several such divisions,
or is partly within and partly without them. Our goal in using partial warps was to find
a more objective basis for dissection. We did not succeed (for reasons discussed below)
but the problem we defined remains a fundamental and unresolved difficulty for character
analysis. Our method had fatal flaws, but so do others that require us to decompose the
organism prior to measurement.
The approach we took is similar to one that is standard in cladistic studies using mor-
phometric data. We defined a set of variables a priori, and compared taxa with respect
to them. A similar tactic is applied to conventional morphometric variables, when a set
of lengths or ratios is defined and measured on taxa, then the values of those lengths or
ratios is compared among them. Most attention has focused on the problem of coding
those variables, but coding is the least of the problems. Such variables do not solve the
problems we had hoped to address, but share with them the flaw that we inadvertently
introduced: they compare arbitrarily selected components of shape one at a time.
Why not to use partial warps as characters
Even though partial warps have a geometric scale, are a function of homologous landmarks,
and do not emphasize differences of large magnitude at the expense of small ones, they
cannot be used as characters for at least two reasons. The first is obvious (in hindsight at
least): partial warps have a spatial scale, but an individual partial warp (PW) describes
only part of a small-scale anatomical feature. Partitioning a change by PWs does not
correspond to partitioning it by anatomy or by characters, because a single PW does
not describe a single, spatially coherent change (although several, taken together, might).
When comparing multiple taxa, a combination of several PWs taken together is usually
needed to describe any change, even one that is anatomically local. Additionally, having a
high score on a localized PW does not mean that there is a localized change. Instead, the
change within that region may be partly described by a PW at a higher spatial scale, and
the localized PW supplements that description. Taken out of context of the larger-scale
PW, we cannot make anatomical sense of the one at smaller scale. Two taxa that have
identical values for a small-scale PW might differ anatomically – differences that cannot
be seen without looking at all PWs.
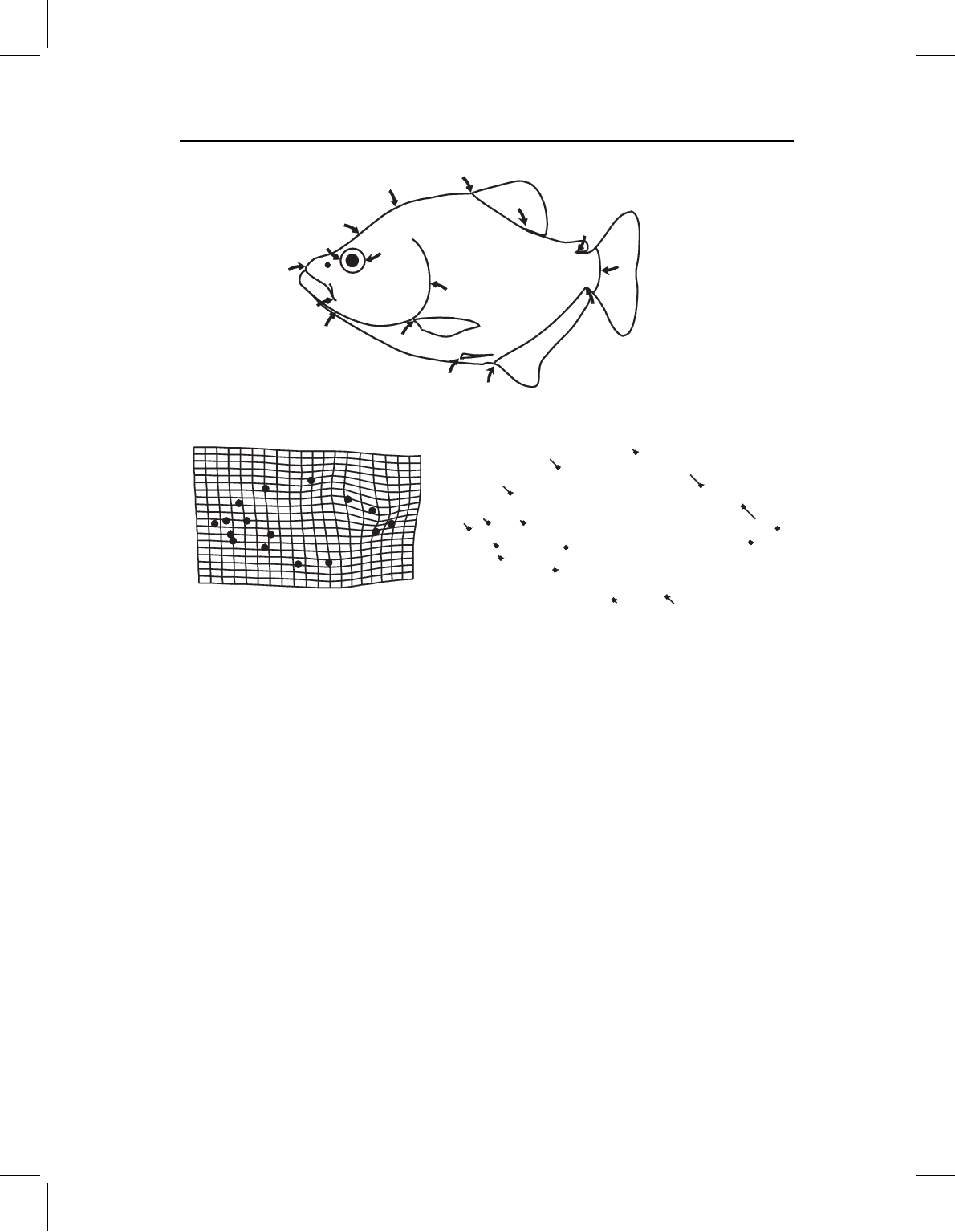
All that may be obvious to readers who have reached this chapter, but to clarify the
point we can re-examine the example that we found most promising at the time – the
ontogenetic change in scores on one PW (Figure 14.2). Two of the taxa, which were
used as outgroups (Pygopristis denticulata and Serrasalmus gouldingi), have statistically
significant ontogenetic change on that PW (in both X and Y directions), whereas the
three Pygocentrus do not. We would not normally be concerned about similarities among
outgroups, but this example shows that similarities implied by individual PWs are not
found in complete descriptions. That P. denticulata and S. gouldingi have anything in
common in their development of that region is not at all obvious when looking at more
complete descriptions of the five ontogenies (Figure 14.3). They are similar to each other,
and differ from the three Pygocentrus, only in that they undergo an ontogenetic change
in the caudal peduncle region that is not fully described by PWs at higher spatial scales.
However, P. denticulata and S. gouldingi are not similar to each other in the changes
described by the higher spatial scales (and neither are the three Pygocentrus). Being similar
chap-14 4/6/2004 17: 29 page 370
370 GEOMETRIC MORPHOMETRICS FOR BIOLOGISTS
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
Figure 14.2 A single PW used to exemplify the procedure for finding systematic characters in
ontogenies of shape in Fink and Zelditch, 1995.
in one PW does not mean being similar in shape (or ontogeny of shape) in a particular
anatomical region. When looking at one PW we lose the context supplied by all the others,
and PWs are all context-dependent. Therefore, we cannot describe what happens within
any one region of the body without placing every PW in context of every other. Even
judged by what the method was supposed to do, it fails; it does not provide an objective,
non-arbitrary method for decomposing changes (except in a purely geometric sense).
The second issue, related to the one above but important in a broader context, is that
interpretations based on individual variables violate the fundamental principles of geo-
metric shape analysis – that results be invariant to the selection of variables. Obviously,
a result that depends on using partial warps is invalid (even if the phylogenetic inference
based on it happens to be valid). A partial warp score is a single variable, a one-dimensional
projection onto a particular basis, and our results cannot depend on that choice. Adhering
to that basic principle does not mean that our phylogenetic results will be invariant to our
choice of characters – the results of a phylogenetic analysis always depend on the charac-
ters. Rather, it means that our recognition of characters must be invariant to the selection
of variables – and for that reason, a morphometric variable cannot be a character in its
own right.
The obvious question is: how can we discover characters when we cannot look at
individual variables? If variables do not provide a legitimate basis for subdividing the
organism, and if conventional anatomical lines of dissection are also viewed as biologically
chap-14 4/6/2004 17: 29 page 371
MORPHOMETRICS AND SYSTEMATICS 371
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
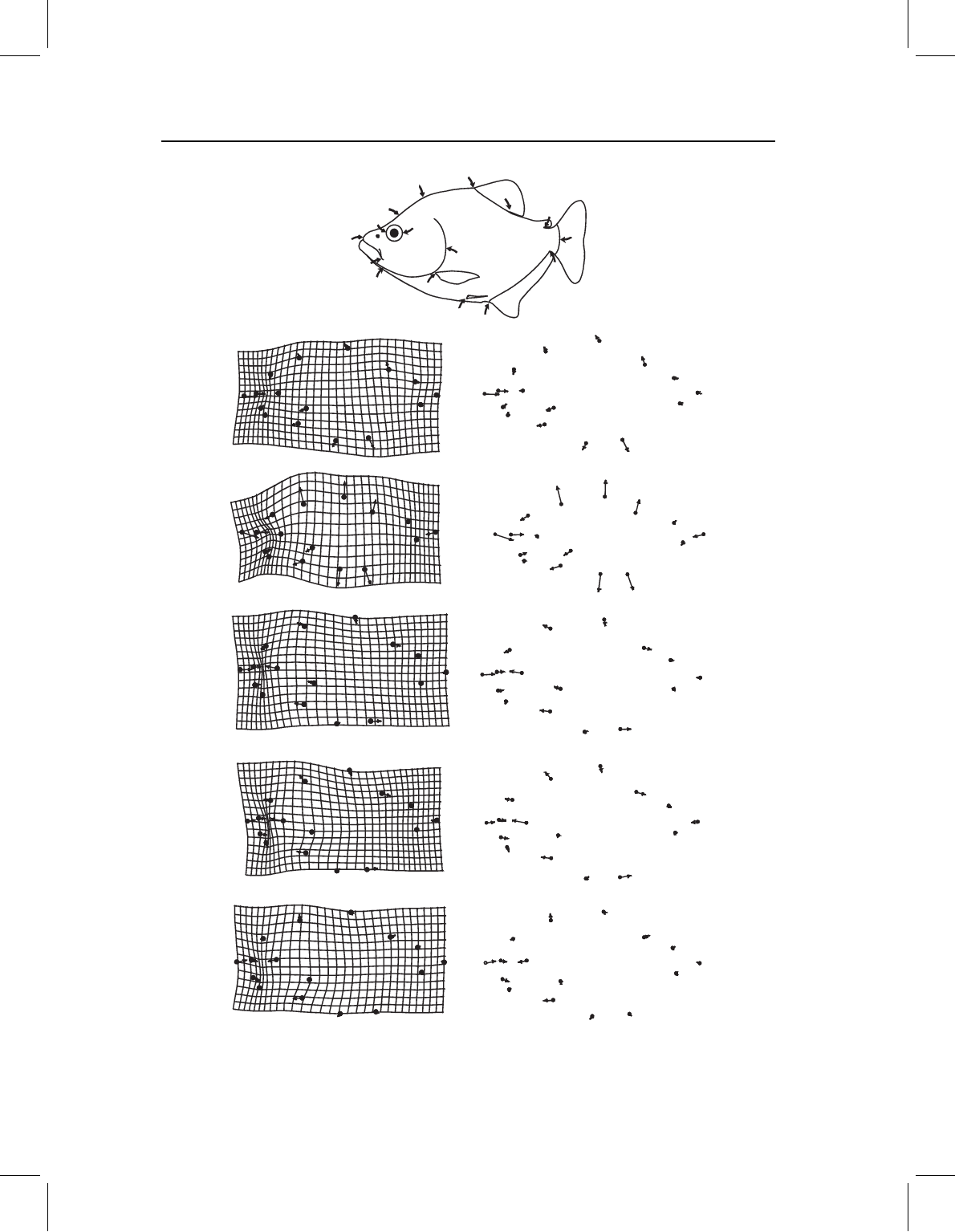
Pygopristis denticulata
Serrasalmus gouldingi
P. cariba
Pygocentrus piraya
P. nattereri
Figure 14.3 Ontogenies of shape for the species analyzed in Fink and Zelditch, 1995. The inference
drawn from the PW shown in Figure 14.2 is that the outgroup species P. denticulata and S. gouldingi
have a localized ontogenetic change in the length and depth of the caudal peduncle relative to the
region between dorsal and adipose fins, whereas the three Pygocentrus do not.

chap-14 4/6/2004 17: 29 page 372
372 GEOMETRIC MORPHOMETRICS FOR BIOLOGISTS
arbitrary, then where can we look for characters? We end this section with that question
because we have no satisfying answer. In the next section, we discuss two possible lines
of attack. One uses a standard multivariate ordination method, principal components
analysis (PCA), to explore similarities and differences, the other uses pairwise contrasts to
find differences, which are then compared to find similarities among taxa in their differ-
ences from others. Neither method is tailored to the problem, but both represent feasible
approaches that can be used in the interim, until we have a more satisfying method.
Using PCA to find characters
PCA provides a coordinate system for shape analysis, and may be useful for finding char-
acters, but individual PCs (like individual PWs) cannot be viewed as characters in their
own right. Just as a partial warp score is a projection onto a single axis, so is a princi-
pal component score, and just as a similarity on one PW does not indicate a similarity in
shape, similarity on a single PC might not demonstrate a sufficiently general (or detailed)
similarity.
Like PWs, PCs are context-dependent, and thus we would not expect a PC to be a
character any more than a partial warp is. That is not to say that the two methods are
strictly comparable – there is a major difference between them. Principal components are
orthogonal directions of variation rather than orthogonal components of bending energy,
and variation is biologically relevant to the problem at hand while bending energy is useful
only in that it is used by the method for depicting the results. PCs have a biological meaning,
as orthogonal dimensions of variance, even though that is not equivalent to the meaning
of a character. They are not likely to be characters in their own right because they are
directions of variation that are constrained to be orthogonal (by definition), not directions
of evolutionary change. Directions of evolutionary change are likely to be oblique to the
PCs – they are within the space spanned by the PCs, but they need not lie along an axis
nor must they be orthogonal.
Although PCs are not likely to be characters, we may still find PCA useful for exploring
similarities and differences. The scatter plots allow us to see the variation among taxa, and
their overlap, and both are important for finding characters. However, just as we need to
interpret partial warps in combination, so we also need to interpret PCs in combination.
Just because two or more species overlap in their PC1 scores does not mean that they
are similar with respect to all features described by PC1. They may differ in some, so
that PC1 splits the difference between them and the other PCs describe what is specific
to their deviations from PC1. Taxa located in different quadrants of a scatter plot may
differ considerably in shape, depending on the proportion of the variance described by
each PC and on how the PCs overlap in their descriptions of variation within the same
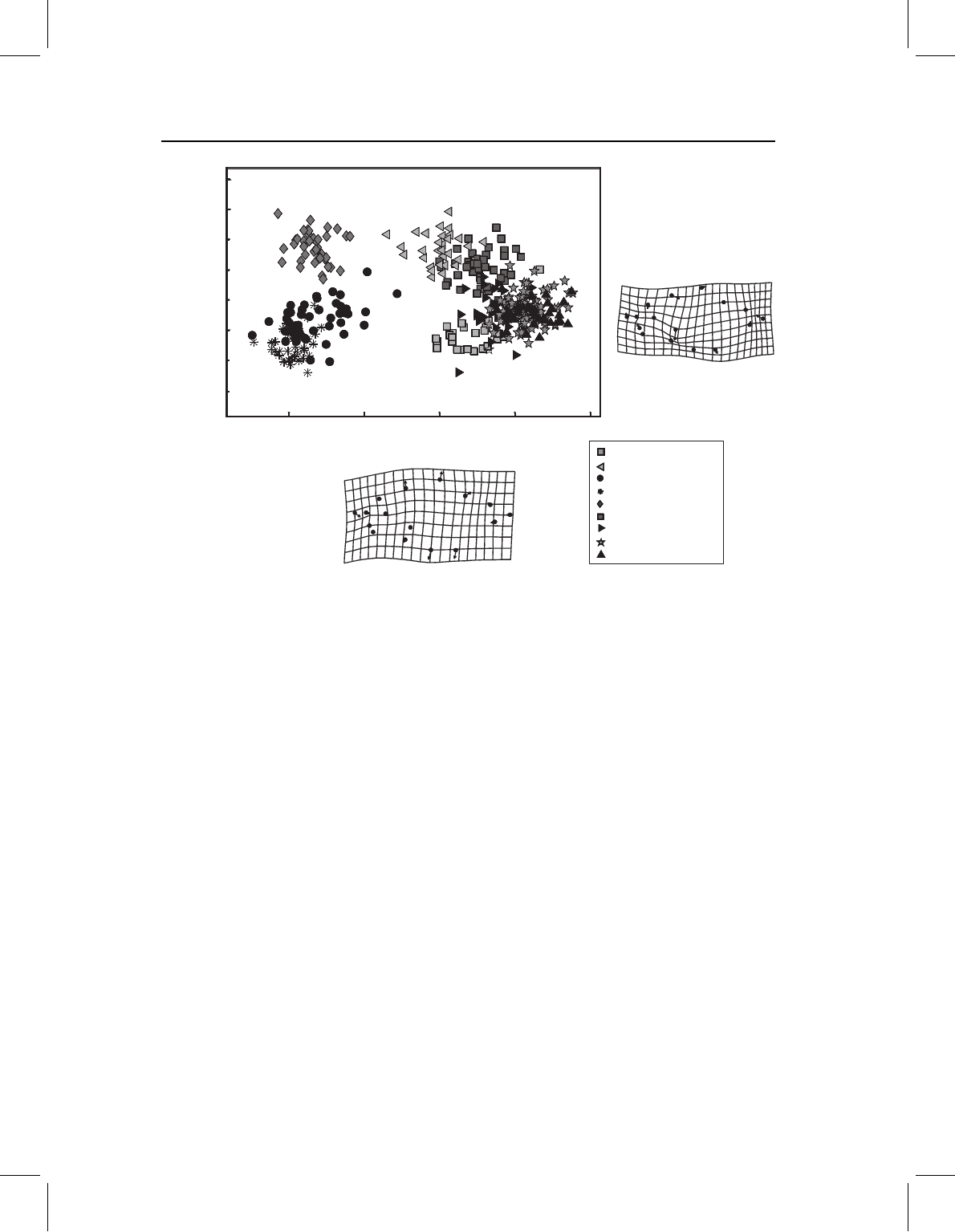
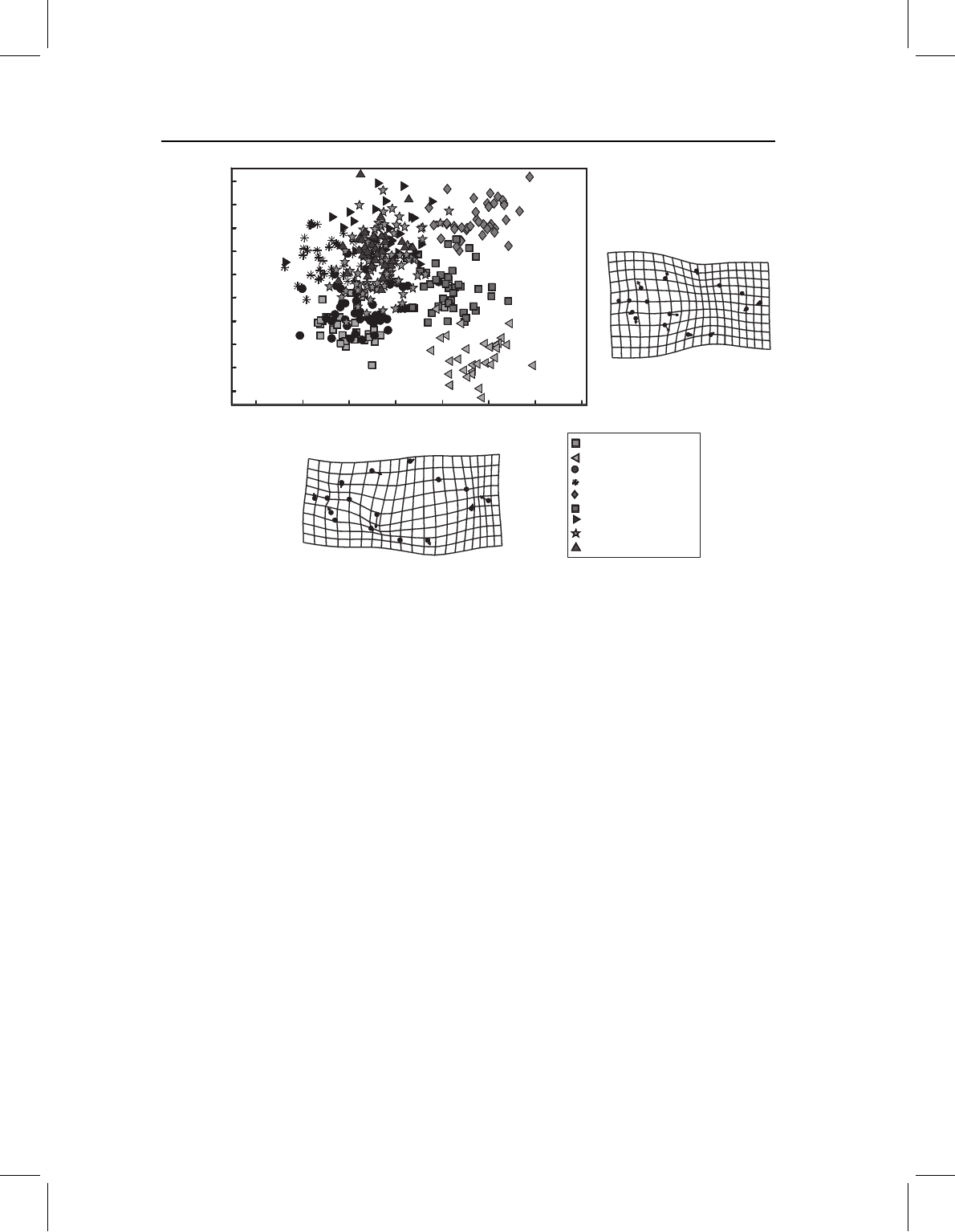
regions. For example, we can look at a case that should be familiar by this point – the
first two PCs of piranha juvenile body shape. The first, which accounts for 62% of the
variance, clearly distinguishes three species (S. manueli, S. elongatus and S. gouldingi)
from all others (Figure 14.4). Looking at the deformation that depicts the direction of
greatest variance, we can see that body depth contributes heavily to it. However, PC1
is not only body depth; it also describes differences in proportions of the posterior body
correlated with body depth. Species with high scores on this axis have relatively long
chap-14 4/6/2004 17: 29 page 373
MORPHOMETRICS AND SYSTEMATICS 373
PC2
PC1
⫺0.1 ⫺0.05 0 0.05 0.1
⫺0.06
⫺0.04
⫺0.02
0
0.02
0.04
0.06
0.08
Pygopristis denticulata
Serrasalmus altuvei
S. elongatus
S. gouldingi
S. manueli
S. spilopleura
Pygocentrus cariba
P. piraya
P. nattereri
Figure 14.4 Principal components of body shape of nine species of piranhas; data were standardized
and variation is examined among juvenile shapes (at the transition from larval to juvenile phases).
caudal peduncles compared to the region between dorsal and adipose fins, as well as deep
bodies, but we cannot necessarily say that species with high scores on PC1 have long caudal
peduncles if other PCs also describe variation in posterior body proportions and scores
on those PCs differ among species with similar scores on PC1. PC2, which accounts for
only 8.3% of the variance, also describes variation in caudal body proportions and, on
this component, species with high scores have very short caudal peduncles relative to more
anterior region. Consequently, species with high scores on both components have a short
caudal peduncle relative to other species with equally high scores on PC1. In effect, PC2
partially “compensates” for PC1.
When interpreting PCs, it is also important to consider that they describe variation
around an average shape. However, the average is not a “typical” piranha; rather, it is
the shape of the consensus, the point having the coordinates 0, 0 (on all PCs). Obviously,
the consensus is not a typical piranha since there are no specimens at the 0, 0 point. The
outgroup (P. denticulata) is fairly near it, but if we want to describe differences between
P. denticulata and other species (or to make any other comparisons among species) we
cannot describe changes along one PC, then along another. The direction of the difference
between particular species is often oblique to several PCs.
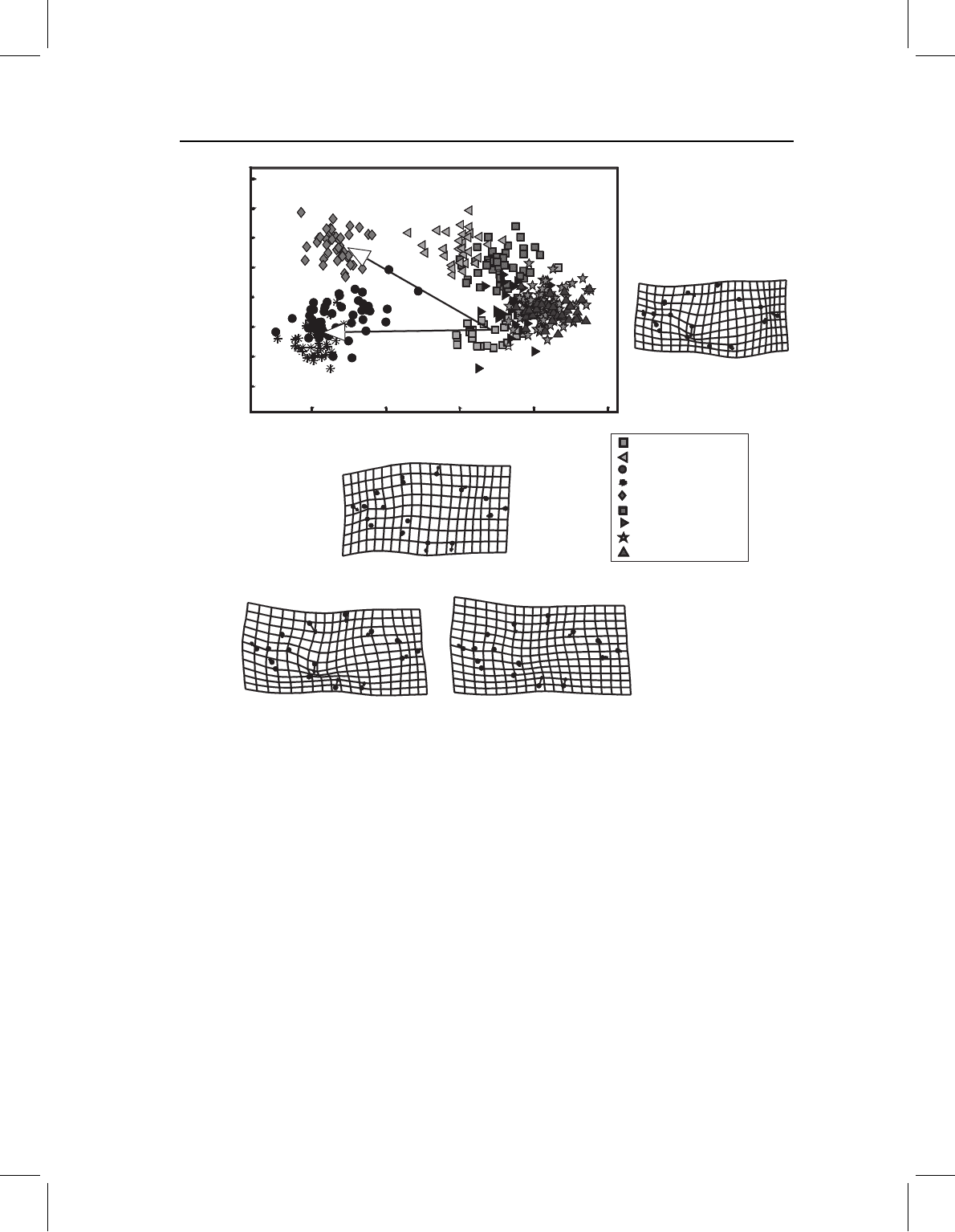
The importance of considering scores on several PCs becomes evident when comparing
the three shallow-bodied species. All three have high scores on PC1, but they differ in
scores on PC2. One of the three, S. manueli, has high scores on PC2 (as do S. altuvei and
S. spilopleura). To see how S. manueli differs from S. gouldingi and S. elongatus with
respect to their differences from other taxa, we can draw the vector extending from
chap-14 4/6/2004 17: 29 page 374
374 GEOMETRIC MORPHOMETRICS FOR BIOLOGISTS
(A)
(B)
PC2
PC1
⫺0.1 ⫺0.05 0 0.05 0.1
⫺0.06
⫺0.04
⫺0.02
0
0.02
0.04
0.06
0.08
A
B
Pygopristis denticulata
Serrasalmus altuvei
S. elongatus
S. gouldingi
S. manueli
S. spilopleura
Pygocentrus cariba
P. piraya
P. nattereri
Figure 14.5 Analyzing direction in which species differ from P. denticulata in juvenile shape, to
determine whether species with overlapping scores on PC1, but different scores on PC2, are similar
with respect to features varying along PC1. (A) The direction of difference from P. denticulata to
S. manueli; (B) the direction of difference from P. denticulata to S. gouldingi and S. elongatus.
those other taxa, e.g. P. denticulata to S. manueli (Figure 14.5A) and to S. elongatus
and S. gouldingi (Figure 14.5B). The reason for doing this is to determine what differences
from other taxa are shared by S. elongatus, S. gouldingi and S. manueli. We will then
eliminate from the character the features peculiar to one species. Although we are making
this comparison to the outgroup, we are not assuming that it has the primitive body shape.
Comparisons to other species will also be necessary, and no decisions about polarity are
made at this point. Based upon the similarities between the two vectors (Figures 14.5A,
14.5B), what all three taxa share is their shallow body. There also may be a second similar-
ity not described by either PC – a shortening of the midbody relative to the head and poste-
rior body. We could include that in the description of the character, but we would exclude
the proportions of the caudal peduncle from that character description because S. manueli
differs from the other two species in that. Clearly, the character is not equivalent to a PC.
chap-14 4/6/2004 17: 29 page 375
MORPHOMETRICS AND SYSTEMATICS 375
PC2
PC3
⫺0.06 ⫺0.04 ⫺0.02 0 0.02 0.04 0.06 0.08
⫺0.05
⫺0.04
⫺0.03
⫺0.02
⫺0.01
0
0.01
0.02
0.03
0.04
Pygopristis denticulata
Serrasalmus altuvei
S. elongatus
S. gouldingi
S. manueli
S. spilopleura
Pygocentrus cariba
P. piraya
P. nattereri
Figure 14.6 Scatter plot of PC3 on PC2, and the deformations depicting these two dimensions of
variation.
The reason for not treating PC2 as a character in its own right is the same as the one
we used to rule out treating individual PWs as characters. S. manueli, S. altuvei and also
S. spilopleura have high scores on this one, which primarily describes a displacement of
the opercle landmark towards the pectoral fin and a shortening of the caudal peduncle
relative to the anal fin. However, S. manueli and S. altuvei differ along PC1 and are not
similar in caudal peduncle proportions; they also differ along PC3 (Figure 14.6). Differ-
ences along PC3, as well as those along PC1, might belie the inference of morphological
similarity implied by similar PC2 scores. Like PC2, PC3 accounts for only a small portion
of the variance (5.6%), but like PC2 it describes a change in location of the pectoral fin
relative to the opercle. S. manueli and S. altuvei have the highest and lowest scores on
PC3, respectively, which means that their pectoral fins are displaced in opposite directions
relative to the opercle, which needs to be taken into account when assessing their similar-
ity on PC2. Despite their similar scores on PC2, a feature that might have been judged a
morphological similarity might not be similar, by virtue of the differences along PC1 and
PC3. Because of their different scores on PC1, we would also avoid construing their caudal
peduncle proportions as similar, despite their similar values on PC2. Because species can
be similar along one component and differ substantially along others, we cannot interpret
one component at a time.
The strategy for combining PCs, outlined above, is undeniably tedious, but it might be
successful at finding the features shared by two or more taxa. In cases like our example,
when over 60% of the variation is along a single PC, two or more taxa have high scores
and two or more have low ones, and there is virtually no overlap among the low and high

chap-14 4/6/2004 17: 29 page 376
376 GEOMETRIC MORPHOMETRICS FOR BIOLOGISTS
scores, PC1 points to a character. When the variation is more evenly spread out across
components, it will be necessary to combine many more of them because similarities on
one may be outweighed by differences on the others. An obvious problem is that the
comparison of the vectors in a single plane, such as we used to compare the similarities
among the three shallow-bodied species with respect to their difference from the outgroup,
examines only some of the differences among them in a single plane. We might prefer to
look at all the differences between each species and the outgroup (or any other species
taken as a standard), comparing those vectors among taxa.
Using comparisons between interspecific vectors to find characters
The basic idea of this approach is to compare all species to one other species (which is held
constant). These pairwise contrasts can then be examined for similarities. By comparing the
differences between one species and each of the others, we can then inspect the differences
for similarities. The logic of the method is that we are looking for similarities in the
differences – i.e. similarities among taxa in features specific to them. To exemplify this
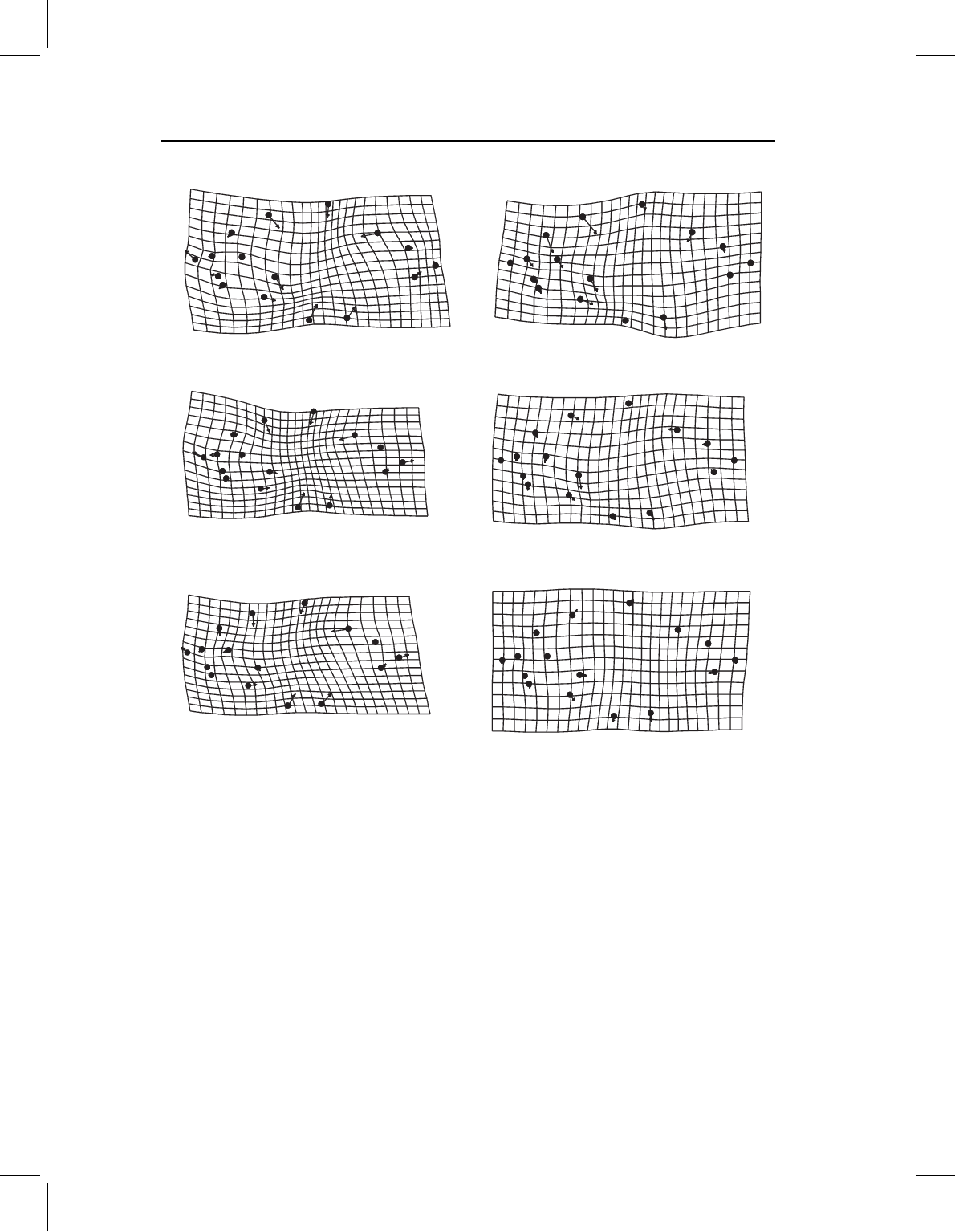
approach, we will continue the analysis of piranha juvenile body shape, comparing each
species to the outgroup (Figure 14.7). Of course it is not necessary to use the outgroup in
these comparisons; any species could be used as that “other,” and it may be useful to use
more than one before drawing conclusions.
From these comparisons, it is obvious that S. elongatus, S. gouldingi and S. manueli are
shallow-bodied compared to all other piranhas. They differ profoundly from P. denticulata
in this, whereas none of the other species do. This is the feature that dominated the PCA
(and it is obvious by qualitative visual inspection as well). Additionally, these three species
have a relatively short mid-body relative to the more posterior body, a feature hinted
at but not so clearly presented when the vectors were drawn from P. denticulata to the
species in the PC1–PC2 plane (Figure 14.5). This particular feature might reflect a decrease
in the length of the dorsal fin relative to the posterior body (dorsally) and the posterior
displacement of the pectoral fin and pelvic fins (rather than changes in proportions of body
between them). The three shallow-bodied species appear to vary in the degree of “midbody
contraction,” but they appear to be consistently more contracted than the others. The
possibility that these three species are similar in having a relatively shortened midbody
is worth examining further because, unlike their shallow body, it is not obvious from a
purely qualitative analysis.
To pursue that possibility further, we can compare the vectors of pairwise contrasts to
each other, asking if a more contracted midbody (compared to that of P. denticulata)is
characteristic of the shallow-bodied species but not of the others. This is done by subtract-
ing one of the pairwise vectors from another; where species are identical to S. gouldingi (in
the differences from P. denticulata) the grid is square (Figure 14.8). Large differences indi-
cate that the direction of change from P. denticulata to S. gouldingi is not shared by another
taxon. Subtracting each contrast from the contrast between P. denticulata to S. gouldingi
shows that S. gouldingi is not much shallower or deeper than either S. elongatus or
S. manueli. All three differ from P. denticulata by nearly the same degree, and in that
same direction. Some differences are evident in the relative length of the midbody, how-
ever. The grid is slightly more contracted in that region, indicating that S. gouldingi is
chap-14 4/6/2004 17: 29 page 377
MORPHOMETRICS AND SYSTEMATICS 377
P. denticulata vs S. elongatus
P. denticulata vs S. gouldingi
P. denticulata vs S. manueli
P. denticulata vs S. altuvei
P. denticulata vs S. spilopleura
P. denticulata vs P. piraya
Figure 14.7 Pairwise comparisons between mean juvenile body shapes of P. denticulata and six
other species. Comparisons to P. nattereri and P. cariba are not distinct from the comparison to
P. piraya.
more extreme than the others in that feature. However, the differences are slight. In strik-
ing contrast, the comparisons to the other species indicate not only that S. gouldingi is far
shallower than the others, but also that all differ from S. gouldingi in either the degree
or the location of midbody contraction. We could either take these results to mean that
S. elongatus, S. gouldingi and S. manueli are all shallow-bodied and contracted in the mid-
body compared to the other species, or we could continue the analysis, doing additional
pairwise contrasts – this time between P. denticulata and S. elongatus, and also between
P. denticulata and S. manueli – to determine that all three species are similarly different
from the others. Of course we would need additional comparisons to find features that are
more widely shared, or specific to some of the deeper-bodied species.
Unlike the shallow body, which is so evident visually that it requires no detailed quanti-
tative study, the midbody contraction discerned in these comparisons is the kind of subtle
feature that justifies the effort of a morphometric analysis.
