Voet D., Voet Ju.G. Biochemistry
Подождите немного. Документ загружается.

Time
Mitochondria + β-hydroxybutyrate added,
NAD
+
-linked oxidation commences
Rotenone or amytal added,
inhibiting NAD
+
-linked oxidation
Succinate added, FAD-linked
oxidation begins
Antimycin added, inhibiting
FAD-linked oxidation
TMPD/ascorbate added,
oxidation resumes
CN
–
added,
oxidation terminates
[O
2
]
1
32
54
6
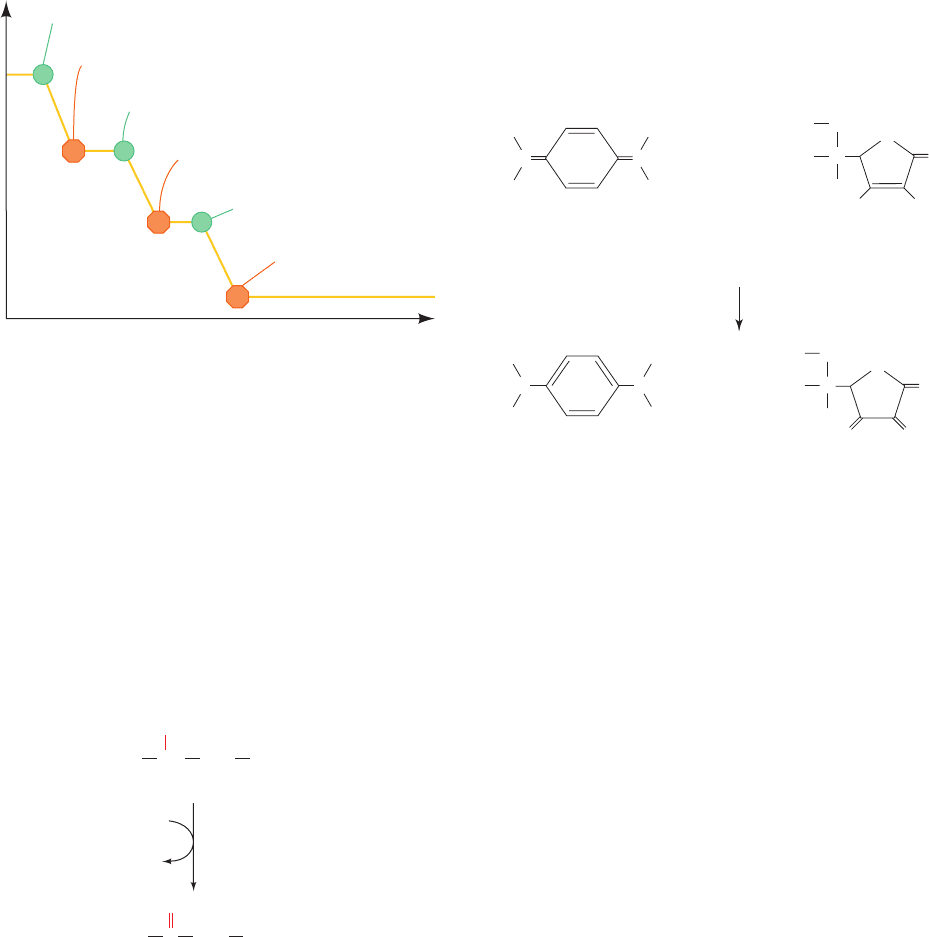
Reagents are then injected into the chamber and the O
2
consumption recorded (Fig. 22-11):
1. Mitochondria and -hydroxybutyrate are injected
into the chamber. Mitochondria mediate the NAD
⫹
-linked
oxidation of -hydroxybutyrate (Section 25-3):
As the resulting NADH is oxidized by the electron-
transport chain with O
2
as the terminal electron acceptor,
the O
2
concentration in the reaction mixture decreases.
2. Addition of rotenone or amytal completely stops the
-hydroxybutyrate oxidation.
3. Addition of succinate, which undergoes FAD-linked
oxidation, causes the [O
2
] to resume its decrease. Electrons
from FADH
2
are therefore still able to reduce O
2
in the
presence of rotenone; that is, electrons from FADH
2
enter
the electron-transport chain after the rotenone-blocked step.
4. Addition of antimycin inhibits electron transport
from FADH
2
.
5. Although NADH and FADH
2
are the electron-
transport chain’s two physiological electron donors,
-Hydroxybutyrate
Acetoacetate
CH
2
CO
–
H
⫹
NAD
⫹
O
OH
CHCH
3
CH
3
CH
2
C
-hydroxybutyrate
dehydrogenase
NADH
⫹
2
CO
–
2
nonphysiological reducing agents can also be used to probe
the flow of electrons. Tetramethyl-p-phenylenediamine
(TMPD) is an ascorbate-reducible redox carrier that
transfers electrons directly to cytochrome c:
Addition of TMPD and ascorbate to the antimycin-
inhibited reaction mixture results in resumption of oxygen
consumption; evidently there is a third point at which elec-
trons can enter the electron-transport chain.
6. The addition of CN
⫺
completely inhibits oxidation
of all three electron donors, indicating that it blocks the
electron-transport chain after the third point of entry of
electrons.
Experiments such as these established the order of elec-
tron flow through the electron-transport chain complexes
and the positions blocked by various electron-transport in-
hibitors (Fig. 22-9).This order was confirmed and extended
by observations that the standard reduction potentials of
the redox carriers forming the electron-transport chain
complexes are very close to the standard reduction poten-
tials of their electron donor substrates (Table 22-1). The
three jumps in reduction potential between NADH, CoQ,
cytochrome c, and O
2
are each of sufficient magnitude to
drive ATP synthesis. Indeed, these redox potential jumps
correspond to the points of inhibition of rotenone (or amy-
tal), antimycin, and CN
⫺
.
b. Phosphorylation and Oxidation
Are Rigidly Coupled
The foregoing thermodynamic studies suggest that oxi-
dation of NADH, FADH
2
, and ascorbate by O
2
is associ-
ated with the synthesis of about 2.5, 1.5, and 1 ATP, respec-
tively. This stoichiometry, called the P/O ratio [the ratio of
ATP synthesized to O atoms reduced (electron pairs taken
up)], has been confirmed experimentally through measure-
ments of O
2
uptake by resting and active mitochondria.An
example of a typical experiment used to determine the P/O
ratio is as follows: A suspension of mitochondria (isolated
by differential centrifugation after cell disruption; Section
6-1B) containing an excess of P
i
but no ADP is incubated in
an oxygen electrode reaction chamber. Oxidation and
+
NN
+
CH
3
CH
3
CH
2
H
HO
OH
C
O
O
H
3
C
H
3
C
Tetramethyl-p-phenylenediamine
(TMPD), oxidized form
Ascorbic acid
+
HO
NH
+
+
HN
TMPD, reduced form Dehydroascorbic acid
+
HO
CH
3
CH
3
CH
2
H
C
H
3
C
H
3
C
HO
HO
O
O
O
O
Section 22-2. Electron Transport 831
Figure 22-11 Effect of inhibitors on electron transport. This
diagram shows an idealized oxygen electrode trace of a
mitochondrial suspension containing excess ADP and P
i
.At the
numbered points, the indicated reagents are injected into the
sample chamber and the resulting changes in [O
2
] are recorded.
The numbers refer to the discussion in the text. [After Nicholls,
D.G., Bioenergetics, p. 110, Academic Press (1982).]
JWCL281_c22_823-870.qxd 3/19/10 10:59 AM Page 831

phosphorylation are closely coupled in well-functioning mi-
tochondria, so electron transport can occur only if ADP is
being phosphorylated (Section 22-3). Indeed, mitochon-
drial metabolism is so tightly regulated that even the appear-
ances of actively respiring and resting mitochondria are
greatly different (Fig. 22-12). Since no ADP is present in
the reaction mixture, the mitochondria are resting and the
O
2
consumption rate is minimal (Fig. 22-13; Region 1). The
system is then manipulated as follows:
(a) ADP (75 mol) and an excess of -hydroxybu-
tyrate (an NAD
⫹
-linked substrate) are added. The
mitochondria immediately enter the active state
and the rate of oxygen consumption increases (Fig.
22-13, Region 2) and is maintained at this elevated
level until all the ADP is phosphorylated. The mito-
chondria then return to the resting state (Fig. 22-13,
Region 3). Phosphorylation of 75 mol of ADP un-
der these conditions consumes 15 mol of O
2
. Since
the oxidation of NADH by O
2
consumes twice as
many moles of NADH (i.e., electron pairs) as of O
2
,
the P/O ratio for NADH reoxidation at Region 2 is
75 mol of ADP/(2 ⫻ 15 mol of O
2
) ⫽ 2.5; that is,
2.5 mol of ADP are phosphorylated per mole of
NADH oxidized.
(b) The experiment is continued by inhibiting electron
transfer from NADH by rotenone and adding an
additional 75 mol of ADP (Fig. 22-13, Region 4),
this time together with an excess of the FAD-linked
substrate succinate. Oxygen consumption again con-
tinues until all the ADP is phosphorylated, and the
system again returns to the resting state (Fig. 22-13,
Region 5). Calculation of the P>O ratio for FADH
2
oxidation yields the value 1.5; that is, 1.5 mol of ADP
are phosphorylated per mole of FADH
2
oxidized.
(c) In the same manner, the oxidation of ascorbate/TMPD
yields a P/O ratio of 1 (Fig. 22-13, Regions 6 and 7).
These conclusions agree with the inhibitor studies indicat-
ing that there are three entry points for electrons into the
electron-transport chain and with the standard reduction
potential measurements exhibiting three potential jumps,
each sufficient to provide the free energy for ATP synthe-
sis (Fig. 22-9).
c. The P/O Ratios Are Difficult to
Measure Accurately
Measurements of P兾O ratios are subject to systematic
experimental errors for which it is difficult to correct, such
as inaccuracies in the measurement of the oxygen concen-
tration, the presence of AMP, and proton leakage through
the inner mitochondrial membrane. In the older literature,
these values were taken to be 3, 2, and 1. However, careful
measurements by Peter Hinkle have yielded values close
to 2.5, 1.5, and 1 for these quantities (we shall see in Section
22-3 that the mechanism of oxidative phosphorylation does
not require that P兾O ratios have integer values). If these
latter values are correct, then the number of ATP molecules
that are synthesized per molecule of glucose oxidized is 2.5
ATP/NADH ⫻ 10 NADH/glucose ⫹ 1.5 ATP/FADH
2
⫻
2 FADH
2
/glucose ⫹ 2 ATP/glucose from the citric acid
cycle ⫹ 2 ATP/glucose from glycolysis ⫽ 32 ATP/glucose
(rather than the value of 38 ATP/glucose implied by P/O
ratios of 3, 2, and 1).
The determination of the in vivo yield of ATP/glucose is
even more problematic.The reducing equivalents from the
GAPDH-generated NADH may be imported into the mi-
tochondrion via the glycerophosphate shuttle, which
yields FADH
2
, or via the malate–aspartate shuttle, which
yields NADH but passes a proton into the matrix (Section
22-1Bc).The mix of these two shuttle systems varies from
tissue to tissue and hence so does the ATP yield. In addi-
tion, the rate of proton leakage back across the inner mi-
tochondrial membrane, which is significant, may vary with
832 Chapter 22. Electron Transport and Oxidative Phosphorylation
Table 22-1 Reduction Potentials of Electron-Transport
Chain Components in Resting Mitochondria
Component e°¿(V)
NADH ⫺0.315
Complex I (NADH:CoQ oxidoreductase; ⬃900 kD,
45 subunits):
FMN ⫺0.380
[2Fe–2S]N1a ⫺0.370
[2Fe–2S]N1b ⫺0.250
[4Fe–4S]N3, 4, 5, 6a, 6b, 7 ⫺0.250
[4Fe–4S]N2 ⫺0.150
Succinate 0.031
Complex II (succinate–CoQ oxidoreductase; ⬃140 kD,
4 subunits):
FAD ⫺0.040
[2Fe–2S] ⫺0.030
[4Fe–4S] ⫺0.245
[3Fe–4S] ⫺0.060
Heme b
560
⫺0.080
Coenzyme Q 0.045
Complex III (CoQ–cytochrome c oxidoreductase; ⬃450 kD,
9–11 subunits):
Heme b
H
(b
562
) 0.030
Heme b
L
(b
566
) ⫺0.030
[2Fe–2S] 0.280
Heme c
1
0.215
Cytochrome c 0.235
Complex IV (cytochrome c oxidase; ⬃410 kD, 8–13 subunits):
Heme a 0.210
Cu
A
0.245
Cu
B
0.340
Heme a
3
0.385
O
2
0.815
Source: Mainly Wilson, D.F., Erecinska, M., and Dutton, P.L., Annu. Rev.
Biophys. Bioeng. 3, 205 and 208 (1974); and Wilson, D.F., In Bittar, E.E.
(Ed.), Membrane Structure and Function, Vol. 1, p. 160, Wiley (1980).
JWCL281_c22_823-870.qxd 3/19/10 10:59 AM Page 832
Time
Δ[O
2
]
(P/O = 1.5)
Δ[O
2
]
(P/O = 2.5)
Δ[O
2
]
(P/O = 1)
[O
2
]
(a) ADP + β-Hydroxybutyrate
1
2
3
4
5
6
7
(b) ADP + Succinate + rotenone
(c) ADP + TMPD/ascorbate
+ antimycin
conditions and cellular identity. Hence, the in vivo yield of
ATP per glucose is likely to be significantly less than the
above figures of 32 ATP/glucose.
How the free energy of electron transport is actually
coupled to ATP synthesis, a subject of active research, is
discussed in Section 22-3.We first examine the structures of
the four respiratory complexes in order to understand how
they are related to the function of the electron-transport
chain. Keep in mind, however, that as in most areas of bio-
chemistry, this field is under intense scrutiny and much of
the information we need for a complete understanding of
these relationships has yet to be elucidated.
C. Components of the Electron-Transport Chain
Many of the proteins embedded in the inner mitochondrial
membrane are organized into the four respiratory com-
plexes of the electron-transport chain. Each complex con-
sists of several protein components that are associated with
a variety of redox-active prosthetic groups with successively
Section 22-2. Electron Transport 833
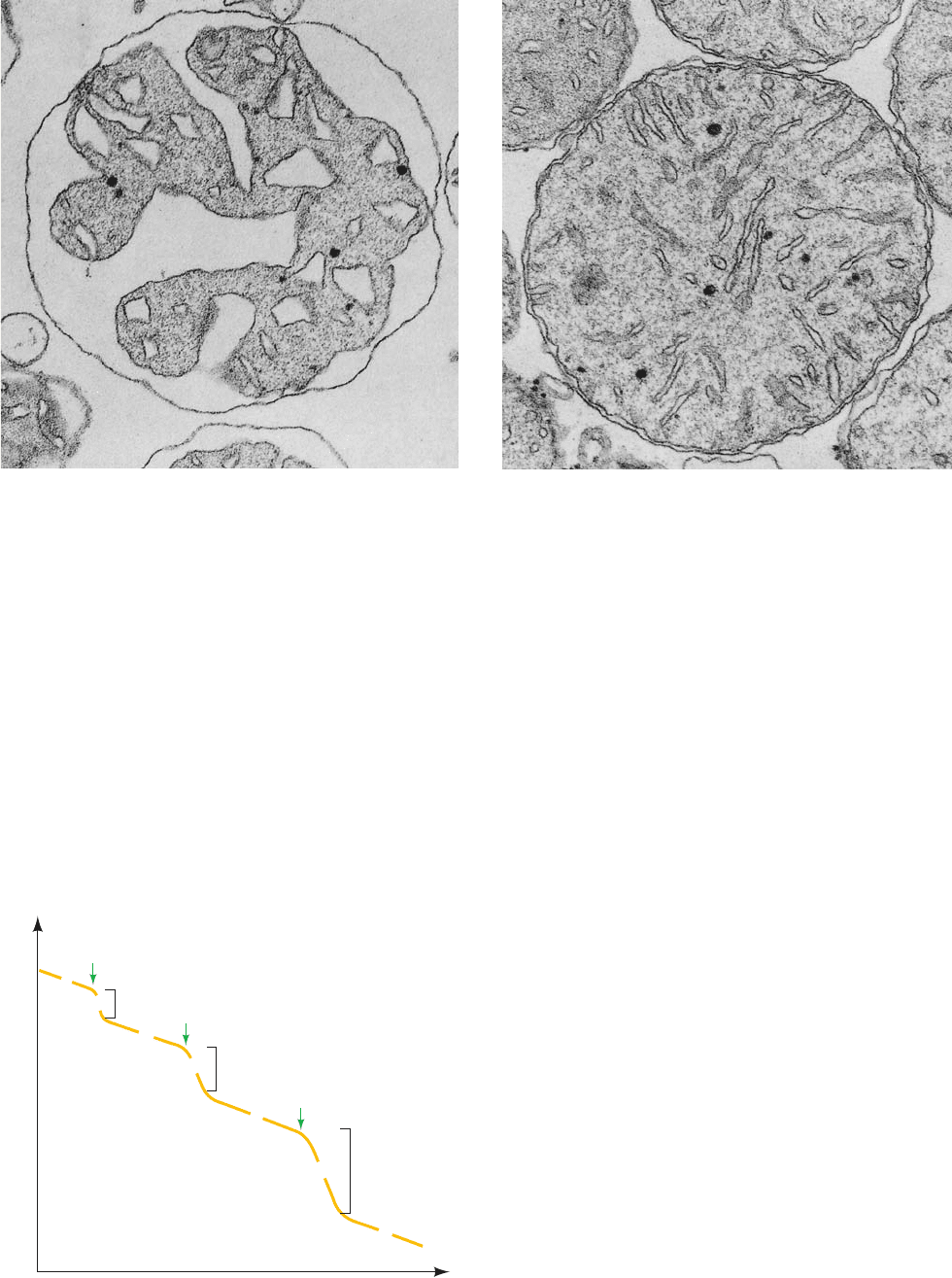
Figure 22-13 Determination of the stoichiometry of coupled
oxidation and phosphorylation (the P/O ratio) with different
electron donors. Mitochondria are incubated in excess phosphate
buffer in the sample chamber of an oxygen electrode. (a) Next,
75 mol of ADP and excess -hydroxybutyrate are added.
Respiration continues until all the ADP is phosphorylated. ⌬O
2
in Region 2 is 15 mol, corresponding to 30 mol of NADH
oxidized; thus, P>O ⫽ 75>30 ⫽ 2.5. (b) Then 75 mol of ADP and
excess succinate are added together with rotenone to inhibit
electron transfer from NADH. ⌬O
2
in Region 4 is 25 mol,
corresponding to 50 mol of FADH
2
oxidized; P>O ⫽ 75>50 ⫽ 1.5.
(c) Finally, 75 mol of ADP and excess TMPD/ascorbate are
added with antimycin to inhibit electron transfer from FADH
2
.
⌬O
2
in Region 6 is 37.5 mol, corresponding to 75 mol of
ascorbate oxidized; P>O ⫽ 75>75 ⫽ 1.
Figure 22-12 Electron micrographs of mouse liver
mitochondria. (a) In the actively respiring state and (b) in the
resting state. The cristae in actively respiring mitochondria are
far more condensed than they are in resting mitochondria.
[Courtesy of Charles Hackenbrock, University of North Carolina
Medical School.]
(a)
(b)
JWCL281_c22_823-870.qxd 6/8/10 9:19 AM Page 833
Cys
[2Fe –2S]
[4Fe – 4S]
S
2–
S
2–
S
2–
S
2–
S
2–
Fe
Fe
Fe
Fe
Fe
Fe
Cys
Cys
Cys
Cys
Cys
Cys
Cys
Cys
S
2–
[Fe–S]
Cys
Cys
Fe
(a)
(b)
(c)
Cys
increasing reduction potentials (Table 22-1). In the follow-
ing paragraphs, we examine their structures and the agents
that transfer electrons between them. Their relationships
are summarized in Fig. 22-14.
1. Complex I (NADH Dehydrogenase or
NADH–Coenzyme Q Oxidoreductase)
Complex I (also called NADH dehydrogenase) passes
electrons from NADH to CoQ. This probably largest pro-
tein component of the inner mitochondrial membrane
(⬃980 kD in mammals and ⬃700 kD in Neurospora crassa)
contains one molecule of flavin mononucleotide (FMN; a
redox-active prosthetic group that differs from FAD only
by the absence of the AMP group) and eight or nine
iron–sulfur clusters that participate in the electron-
transport process (Table 22-1). In mammals, 7 of its 45 sub-
units, its most hydrophobic subunits, which form the core of
its transmembrane region, are encoded by mitochondrial
genes, with the remainder encoded by nuclear genes. Most
of its subunits are homologous to soluble redox center-
containing proteins, which strongly suggests that Complex
I arose through the evolutionary aggregation of these pre-
viously existing proteins.
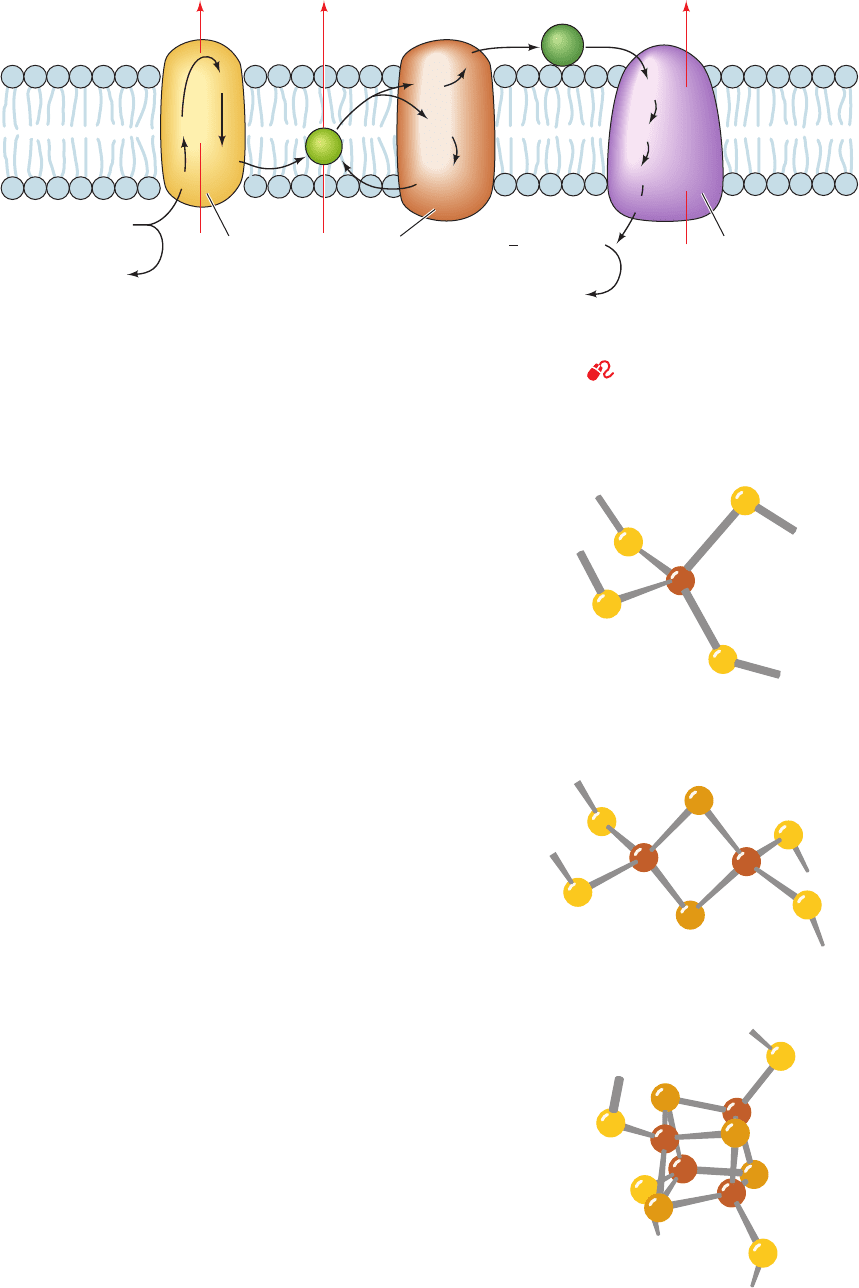
a. Iron–Sulfur Clusters Are Redox Active
Iron–sulfur clusters, first discovered by Helmut Beinert,
commonly occur as prosthetic groups in iron–sulfur pro-
teins. There are four common types of iron–sulfur clusters
(Fig. 22-15). Those designated [2Fe–2S] and [4Fe–4S]
834 Chapter 22. Electron Transport and Oxidative Phosphorylation
Figure 22-14 The mitochondrial electron-transport chain. The
pathways of electron transfer (black) and proton pumping (red)
are indicated. Electrons are transferred between Complexes I
and III by membrane-soluble CoQ (Q) and between Complexes
FeS
Intermembrane
space
Inner
mitochondrial
membrane
Matrix
FeS
FMN
4H
+
NADH
Cyt c
1
Cyt b
L
FeS
Cyt b
H
Cyt c
Cu
A
Cyt a
Complex IIIComplex I Complex IV
Cyt a
3
– Cu
B
H
2
O
2H
+
2e
–
2e
–
+
O
2
2H
+
4H
+
Q
2
1
H
+
+ NAD
+
III and IV by the peripheral membrane protein cytochrome c
(Cyt c). Complex II (not shown) transfers electrons from
succinate to CoQ.
See the Animated Figures
Figure 22-15 Structures of the common iron–sulfur clusters. (a) An [Fe–S] cluster,
(b) a [2Fe–2S] cluster, and (c) a [4Fe–4S] cluster. The [3Fe–4S] cluster resembles the
[4Fe–4S] cluster with one of its Fe ions removed. Note that, whereas the Fe and
S
2⫺
ions of [4Fe–4S] clusters form what appears to be a distorted cube, the structure
is really two interpenetrating tetrahedra of Fe ions and S
2⫺
ions.
JWCL281_c22_823-870.qxd 7/2/10 11:09 AM Page 834

clusters consist of equal numbers of iron and sulfide ions
and are both coordinated to four protein Cys sulfhydryl
groups. The [3Fe–4S] cluster is essentially a [4Fe–4S] clus-
ter that lacks one Fe. One means of identifying these clus-
ters utilizes the fact that their sulfide ions are acid labile:
They are released as H
2
S near pH 1. The [Fe–S] cluster,
which occurs only in bacteria, consists of a single Fe atom
liganded to four Cys residues. Note that the Fe ions in all
four types of clusters are each coordinated by four S atoms,
which are more or less tetrahedrally disposed around the
Fe. However, in Rieske iron–sulfur proteins (named after
their discoverer, John Rieske), one of the Fe atoms in a
[2Fe–2S] cluster is coordinated by two His residues rather
than two Cys residues.
The oxidized and reduced states of all iron–sulfur clus-
ters differ by one formal charge regardless of their number
of Fe ions. This is because the Fe ions in each cluster form a
conjugated system and thus can have oxidation states be-
tween the ⫹2 and ⫹3 values possible for individual Fe ions.
For example, each of the two [4Fe–4S] clusters in the pro-
tein ferredoxin (Fig. 22-16) contains one Fe(II) and three
Fe(III) in its oxidized form and two Fe(II) and two Fe(III)
in its reduced form. The standard reduction potential of a
given type of iron–sulfur cluster depends on its interaction
with its associated protein as well as on its oxidation state.
Iron–sulfur proteins also occur in the photosynthetic
electron-transport chains of plants and bacteria (Section
24-2); indeed, photosynthetic electron-transport chains are
thought to be the evolutionary precursors of oxidative
electron-transport chains (Section 1-5Cb).
b. The Coenzymes of Complex I
FMN and ubiquinone (CoQ), the coenzymes of Com-
plex I, can each adopt three oxidation states (Fig. 22-17).
Although NADH can only participate in a two-electron
transfer, both FMN and CoQ are capable of accepting and
donating either one or two electrons because their semi-
quinone forms are stable. In contrast, the cytochromes of
Complex III (see below), to which reduced CoQ passes its
electrons, are only capable of one-electron reductions.
FMN and CoQ therefore provide an electron conduit be-
tween the two-electron donor NADH and the one-electron
acceptors, the cytochromes.
CoQ’s hydrophobic tail makes it soluble in the inner mi-
tochondrial membrane’s lipid bilayer. In mammals, this tail
consists of ten C
5
isoprenoid units and hence the coenzyme
is designated Q
10.
In other organisms, CoQ may have only
six (Q
6
) or eight (Q
8
) isoprenoid units.
c. Electrons Follow a Multistep Path
through Complex I
Complex I from prokaryotes consists of 13 to 15 sub-
units with an aggregate molecular mass of ⬃550 kD. Ho-
mologs of all of its conserved subunits occur in the mito-
chondrial enzyme and they contain equivalent redox
components. Evidently, the prokaryotic enzyme is a
stripped down version of mitochondrial Complex I. The
functions of the 30 to 32 additional “accessory” subunits
that are components of mitochondrial Complex I are
largely unknown.
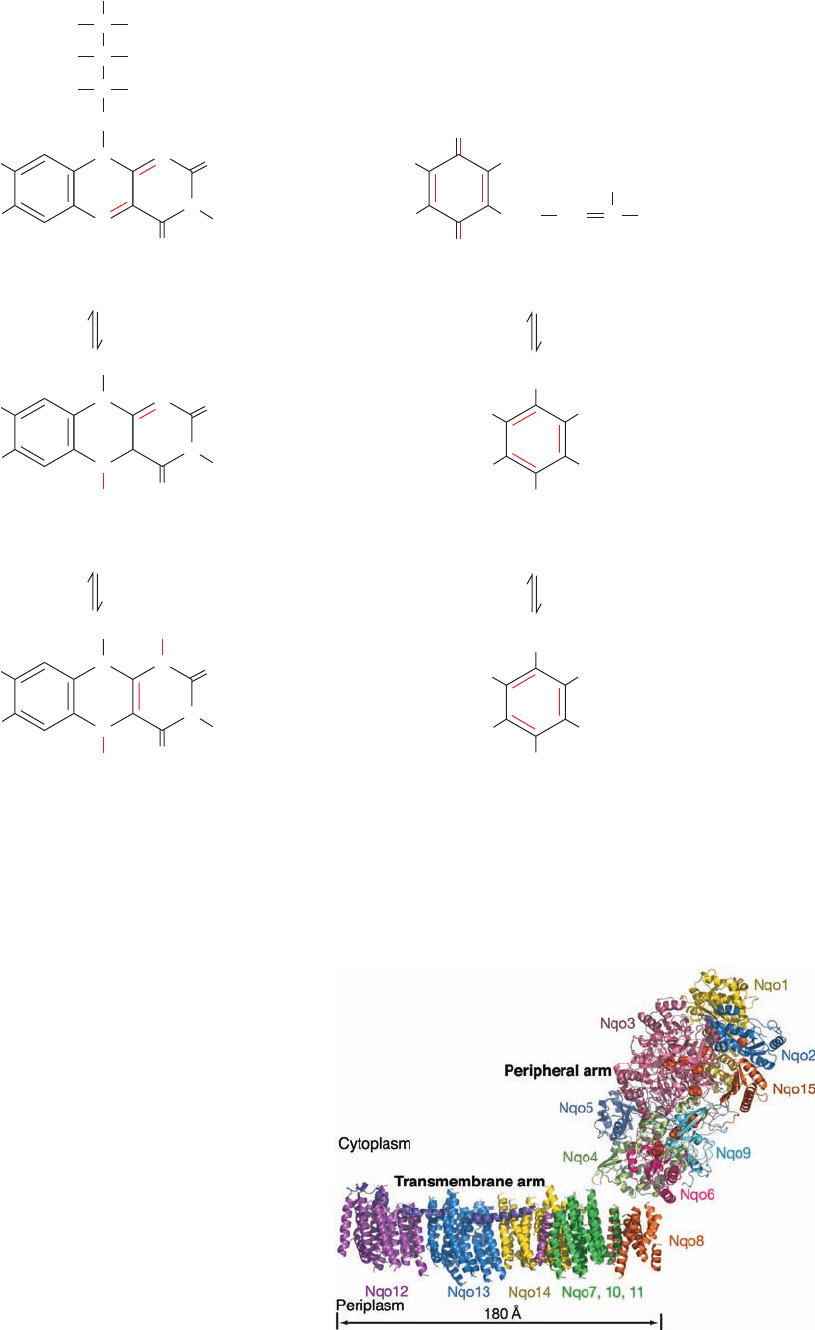
The low resolution X-ray structure of the 15-subunit
Complex I from the thermophilic bacterium Thermus ther-
mophilus (Fig. 22-18), determined by Leonid Sazanov,
reveals an L-shaped protein with one arm of the L, the
transmembrane arm, immersed in the plasma membrane
(the inner mitochondrial membrane for eukaryotes) and
the other, the peripheral arm, extending into the cytosol
(the matrix for eukaryotes).The transmembrane arm, con-
sists of seven subunits that collectively have 63 transmem-
brane helices. The three largest transmembrane subunits,
Nqo12, 13, and 14, are structurally similar to each other and
to previously determined structures of Na
⫹
/H
⫹
antiporters.
However, the most unusual feature of the transmembrane
portion is a 110-Å long amphipathic ␣ helix that extends
from Nqo12 parallel to the plane of the membrane so as to
span Nqo13 and 14.
The high resolution X-ray structure of the 8-subunit,
hydrophilic, peripheral arm of Complex I from the ther-
mophilic bacterium. Thermophilus, also determined by
Sazanov, reveals a Y-shaped assembly that is 140 Å high
(Fig. 22-19a).This subcomplex contains all of the enzyme’s
redox centers: an FMN, seven [4Fe–4S] clusters, and two
[2Fe–2S] clusters. The FMN is located at the end of a
solvent-exposed cavity that presumably forms the NADH
Section 22-2. Electron Transport 835
Figure 22-16 X-ray structure of ferredoxin from Peptostrepto-
coccus asaccharolyticus. This monomeric 55-residue protein is
drawn in ribbon form colored in rainbow order from its N-terminus
(blue) to its C-terminus (red). Its two [4Fe–4S] clusters are drawn
in ball-and-stick form with S yellow and Fe red-brown. The Cys
side chains that ligand each Fe atom are drawn in stick form with
C green and S yellow. [Based on an X-ray structure by Elinor
Adman, Larry Sieker, and Lyle Jensen, University of Washington.
PDBid 1DUR.]
See Interactive Exercise 16
JWCL281_c22_823-870.qxd 10/19/10 8:07 AM Page 835
(a)
HCHO
CHO
H
CH
2
OPO
3
2–
HCHO
CH
2
N
N
O
N
N
O
H
O
O
(CH
2
CH
2
)
n
HCCH
Isoprenoid units
Coenzyme Q (CoQ) or ubiquinone
(oxidized or quinone form)
Flavin mononucleotide (FMN)
(oxidized or quinone form)
[H•]
[H•] [H•]
[H•]
N
N
O
N
N
O
H
Coenzyme QH• or ubisemiquinone
(radical or semiquinone form)
FMNH• (radical or semiquinone form)
CH
3
CH
3
CH
3
R
O•
OH
•
H
N
N
O
N
N
O
H
Coenzyme QH
2
or ubiquinol
(reduced or hydroquinone form)
FMNH
2
(reduced or hydroquinone form)
CH
3
R
OH
H
R
R H
OH
(b)
H
3
C
H
3
C
H
3
C
H
3
C
H
3
C
H
3
C
H
3
CO
H
3
CO
H
3
CO
H
3
CO
H
3
CO
H
3
CO
836 Chapter 22. Electron Transport and Oxidative Phosphorylation
Figure 22-17 Oxidation states of the organic coenzymes of complex I. (a) FMN and (b) CoQ.
Both coenzymes form stable semiquinone free radical states.
Figure 22-18 Low (4.5-Å) resolution X-ray structure of
Complex I from T. thermophilus. The structure is viewed parallel
to the plane of the plasma membrane with the cytoplasm above
and the periplasm below. Its 15 subunits are drawn in ribbon
form in different colors with the 110-Å-long horizontal helix of
Nqo12 purple. The nine Fe–S clusters in the peripheral arm are
drawn in space-filling form with S yellow and Fe red-brown. At
the structure’s low resolution, only the transmembrane helices of
the transmembrane arm can be seen; their connecting loops are
not visible, which accounts for the apparent gap between the
transmembrane and peripheral arms. However, the loops of the
peripheral arm are visible because that part of the structure was
based on the higher resolution X-ray structure of the peripheral
arm alone (see Fig. 22-19). [Courtesy of Leonid Sazanov, Medical
Research Council, Cambridge, U.K. PDBid 3M9S.]
JWCL281_c22_823-870.qxd 7/8/10 8:31 AM Page 836
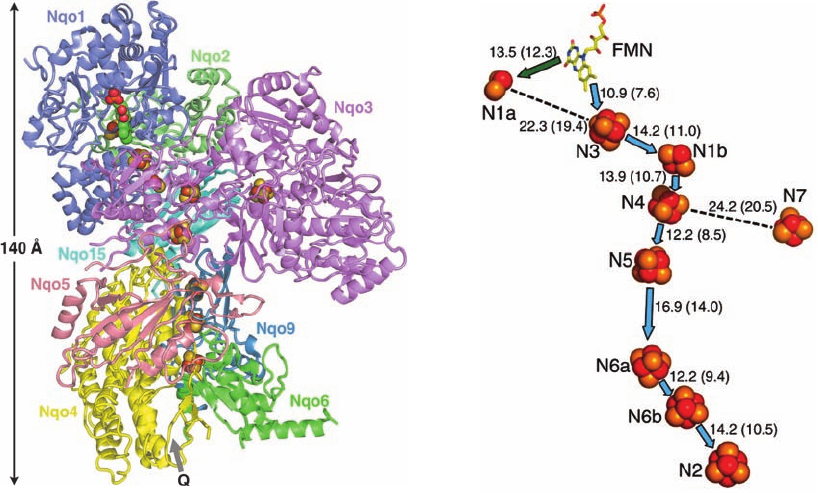
binding site. Model building suggests that bound NADH
protects the FMN from solvent. The CoQ binding site
appears to be in a cavity below the [4Fe–4S] cluster N2
that is located at the interface with Complex I’s transmem-
brane arm.
The transit of electrons from NADH to CoQ in Com-
plex I begins with a two-electron reduction in the form of a
hydride ion (H
⫺
) transfer from NADH to FMN to yield
NAD
⫹
and FMNH
⫺
(similar to the reverse of the final step
of the dihydrolipoyl dehydrogenase reaction; Section 21-
2Bb). The electrons are then transferred, one at a time, to
Fe–S cluster N3 and then between Fe–S clusters along a
gradient of increasing redox potential (Table 22-1) until
they reach Fe–S cluster N2. This “downhill” process in-
volves the transient reduction of each Fe–S cluster as it
binds electrons and its reoxidation when it passes the elec-
trons to the next cluster. Their spatial arrangement indi-
cates the probable path of the electrons (Fig. 22-19b). Note
that redox centers need not be in contact in order to trans-
fer electrons. Finally, the electrons are passed from N2 to
CoQ, which is sequentially reduced to CoQH⋅ and then to
CoQH
2
, which is released into the membrane and replaced
by CoQ, thereby completing the catalytic cycle.
In translocating an electron pair from NADH to CoQ,
Complex I pumps four protons out of the mitochondrial
Section 22-2. Electron Transport 837
matrix (the bacterial cell). How complex I accomplishes
this task is discussed in Section 22-3Bj.
2. Complex II [Succinate Dehydrogenase or
Succinate–Coenzyme Q Oxidoreductase (SQR)]
Complex II, which is also the citric acid cycle enzyme
succinate dehydrogenase (Section 21-3F), passes electrons
from succinate to CoQ to yield fumarate and CoQH
2
. It
does so with the participation of a covalently bound FAD,
a [2Fe–2S] cluster, a [4Fe–4S] cluster, a [3Fe–4S] cluster,
and one heme b
560
(Table 22-1). All of its four subunits are
encoded by nuclear genes.
The standard redox potential difference for electron
transfer from succinate to CoQ (Fig. 22-9) is insufficient to
provide the free energy necessary to drive ATP synthesis.
Complex II is, nevertheless, important because it injects
these relatively high-potential electrons into the electron-
transport chain. Two other enzymes, in addition to Com-
plexes I and II, synthesize and release CoQH
2
in the inner
mitochondrial membrane and thereby power oxidative
phosphorylation via the actions of Complexes III and IV.
They are glycerol-3-phosphate dehydrogenase of the glyc-
erophosphate shuttle (Fig. 22-8) and ETF–ubiquinone oxi-
doreductase, which participates in fatty acid oxidation (Sec-
tion 25-2Ca; ETF stands for electron-transfer flavoprotein).
Figure 22-19 X-ray structure of the peripheral arm of
Complex I from T. thermophilus. (a) The protein is viewed
parallel to the plane of the plasma membrane but oriented
differently to that in Fig. 22-18. Its eight subunits are drawn in
semitransparent ribbon form in different colors (which differ
from those in Fig. 22-18). The FMN (upper left) and the nine
Fe–S clusters are shown in space-filling form with C green,
N blue, O red, P orange, S yellow, and Fe red-brown.The likely
CoQ binding site (Q) is indicated by a gray arrow. (b) The
arrangement of redox groups viewed similarly to Part a.The
FMN (stick model with C yellow and the two [Fe–S] and seven
[4Fe–4S] clusters (space-filling models) are shown together with
their center-to-center distances in angstroms (shortest
edge-to-edge distances are indicated in parentheses). Blue
arrows represent the ⬃94-Å-long main path of electrons after
their transfer from NADH to FMN. Fe–S clusters N1a and N7 lie
too far off this path for electrons to pass through them but
perhaps they function to fine-tune its electronic properties.
Spectroscopic measurements suggest that the CoQ binding site is
⬃12 Å distant from N2. [Part a based on an X-ray structure by
and Part b courtesy of Leonid Sazanov, Medical Research
Council, Cambridge, U.K. PDBid 2FUG.]
(a)
(b)
JWCL281_c22_823-870.qxd 7/20/10 6:14 PM Page 837
a. Cytochromes Are Heme Proteins That
Transport Electrons
Cytochromes, whose function was elucidated in 1925 by
David Keilin, are redox-active proteins that occur in all or-
ganisms except a few types of obligate anaerobes. These
proteins contain heme groups that reversibly alternate be-
tween their Fe(II) and Fe(III) oxidation states during elec-
tron transport.
The heme groups of the reduced [Fe(II)] cytochromes
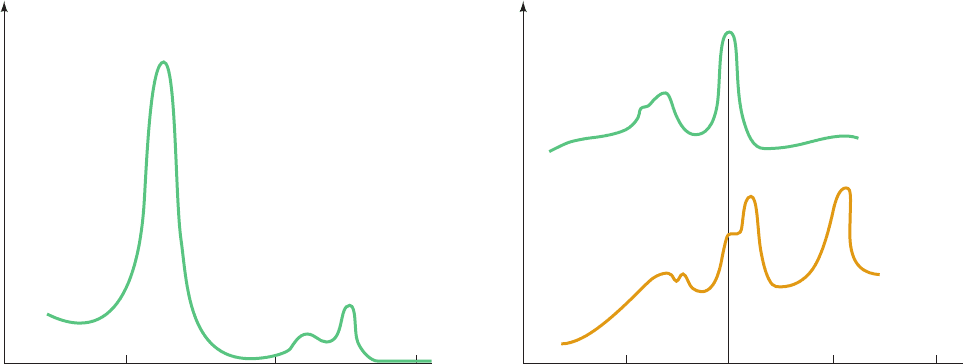
have prominent visible absorption spectra consisting of
three peaks: the ␣, , and ␥ (Soret) bands (Fig. 22-20a).The
wavelength of the ␣ peak, which varies characteristically
with the particular reduced cytochrome species (it is absent
in oxidized cytochromes), is useful for differentiating the
various cytochromes.Accordingly, the spectra of mitochon-
drial membranes (Fig. 22-20b) indicate that they contain
three cytochrome types, cytochromes a, b, and c.
Within each type of cytochrome, different heme group
environments may be characterized by slightly different ␣
peak wavelengths. For example, Complex III (see below)
has two b-type hemes:That absorbing maximally at 562 nm
is referred to as heme b
562
or b
H
(for high potential),
whereas that absorbing maximally at 566 nm is referred to
as heme b
566
or b
L
(for low potential).
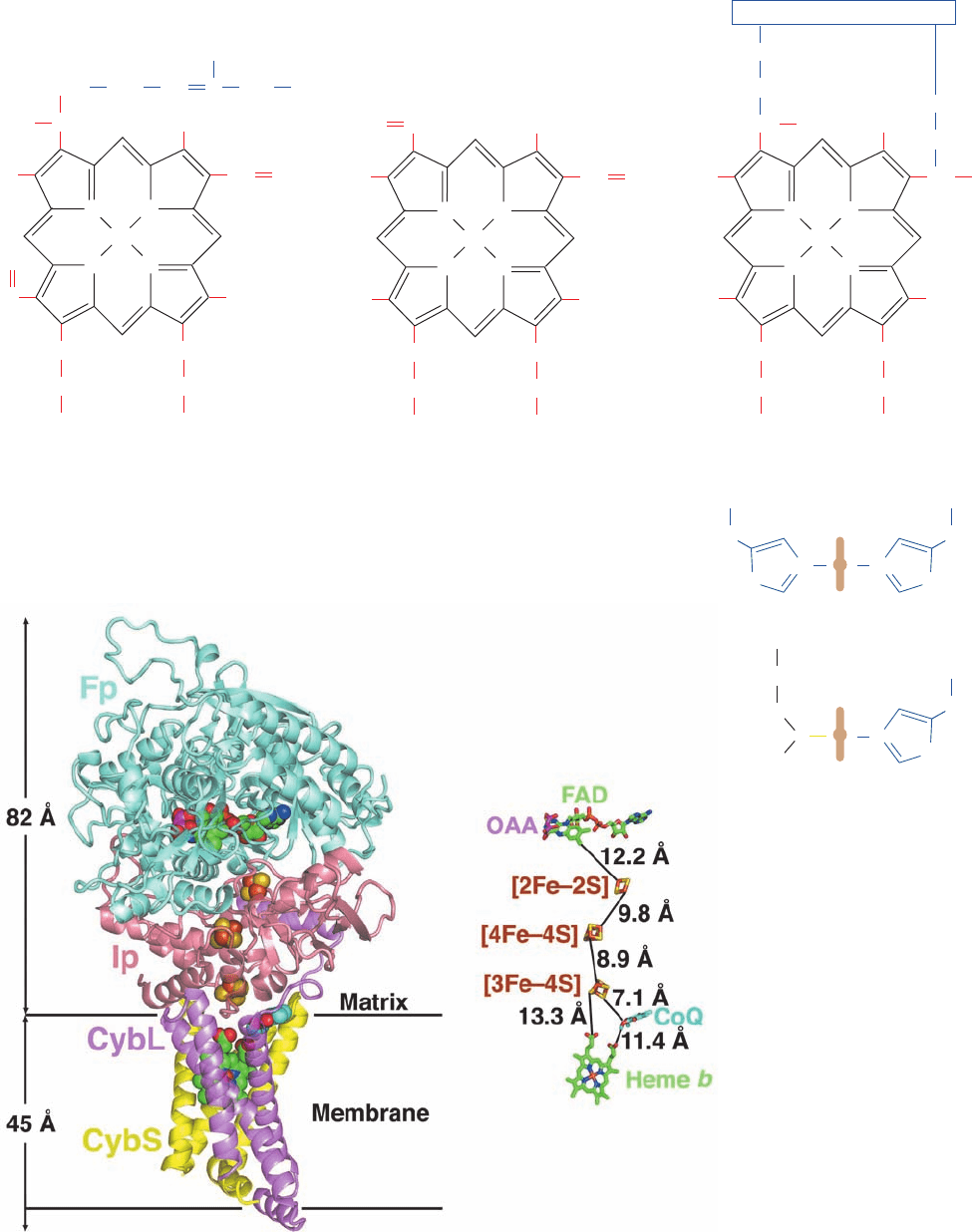
Each type of cytochrome contains a differently substi-
tuted porphyrin ring (Fig. 22-21a) coordinated with the
redox-active iron atom.A b-type cytochrome contains pro-
toporphyrin IX, which also occurs in hemoglobin and
myoglobin (Section 10-1A).The heme group of a c-type cy-
tochrome differs from protoporphyrin IX in that its vinyl
groups have added the Cys sulfhydryls in the sequence
Cys-X-Y-Cys-His across their double bonds to form
thioether linkages to the protein (Fig. 9-39). Heme a con-
tains a long hydrophobic tail of three isoprene units (a far-
nesyl group) linked to the porphyrin via a hydroxyethyl
group, as well as a formyl group in place of a methyl
substituent.
The axial ligands of the heme iron also vary with the cy-
tochrome type. In cytochromes a and b, both ligands are
His residues, whereas in cytochrome c, one is the His in the
Cys-X-Y-Cys-His sequence and the other is Met (Fig.
22-21b). Note that the designation of a given cytochrome
type refers only to the identity of the cytochrome’s heme
prosthetic group(s); a given cytochrome type may have any
of several unrelated protein folds.
b. Complex II Contains a Linear Chain
of Redox Cofactors
Both bacterial and mitochondrial Complex II’s consist
of four subunits: two water-soluble subunits, a flavoprotein
(Fp, ⬃665 residues) that binds the enzyme’s substrate suc-
cinate and FAD (which is covalently linked to a specific
His side chain via a bond to FAD atom C8a; Section
21-3F), and an iron–sulfur protein (Ip, ⬃290 residues) that
binds the three Fe–S clusters; and two transmembrane sub-
units, CybL (⬃170 residues) and CybS (⬃160 residues)
that collectively bind the heme b
560
and the CoQ. Fp and Ip
are highly conserved, whereas the sequences of CybS and
CybL vary among different organisms.
The X-ray structures of porcine, chicken, and E. coli
Complex II were independently determined by Zihe Rao,
Edward Berry, and So Iwata. In all cases, it has the shape of
the letter “q,” with its top lobe containing Fp and Ip and its
tail composed of the transmembrane subunits (Fig. 22-22a).
The complex is oriented in the inner mitochondrial mem-
brane such that Fp and Ip extend into the matrix. The
838 Chapter 22. Electron Transport and Oxidative Phosphorylation
Figure 22-20 Visible absorption spectra of cytochromes.
(a) Absorption spectrum of reduced cytochrome c showing its
characteristic ␣, , and ␥ (Soret) absorption bands. The
absorption maxima for cytochromes a, b, c, and c
1
are listed.
(b) The three separate ␣ bands in the visible absorption
Absorbance
Wavelength (nm)
400300 500 600
Cytochrome a
Cytochrome b
Cytochrome c
Cytochrome c
1
439
429
415
418
600
563
550
554
532
521
524
(a)(b)
Cytochrome c
α
β
γ
αβγ
Absorbance
Wavelength (nm)
500 550450 600 650
Cytochrome c
Beef heart
mitochondrial
membranes
c α
b α
α
β
a α
spectrum of beef heart mitochondrial membranes (below)
indicate the presence of cytochromes a, b, and c.The spectrum of
purified cytochrome c (above) is provided for reference. [After
Nicholls, D.G. and Ferguson, S.J., Bioenergetics 3, p. 96,Academic
Press (2002).]
JWCL281_c22_823-870.qxd 3/19/10 11:00 AM Page 838
Section 22-2. Electron Transport 839
Fe
3
+
N
NN
N
CH
3
CH
3
CH
3
CH
2
CH
2
CH
2
( CH
2
CH
2
)
3
CH
2
COO
–
CH
2
COO
–
HC
CH
CH
CH
HO
HC
CH
2
O
H
3
C
1
2
3
4
5
67
8
Heme a
(a)
⫹
N
NN
N
Fe
3
+
CH
3
CH
3
CH
2
CH
2
CH
2
COO
–
CH
2
COO
–
CH
CH
CH
2
CH
2
H
3
C
H
3
C
Heme b
(iron–protoporphyrin IX)
⫹
N
NN
N
Fe
3
+
CH
3
CH
3
CH
2
CH
2
CH
2
COO
–
CH
2
COO
–
CH
CH
CH
3
CH
3
H
3
C
H
3
C
Heme c
S
Cys
S
Cys
Protein
⫹
Figure 22-21 Porphyrin rings in cytochromes. The (a) chemical structures and
(b) axial liganding of the heme groups contained in cytochromes a, b, and c are shown.
Figure 22-22 X-ray structure of chicken Complex II in complex
with its inhibitor oxaloacetate and CoQ. (a) A semitransparent
ribbon diagram viewed parallel to the inner mitochondrial
membrane with the matrix above and in which the enzyme’s four
subunits are drawn in different colors.The oxaloacetate (OAA),
H
2
C
H
2
C
H
2
C
H
3
C
(b)
HN
N
His
Hemes a and b
Heme c
S
⫹
Met His
CH
2
NH
N
His
CH
2
NH
N
FAD, three Fe–S clusters, CoQ, and heme b are shown in
space-filling form with OAA C magenta, FAD and heme b C
green, CoQ C cyan, N blue, O red, P orange, S yellow, and Fe
red-brown.The inferred position of the inner mitochondrial
membrane is indicated. (b) The ligands and redox cofactors
viewed and colored as in Part a but drawn in stick form.Their
closest edge-to-edge distances are indicated. [Based on an X-ray
structure by Edward Berry, Lawrence Berkeley National
Laboratory, Berkeley, California. PDBid 1YQ3.]
(a)
(b)
JWCL281_c22_823-870.qxd 3/19/10 11:00 AM Page 839
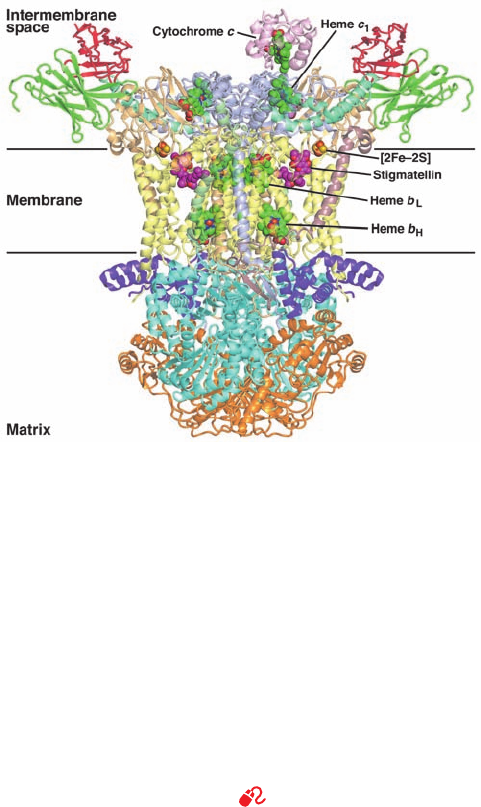
substrate and CoQ binding sites are connected by a nearly
linear ⬃40-Å-long chain of redox centers with the sequence
substrate–FAD—[2Fe–2S]—[4Fe–4S]—[3Fe–4S]—CoQ
(top to bottom in Fig. 22-22b). Thus, despite the measured
–0.245 V reduction potential of the [4Fe–4S] cluster (Table
22-1), which would appear to be too low for it to accept
electrons from succinate in the succinate S fumarate reac-
tion, the [4Fe–4S] cluster probably participates in the
electron-transfer process.
c. Complex II Suppresses the Formation
of Reactive Oxygen Species
The bacterial respiratory complex quinol–fumarate re-
ductase (QFR) is a homolog of Complex II that functions
in anaerobic organisms that use fumarate as a terminal
electron acceptor. There it catalyzes the same reaction as
does Complex II but in the opposite direction, that is, it
uses a quinol to reduce fumarate to succinate.
Even though QFR and Complex II have similar struc-
tures, organisms such as E. coli that are capable of both
aerobic and anaerobic metabolism employ the two differ-
ent complexes for these different purposes. This appears
to be because under aerobic conditions, QFR produces 25
times as much superoxide radical ( ) as does E. coli
Complex II as well as H
2
O
2
, which Complex II does not
produce, presumably by leaking electrons to O
2
.As we dis-
cuss in Section 22-4Cg,these reactive oxygen species (ROS)
are highly destructive. Comparison of the X-ray structure
of QFR with that of the closely similar E. coli Complex II
indicates that the electron distributions about their various
redox centers greatly favors the ROS-generating side re-
actions of O
2
with the flavin ring of QFR relative to that
of Complex II. Indeed, the heme b
560
of Complex II,
which QFR lacks and which is not in Complex II’s direct
electron transfer pathway (Fig. 22-22b), appears to fine-
tune its electronic properties so as to suppress the side
reactions that generate the ROS.This suggests that muta-
tions of the genes encoding human Complex II, which
cause a wide variety of disorders including tumor forma-
tion, neurological defects, and premature aging, result
from ROS generation.
3. Complex III (Coenzyme Q–Cytochrome
c Oxidoreductase or Cytochrome bc
1
Complex)
Complex III passes electrons from reduced CoQ to cy-
tochrome c. It contains four redox cofactors: two b-type
hemes, a c-type heme, and one [2Fe–2S] cluster (Table 22-1).
a. X-Ray Structure of the Cytochrome bc
1
Complex
All known cytochrome bc
1
complexes contain three com-
mon subunits: cytochrome b, which binds both the b
H
and b
L
hemes, cytochrome c
1
, which contains a single c-type heme,
and a Rieske iron–sulfur protein (ISP), which contains a
[2Fe–2S] cluster. The bovine bc
1
complex contains 8 addi-
tional subunits for a total of 11 different subunits that
combine to form a 2166-residue (243-kD) protomer that
dimerizes. Of these,only cytochrome b is encoded by a mito-
chondrial gene.
O
2
⫺
ⴢ
The X-ray structures of bovine, chicken, and yeast cy-
tochrome bc
1
complexes were independently determined
by Johann Deisenhofer, by Iwata and Bing Jap, by Berry,
Antony Crofts, and Sung-Hou Kim, and by Hartmut
Michel. All of these structures reveal a 2-fold symmetric
pear-shaped molecule of maximum diameter ⬃130 Å and
height ⬃150 Å, whose wide end extends ⬃75 Å into the
matrix space and whose narrow end extends ⬃35 Å into
the intermembrane space (Fig. 22-23). Its membrane-
spanning region is ⬃40 Å thick and consists of 13 trans-
membrane helices in each protomer (12 in yeast, which
consists of 9 different subunits). Eight of these transmem-
brane helices are contributed by the cytochrome b sub-
unit, which binds both heme b
H
and heme b
L
within its
transmembrane region, with heme b
L
being closest to the
intermembrane space. One of the remaining transmem-
brane helices is the membrane anchor of cytochrome c
1
,
the rest of which is a globular domain that extends into the
intermembrane space. This is the portion of the complex
840 Chapter 22. Electron Transport and Oxidative Phosphorylation
Figure 22-23 X-ray structure of the yeast Complex III
homodimer in complex with cytochrome c and the inhibitor
stigmatellin. The view is perpendicular to its 2-fold axis and
parallel to the membrane with the matrix below. The nine
different subunits in each protomer of Complex II, which
collectively have 12 transmembrane helices, are drawn in
semitransparent ribbon form and are differently colored with
cytochrome b pale yellow, cytochrome c
1
light purple, the ISP
tan, Core 1 cyan, and Core 2 orange. The bound cytochrome c is
pink.The four different heme groups, the [2Fe–2S] cluster, and
stigmatellin are drawn in space-filling form with heme C green,
stigmatellin C magenta, N blue, O red, S yellow, and Fe red-
brown.The horizontal lines delineate the inferred position of the
membrane. Note that only one cytochrome c is bound to the
homodimeric Complex III. [Based on an X-ray structure by
Carola Hunte, Max Planck Institute for Biophysics, Frankfurt am
Main, Germany. PDBid 1KYO.]
See Interactive Exercise 17.
JWCL281_c22_823-870.qxd 10/19/10 8:07 AM Page 840
