Voet D., Voet Ju.G. Biochemistry
Подождите немного. Документ загружается.

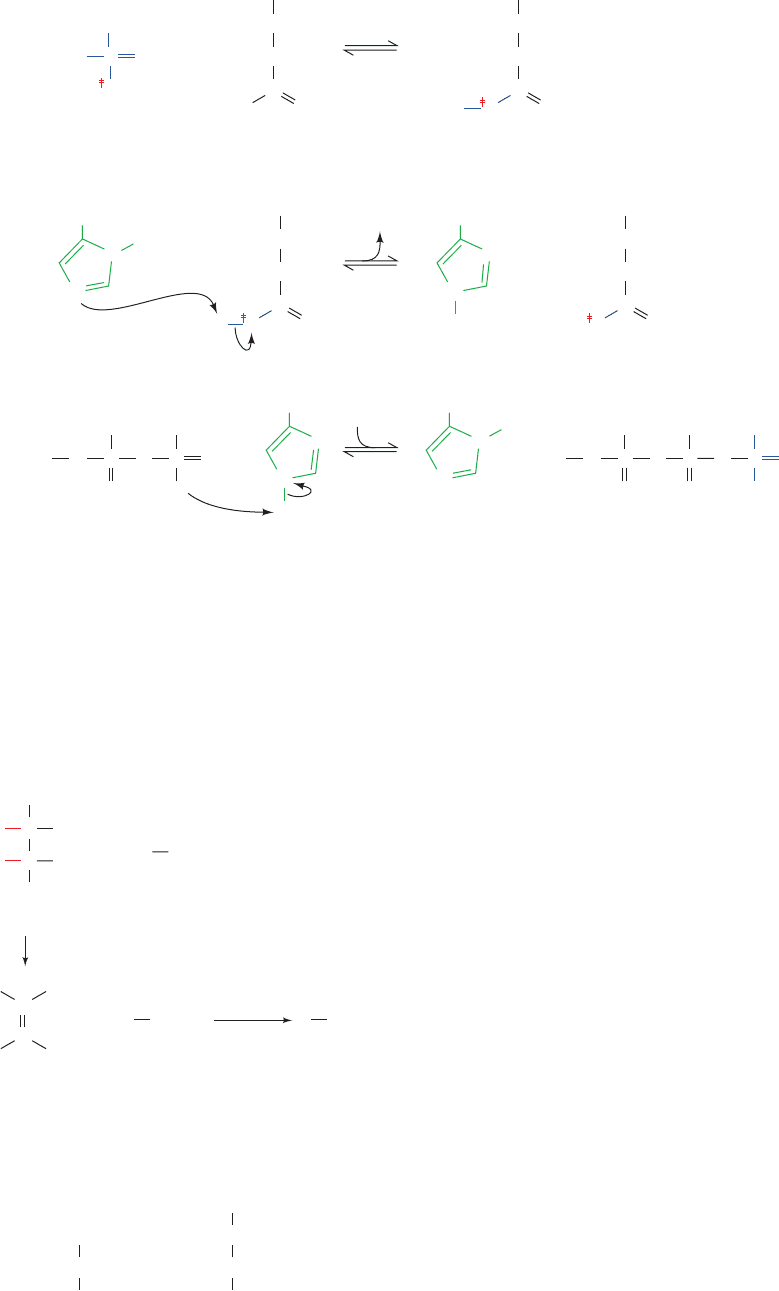
F. Succinate Dehydrogenase
Succinate dehydrogenase catalyzes stereospecific dehydro-
genation of succinate to fumarate (Reaction 6 of Fig. 21-1):
The enzyme is strongly inhibited by malonate, a structural
analog of succinate and a classic example of a competitive
inhibitor:
COO
_
COO
_
CH
2
Malonate
COO
_
COO
_
CH
2
CH
2
Succinate
COO
_
_
COO
OOC
_
COO
_
C
E FAD
C
C
C
pro-S
H
pro-R
H
H
pro-R
pro-R⬘
H H
pro-S⬘
H
pro-S⬘
E FAD
+
E FADH
2
+
Succinate
Fumarate
electron
transport
chain
Recall that malonate inhibition of cellular respiration was
one of the observations that led Krebs to hypothesize the
citric acid cycle (Section 21-1B).
Succinate dehydrogenase contains an FAD, the reac-
tion’s electron acceptor. In general, FAD functions bio-
chemically to oxidize alkanes to alkenes, whereas NAD
⫹
oxidizes alcohols to aldehydes or ketones. This is because
the oxidation of an alkane (such as succinate) to an alkene
(such as fumarate) is sufficiently exergonic to reduce FAD
to FADH
2
but not to reduce NAD
⫹
to NADH.Alcohol ox-
idation, in contrast, can reduce NAD
⫹
(Table 16-4).
Succinate dehydrogenase’s FAD is covalently bound via
its C8a atom to an enzyme His residue (Fig. 21-23).A cova-
lent link between FAD and a protein is unusual; in most
cases FAD is noncovalently although tightly bound to its
associated enzyme (e.g., to dihydrolipoyl dehydrogenase;
Section 21-2Ba). How does succinate dehydrogenase’s
FADH
2
become reoxidized? Being permanently linked to
the enzyme, this prosthetic group cannot function as a
metabolite as does NADH. Rather,succinate dehydrogenase
(also known as Complex II) is reoxidized by the membrane-
soluble coenzyme Q in the electron-transport chain, an as-
pect of its function that we discuss in Section 22-2C. This
rationalizes why succinate dehydrogenase is the only citric
acid cycle enzyme that is anchored in the inner mitochon-
drial membrane; all the others are dissolved in the mito-
chondrial matrix (mitochondrial anatomy is described in
Section 22-1A).
Section 21-3. Enzymes of the Citric Acid Cycle 811
Figure 21-22 Reactions catalyzed by succinyl-CoA synthetase.
(1) Formation of succinyl phosphate, a “high-energy” mixed an-
hydride. (2) Formation of phosphoryl–His, a “high-energy” inter-
C
OCoAS
CH
2
COO
–
Succinyl-CoA
CH
2
–
O
OH
O
–
PO
+
P
i
C
O
CH
2
COO
–
Succinyl
phosphate
CH
2
–2
O
3
PO
+
CoASH
1
C
O
CH
2
COO
–
3-PhosphoHis
CH
2
–2
O
3
PO
2
+
E
N
H
N
..
Enzyme-His
C
O
CH
2
COO
–
CH
2
O
–
+
E
N
N
PO
3
2
–
Succinate
O
O
–
O
PO
+
GDP
3
E
N
N
PO
3
2
–
O
–
O
–
PO
G
E
N
N
O
O
–
O
PO
GTP
O
–
O
PO
G+
H
..
O
–
O
–
PO
H
+
H
+
mediate. (3) Transfer of the phosphoryl group to GDP, forming
GTP. The symbol ‡ represents
18
O in isotopic labeling reactions.
JWCL281_c21_789-822.qxd 3/17/10 11:34 AM Page 811
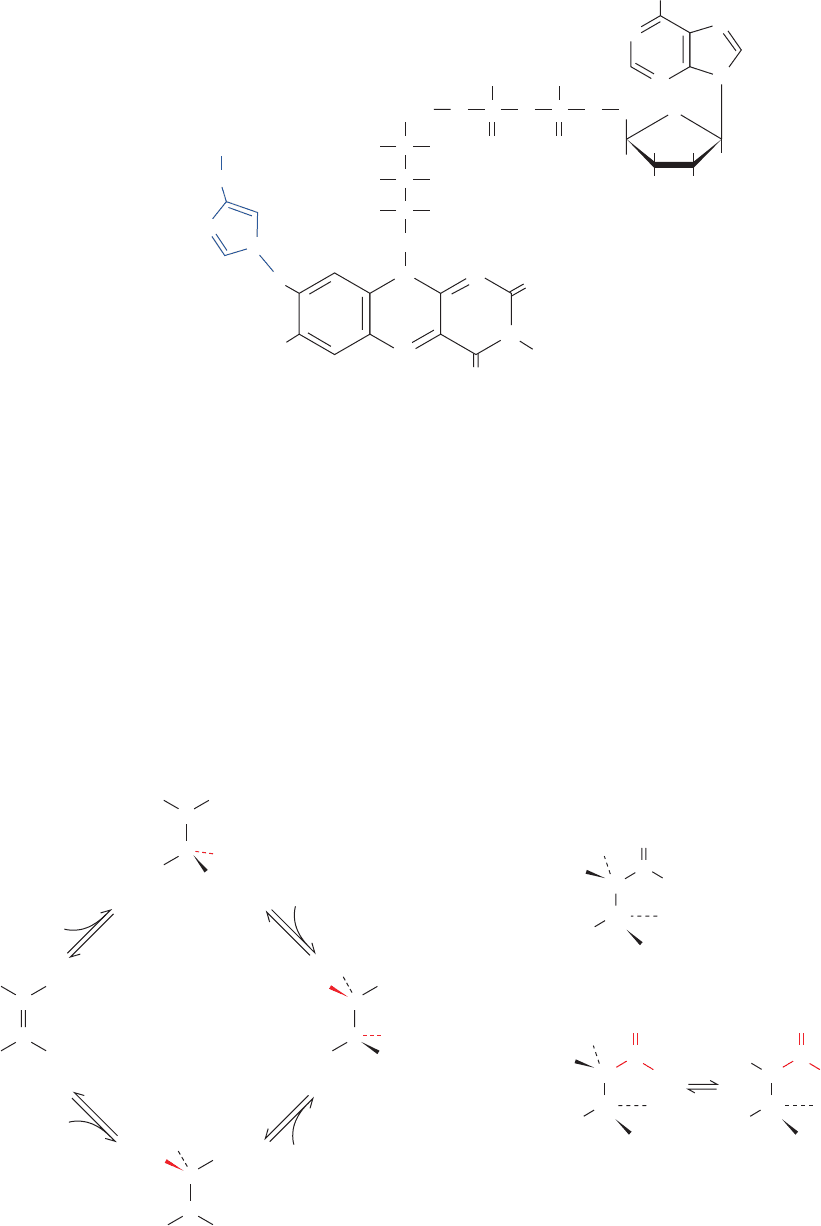
G. Fumarase
Fumarase (fumarate hydratase) catalyzes the hydration of
fumarate’s double bond to form (S)-malate (
L-malate) (Re-
action 7 of Fig. 21-1). Consideration of experiments that
have contributed to our understanding of the fumarase
mechanism illustrates the role played by independent
investigations.
a. Conflicting Mechanistic Evidence: What Is the
Sequence of H
ⴙ
and OH
ⴚ
Addition?
Experiments designed to establish whether the fu-
marase reaction occurs by a carbanion (OH
⫺
addition first)
or carbocation (H
⫹
addition first) mechanism have pro-
vided contradictory information (Fig. 21-24). Evidence fa-
voring the carbocation mechanism was obtained by study-
ing the dehydration of (S)-malate (the fumarase reaction
run in reverse) in H
2
18
O. [
18
O]Malate appears in the reac-
tion mixture more rapidly than it would if the
18
O were in-
corporated via a back reaction of the newly formed fu-
marate. This suggests the rapid formation of a carbocation
intermediate at C2, from which OH
⫺
could exchange with
18
OH
⫺
, followed by slow hydrogen removal at C3 (Fig.
21-24, lower route).
Other observations, however, indicate that the reaction
occurs via the formation of a carbanion intermediate at C3
(Fig. 21-24, upper route). David Porter synthesized 3-nitro-
2-(S)-hydroxypropionate, which sterically resembles (S)-
malate:
The nitro group’s electron-withdrawing character renders
the C3 protons relatively acidic (pK ⬇ 10). The resulting
anion is an analog of the postulated C3 carbanion transi-
tion state of the fumarase reaction but not of the C2 carbo-
cation transition state (Fig. 21-24; transition state analogs
are discussed in Section 15-1Fa). This anion is, in fact, an
H
H
H
OHC
2
C
3
C
O
O
–
–
OOC
(S)-Malate
H
H
H
H
H
OHC
2
C
3
N
O
O
–
+
–
OOC
H
OHC
2
C
3
N
O
O
–
+
+
+
–
–
OOC
3-Nitro-2-
(S)-hydroxypropionate
812 Chapter 21. Citric Acid Cycle
HCHO
CHO
H
CH
O
–
O
2
CH
2
P
CHO
H
CH
2
O
O
–
O OP
O
N
N
N
N
O
H
H
3
C
H
2
C
N
N
CH
2
E
FAD
O
His
75a
9a
7a
8a
8
9
10
4a
10a
1
4
3
2
5
6
O
HH
H
H
OH OH
N
NH
2
N
N
N
Figure 21-24 Possible mechanisms for the hydration of
fumarate as catalyzed by fumarase.
Figure 21-23 Covalent attachment of FAD to a His residue of succinate
dehydrogenase.
Fumarate
H
–
OOC
–
OOC
H COO
–
OH
–
OH
–
H
+
OH
H
+
C
C
Malate
H
H
H
COO
–
C
C
Carbanion
transition state
H
–
OOC
H COO
–
C
C
–
OH
Carbocation
transition state
H
–
OOC
COO
–
C
+
C
H
H
(S)-
JWCL281_c21_789-822.qxd 3/17/10 11:34 AM Page 812

excellent inhibitor of fumarase: It has an 11,000-fold higher
binding affinity for the enzyme than does (S)-malate.
If the fumarase reaction proceeds by a carbanion mech-
anism, how can the rapid OH
⫺
exchange be explained?
Conversely, if it has a carbocation mechanism, why is the
nitro anion such an effective inhibitor? This contradictory
set of observations makes only one thing clear: When
studying enzyme reaction mechanisms, it is always necessary
to approach the problem from several different directions.
One set of experiments should never be taken as proof, and
interpretation should never be taken as fact. Indeed, reinter-
pretation of the
18
O exchange experiment makes it consis-
tent with a carbanion mechanism. It turns out that product
release is the enzyme’s rate-determining step:
Malate binds to fumarase (1), forms a carbanion (2), elim-
inates OH
⫺
to form fumarate (3), and rapidly releases
OH
⫺
from the enzyme surface (4). Release of the other
products (5) is slow.
18
OH
⫺
can therefore exchange with
OH
⫺
to produce [
18
O]malate more rapidly than the overall
rate of the fumarase reaction.
H. Malate Dehydrogenase
Malate dehydrogenase catalyzes the final reaction of the citric
acid cycle,the regeneration of oxaloacetate (Reaction 8 of Fig.
21-1). This occurs through the oxidation of (S)-malate’s hy-
droxyl group to a ketone in an NAD
⫹
-dependent reaction:
The hydride ion released by the alcohol in this reaction is
transferred to the re face of NAD
⫹
, the same face acted on
H
H
H
OHC
C
C
O
O NAD
–
+
+
–
OOC
H
H
C
C
C
O
O
O
NADHH
–
++
–
OOC
(S)-Malate
Oxaloacetate
+
EEEH
+
Mal
E
++
H Fum
Mal Mal
+
+
E H Fum
+
EH
OHFum
+ –
OH
–
–
slow
18
OH
rapid
32
5
4
1
–
Carbanion
intermediate
by lactate dehydrogenase (Section 17-3A) and alcohol de-
hydrogenase (Section 17-3Bc). In fact, X-ray crystallo-
graphic comparisons of the NAD
⫹
-binding domains of
these three dehydrogenases indicate that they are remark-
ably similar and have led to the proposal that all NAD
⫹
-
binding domains have evolved from a common ancestor
(Section 9-3B).
The ⌬G°¿ for the malate dehydrogenase reaction is
⫹29.7 kJ ⴢ mol
⫺1
; the concentration of oxaloacetate formed
at equilibrium is consequently very low. Recall, however,
that the reaction catalyzed by citrate synthase, the first
enzyme in the cycle, is highly exergonic (⌬G°¿ ⫽⫺31.5 kJ ⴢ
mol
⫺1
; Section 21-3A) because of the hydrolysis of citryl-
CoA’s thioester bond. We can now understand the neces-
sity for such a seemingly wasteful process. It allows citrate
formation to be exergonic at even low physiological con-
centrations of oxaloacetate, with the resultant initiation of
another turn of the cycle.
I. Integration of the Citric Acid Cycle
a. The Impact of the Citric Acid Cycle on
ATP Production
The foregoing discussion indicates that one turn of the
citric acid cycle results in the following chemical transfor-
mations (Fig. 21-1):
1. One acetyl group is oxidized to two molecules of
CO
2
, a four-electron pair process (although, as we discuss
below, it is not the carbon atoms of the entering acetyl
group that are oxidized).
2. Three molecules of NAD
⫹
are reduced to NADH,
which accounts for three of the electron pairs.
3. One molecule of FAD is reduced to FADH
2
, which
accounts for the fourth electron pair.
4. One “high-energy” phosphate group is produced as
GTP (or ATP).
The eight electrons abstracted from the acetyl group in the
citric acid cycle subsequently pass into the electron-
transport chain, where they ultimately reduce two mole-
cules of O
2
to H
2
O. The three electron pairs from NADH
each produce ⬃2.5 ATPs via oxidative phosphorylation,
whereas the one electron pair of FADH
2
produces ⬃1.5
ATPs. One turn of the citric acid cycle therefore gives rise to
the generation of ⬃10 ATPs. Electron transport and oxida-
tive phosphorylation are the subject of Chapter 22.
b. Isotopic Tests of Citric Acid Cycle Stereochemistry
The reactions of the citric acid cycle have been con-
firmed through the use of radioactive tracer experiments
that became possible in the late 1930s and early 1940s. At
that time, compounds could be synthesized enriched with
the stable isotope
13
C (detectable at the time by mass spec-
trometry and now also by NMR) or with the radioactive
isotope
11
C, which has a half-life of only 20 min.
14
C, whose
use was pioneered in the late 1940s by Samuel Ruben and
Martin Kamen, has the advantage over
11
C that its half-life
is 5715 years.
Section 21-3. Enzymes of the Citric Acid Cycle 813
JWCL281_c21_789-822.qxd 3/17/10 11:34 AM Page 813

In one landmark experiment, [4-
11
C]oxaloacetate was
generated from
11
CO
2
and pyruvate,
reacted in the citric acid cycle of metabolizing muscle cells,
and the resulting cycle intermediates were isolated. Identi-
fication of the labeled position in the isolated ␣-ketoglu-
tarate caused a furor. As we have seen in Sections 21-3A
and 21-3B, citrate synthase and aconitase catalyze stere-
ospecific reactions in which citrate synthase can distinguish
between the two faces of oxaloacetate’s carbonyl group
and aconitase can distinguish between citrate’s pro-R and
pro-S carboxymethyl groups. In the early 1940s, however,
the concept of prochirality had not been established; it was
assumed that the two halves of citrate were indistinguish-
able (in nonenzymatic systems, this is, in effect, the case). It
was therefore assumed that the radioactivity originally lo-
cated at C4 in oxaloacetate (* in Fig. 21-1) would be scram-
bled in citrate so that its C1 and C6 atoms would be equally
labeled, resulting in ␣-ketoglutarate labeled at both C1 and
C5. In fact, only C1, the carboxyl group ␣ to the keto group
of ␣-ketoglutarate, was found to be radioactive (Fig. 21-1).
This result threw the identity of the condensation product
of oxaloacetate and acetyl-CoA into doubt. How could it
be the symmetrical citrate molecule in light of such “con-
clusive” labeling experiments? This problem of which tri-
carboxylic acid was the cycle’s original condensation prod-
uct resulted in a name change from the citric acid cycle
(proposed by Krebs) to the tricarboxylic acid (TCA) cycle.
In 1948, Alexander Ogston pointed out that citric acid,
while symmetrical, is prochiral and thus can interact asym-
metrically with the surface of aconitase (Section 13-2A).
Even though citrate is now accepted as a cycle intermedi-
ate, the duality of the cycle’s name persists.
While the net reaction of the cycle is oxidation of the
carbon atoms of acetyl units to CO
2
, the CO
2
lost in a given
turn of the cycle is derived from the carbon skeleton of ox-
aloacetate. This can be shown by following the fate of iso-
topically labeled C4 of oxaloacetate (* in Fig. 21-1) through
the citric acid cycle. We can see (Fig. 21-1) that it is lost as
CO
2
at the ␣-ketoglutarate dehydrogenase reaction.
Experiments have been performed using [1-
14
C]acetate,
which cells convert to [1-
14
C]acetyl-CoA:
Tracing the path of this label (‡ in Fig. 21-1) allows the reader
to ascertain that the label is not lost as CO
2
. It becomes
CH
3
C SCoA
O
**
CH
3
COO CoASH
ATP
acetate
thiokinase
AMP PP
i
+
+
Acetate Acetyl-CoA
_
CO
2
CO
2
CH
3
C
O
–
*
+
CO
2
OOC CH
2
C
O
––
*
ATP
pyruvate
carboxylase
ADP P
i
+
Pyruvate
Oxaloacetate
scrambled (1/2‡ in Fig.21-1) during the cycle, but not until the
formation of succinate, the cycle’s first rotationally symmet-
ric (nonprochiral) intermediate.
J. Evolution of the Citric Acid Cycle
The citric acid cycle is ubiquitous in aerobic organisms,
where it has a central role in energy metabolism. However,
an eight-step catalytic cycle such as the citric acid cycle
could not have arisen as a whole and must therefore have
evolved from simpler sets of enzyme-catalyzed reactions.
Clues to the citric acid cycle’s origins can be found by ex-
amining the metabolism of cells that resemble early life-
forms. Such organisms emerged over 3 billion years ago
(Section 1-5), before significant quantities of atmospheric
O
2
were available. These cells probably used sulfur as their
ultimate oxidizing agent, reducing it to H
2
S. Their modern
counterparts are anaerobic autotrophs (cells that synthe-
size all their components from simple molecules such as
H
2
O, CO
2
,NH
3
, and H
2
S) that harvest free energy by path-
ways, which are independent of those that oxidize carbon-
containing compounds. These organisms therefore do not
use the citric acid cycle to generate reduced cofactors such
as NADH that are subsequently oxidized by O
2
.
The task of divining an organism’s metabolic capabili-
ties has been facilitated by bioinformatics. By comparing
the sequences of prokaryotic genomes and assigning func-
tions to various homologous genes, it is possible to recon-
struct their central metabolic pathways. This approach has
been fruitful because the genes that encode the enzymes
that generate free energy and a cell’s most common
metabolites are highly conserved among different species
and hence are relatively easy to recognize.
Many prokaryotes lack the citric acid cycle. However,
these organisms harbor genes for some citric acid cycle en-
zymes. The last three reactions of the cycle, leading from
succinate to oxaloacetate, appear to be the most highly
conserved.This pathway fragment constitutes a mechanism
for accepting electrons that are released during sugar fer-
mentation. For example, the reverse of this pathway could
regenerate NAD
⫹
from the NADH produced by the glyc-
eraldehyde-3-phosphate dehydrogenase step of glycolysis.
The resulting succinate could then be used as a starting
material for the biosynthesis of other compounds.
Many archaeal cells have a pyruvate:ferredoxin oxi-
doreductase that converts pyruvate to acetyl-CoA (but
Oxaloacetate
NADH
NAD
+
NADH
NAD
+
Malate
Fumarate
Succinate
814 Chapter 21. Citric Acid Cycle
JWCL281_c21_789-822.qxd 3/17/10 11:34 AM Page 814
without producing NADH). In a primitive cell, the result-
ing acetyl groups could have condensed with oxaloacetate
(by the action of a citrate synthase), eventually giving rise
to an oxidative sequence of reactions resembling the first
few steps of the modern citric acid cycle.
The ␣-ketoglutarate produced in this way can be converted
to glutamate and other amino acids (Section 26-5Ab).
The reductive and oxidative branches of the citric acid
cycle outlined so far function in modern bacterial cells such
as E. coli cells when they are growing anaerobically, sug-
gesting that similar pathways could have filled the meta-
bolic needs of primordial cells. The evolution of a complete
citric acid cycle in which the two branches are linked
and both proceed in an oxidative direction (clockwise)
would have required an enzyme such as ␣-ketoglutarate:
ferredoxin reductase (a homolog of pyruvate:ferredoxin
oxidoreductase) to link ␣-ketoglutarate and succinate.
Interestingly, a primitive citric acid cycle that operated in
the reverse (counterclockwise) direction could have provided
a pathway for incorporating CO
2
into biological molecules.
Glucose
Oxaloacetate
Acetyl-CoA
Pyruvate
Phosphoenolpyruvate
Citrate
Succinate
α-Ketoglutarate
CO
2
CO
2
CO
2
CO
2
Oxaloacetate
NADH
NAD
+
CO
2
Citrate
Acetyl-CoA
Isocitrate
α-Ketoglutarate
The genes encoding enzymes that catalyze the steps of such
a pathway have been identified in several modern au-
totrophic bacteria.This reductive pathway, which occurs in
some deeply rooted archaeal species, possibly predates the
CO
2
-fixing pathway used in some photosynthetic bacteria
and in the chloroplasts of green plants (Section 24-3A).
4 REGULATION OF THE
CITRIC ACID CYCLE
In this section we consider how metabolic flux through the
citric acid cycle is regulated.In our discussions of metabolic
flux control (Section 17-4), we established that to under-
stand how a metabolic pathway is controlled, we must
identify the enzyme(s) that catalyzes its rate-determining
step(s), the in vitro effectors of these enzymes, and the in
vivo concentrations of these substances. A proposed mech-
anism of flux control must demonstrate that an increase or
decrease in flux is correlated with an increase or decrease in
the concentration of the proposed effector.
a. Citrate Synthase, Isocitrate Dehydrogenase, and
␣-Ketoglutarate Dehydrogenase Are the Citric Acid
Cycle’s Rate-Controlling Enzymes
Establishing the rate-determining steps of the citric acid
cycle is more difficult than it is for glycolysis because most
of the cycle’s metabolites are present in both mitochondria
and cytosol and we do not know their distribution between
these two compartments [recall that identifying a path-
way’s rate-determining step(s) requires determining the
⌬G of each of its reactions from the concentrations of its
substrates and products]. If, however, we assume equilib-
rium between the two compartments, we can use the total
cell contents of these substances to estimate their mito-
chondrial concentrations. Table 21-2 gives the standard
free energy changes for the eight citric acid cycle enzymes
Section 21-4. Regulation of the Citric Acid Cycle 815
Table 21-2 Standard Free Energy Changes (⌬G°¿) and
Physiological Free Energy Changes (⌬G) of
Citric Acid Cycle Reactions
⌬G°¿ ⌬G
Reaction Enzyme (kJ ⴢ mol
⫺1
) (kJ ⴢ mol
⫺1
)
1 Citrate synthase ⫺31.5 Negative
2 Aconitase ⬃5 ⬃0
3 Isocitrate ⫺21 Negative
dehydrogenase
4 ␣-Ketoglutarate ⫺33 Negative
dehydrogenase
multienzyme
complex
5 Succinyl-CoA ⫺2.1 ⬃0
synthetase
6 Succinate ⫹6 ⬃0
dehydrogenase
7 Fumarase ⫺3.4 ⬃0
8 Malate ⫹29.7 ⬃0
dehydrogenase
JWCL281_c21_789-822.qxd 3/17/10 11:34 AM Page 815
and estimates of the physiological free energy changes for
the reactions in heart muscle or liver tissue.We can see that
three of the enzymes are likely to function far from equilib-
rium under physiological conditions (negative ⌬G): citrate
synthase, NAD
⫹
-dependent isocitrate dehydrogenase, and
␣-ketoglutarate dehydrogenase. We shall therefore focus
our discussion on how these enzymes are regulated
(Fig. 21-25).
b. The Citric Acid Cycle Is Largely Regulated by
Substrate Availability, Product Inhibition, and
Inhibition by Other Cycle Intermediates
In heart muscle, where the citric acid cycle functions
mainly to generate ATP for use in muscle contraction, the
enzymes of the cycle almost always act as a functional unit,
with their metabolic flux proportional to the rate of cellu-
lar oxygen consumption. Since oxygen consumption,
NADH reoxidation, and ATP production are tightly cou-
pled (Section 22-4), the citric acid cycle must be regulated by
feedback mechanisms that coordinate its NADH production
with energy expenditure. Unlike the rate-limiting enzymes
of glycolysis and glycogen metabolism, which utilize elabo-
rate systems of allosteric control, substrate cycles, and
covalent modification as flux control mechanisms, the reg-
ulatory enzymes of the citric acid cycle seem to be con-
trolled almost entirely in three simple ways: (1) substrate
availability, (2) product inhibition, and (3) competitive
feedback inhibition by intermediates farther along the cy-
cle. We shall encounter several examples of these straight-
forward mechanisms in the following discussion.
Perhaps the most crucial regulators of the citric acid cycle
are its substrates, acetyl-CoA and oxaloacetate, and its prod-
uct NADH. Both acetyl-CoA and oxaloacetate are present
in mitochondria at concentrations that do not saturate cit-
rate synthase. The metabolic flux through the enzyme
therefore varies with substrate concentration and is subject
to control by substrate availability. The production of
acetyl-CoA from pyruvate is regulated by the activity of
pyruvate dehydrogenase (Section 21-2C). Oxaloacetate is
in equilibrium with malate, its concentration fluctuating
with the [NADH]/[NAD
⫹
] ratio according to the equilib-
rium expression
In the transition from low to high work and respiration
rates, mitochondrial [NADH] decreases. The consequent
increase in [oxaloacetate] stimulates the citrate synthase
reaction, which controls the rate of citrate formation.
The observation that [citrate] invariably falls as the
workload increases indicates that the rate of citrate re-
moval increases more than its rate of formation. The rate of
citrate removal is governed by NAD
⫹
-dependent isocitrate
dehydrogenase (aconitase functions close to equilibrium),
which is strongly inhibited in vitro by NADH (product in-
hibition). Citrate synthase is also inhibited by NADH. Evi-
dently, NAD
⫹
-dependent isocitrate dehydrogenase is more
sensitive to [NADH] changes than citrate synthase.
The decrease in [citrate] that occurs on transition from
low to high work and respiration rates results in a domino
effect:
1. Citrate is a competitive inhibitor of oxaloacetate for
citrate synthase (product inhibition); the fall in [citrate]
caused by increased isocitrate dehydrogenase activity in-
creases the rate of citrate formation.
2. ␣-Ketoglutarate dehydrogenase is also strongly in-
hibited by its products, NADH and succinyl-CoA. Its activ-
ity therefore increases when [NADH] decreases.
3. Succinyl-CoA also competes with acetyl-CoA in the
citrate synthase reaction (competitive feedback inhibition).
This interlocking system serves to keep the citric acid cycle
coordinately regulated.
c. ADP, ATP, and Ca
2ⴙ
Are Allosteric Regulators
of Citric Acid Cycle Enzymes
In vitro studies on the enzymes of the citric acid cycle
have identified a few allosteric activators and inhibitors. In-
creased workload is accompanied by increased [ADP] re-
sulting from the consequent increased rate of ATP hydrol-
ysis. ADP acts as an allosteric activator of isocitrate
K ⫽
[oxaloacetate] [NADH]
[malate] [NAD
⫹
]
816 Chapter 21. Citric Acid Cycle
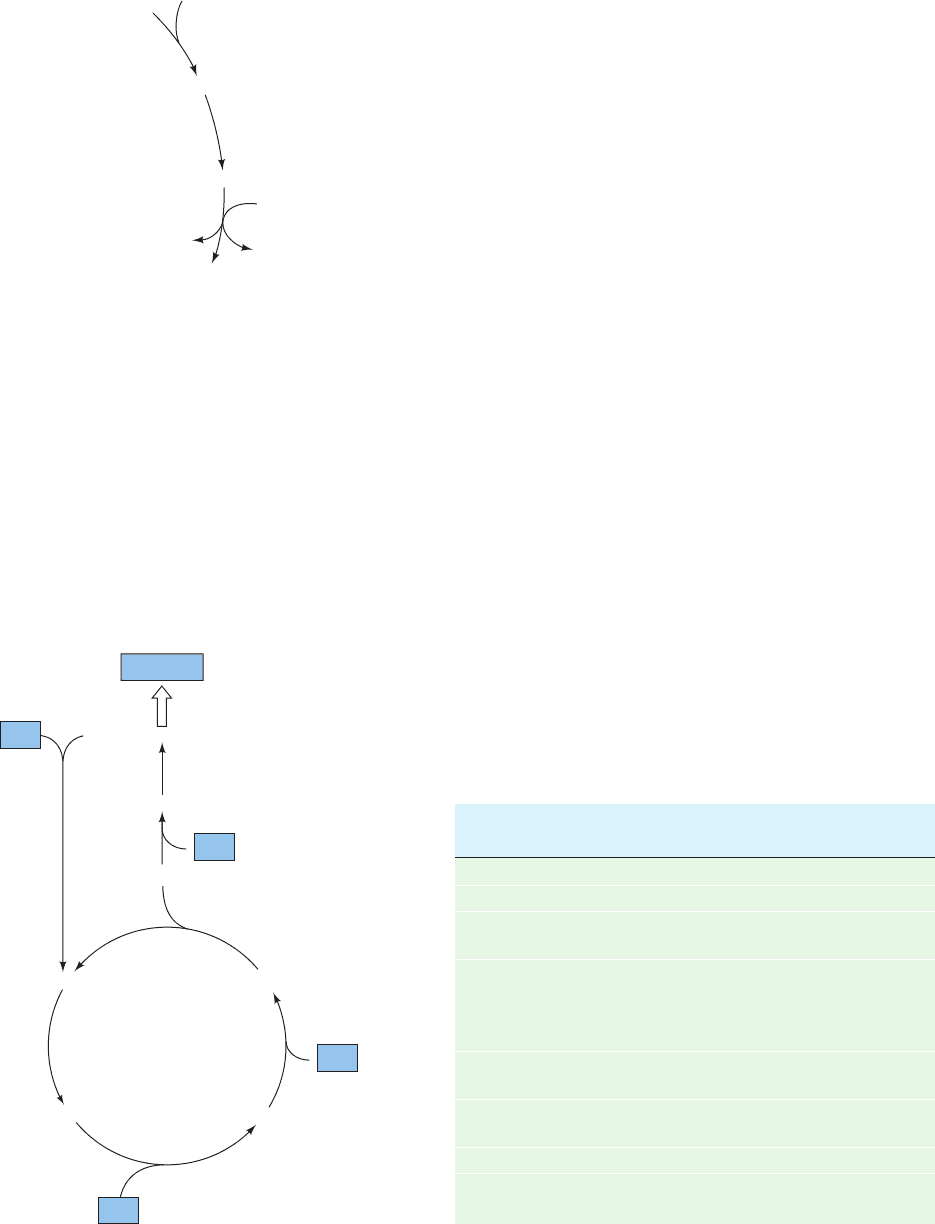
Figure 21-25 Regulation of the citric acid cycle. This diagram
of the citric acid cycle and the pyruvate dehydrogenase reaction
indicates their points of inhibition (red octagons) and the
pathway intermediates that function as inhibitors (dashed red
arrows).ADP and Ca
2⫹
(green dots) are activators. See the
Animated Figures
Oxaloacetate
Fumarate
Malate
Succinate
Succinyl-CoA
α-Ketoglutarate
Isocitrate
Citrate
GTP
ATP
Pyruvate
Ca
2+
Ca
2+
, ADP
Ca
2+
Acetyl-CoA
NADH
JWCL281_c21_789-822.qxd 3/17/10 11:34 AM Page 816
dehydrogenase by decreasing its apparent K
M
for isoci-
trate. ATP, which builds up when muscle is at rest, inhibits
this enzyme.
Ca
2⫹
, among its many biological functions, is an essential
metabolic regulator.It stimulates glycogen breakdown (Sec-
tion 18-3Ce), triggers muscle contraction (Section 35-3Ca),
and mediates many hormonal signals as a second messenger
(Section 19-4A). Ca
2⫹
also plays an important role in the
regulation of the citric acid cycle (Fig. 21-25). It activates
pyruvate dehydrogenase phosphatase and inhibits pyruvate
dehydrogenase kinase, thereby activating the PDC to pro-
duce acetyl-CoA (Fig. 21-17b). In addition, Ca
2⫹
activates
both isocitrate dehydrogenase and ␣-ketoglutarate dehy-
drogenase. Thus, the same signal stimulates muscle contrac-
tion and the production of the ATP to fuel it.
In the liver, the role of the citric acid cycle is more com-
plex than in heart muscle. The liver synthesizes many sub-
stances required by the body including glucose, fatty acids,
cholesterol, amino acids, and porphyrins. Reactions of the
citric acid cycle play a part in many of these biosynthetic
pathways in addition to their role in energy metabolism. In
the next section, we discuss the contribution of the citric
acid cycle to these processes.
d. Are the Enzymes of the Citric Acid Cycle
Organized Into a Metabolon?
Considerable efficiency can be gained by organizing the
enzymes of a metabolic pathway such that the enzymes cat-
alyzing its sequential steps interact to channel intermedi-
ates between them. Indeed, we saw this to be the case with
the PDC (Section 21-2A). The advantages of such an as-
sembly, termed a metabolon, include the protection of la-
bile intermediates and the increase of their local concen-
tration for more efficient catalysis. Considerable effort has
been expended to obtain evidence for such interactions in
the major metabolic pathways including glycolysis and the
citric acid cycle. However, since citric acid cycle intermedi-
ates must be available for use in other metabolic pathways,
any complexes between citric acid cycle enzymes are likely
to be weak and therefore unable to withstand the labora-
tory manipulations necessary to isolate them.
Despite the foregoing, specific interactions have been
demonstrated in vivo between the members of several
pairs of citric acid cycle enzymes, including those between
citrate synthase and malate dehydrogenase. For example,
Paul Srere isolated the gene for a mutant citrate synthase
in yeast that he termed an “assembly mutant” because it
has normal enzymatic activity in vitro but nevertheless
causes a citric acid cycle deficiency in vivo. This mutation
occurs in a highly conserved 13-residue segment of the en-
zyme (Pro 354–Pro 366 in yeast), which forms a solvent-
exposed loop that could interact with other proteins. To
further investigate this phenomenon, Srere constructed a
plasmid expressing a fusion protein (Section 5-5Ga) con-
sisting of the wild-type citrate synthase peptide grafted to
the C-terminal end of green fluorescent protein (GFP; Sec-
tion 5-5Gd). If the peptide actually binds to malate dehy-
drogenase, the expression of this enzymatically inactive fu-
sion protein in yeast would be expected to inhibit the citric
acid cycle. This is, in fact, the case as judged by the severely
decreased ability of such yeast to grow on acetate, a
metabolite that can only be metabolized via the citric acid
cycle. Moreover, replacing the citrate synthase peptide in
the fusion protein with an unrelated peptide only slightly
decreases the normal growth rate on acetate, thereby
demonstrating that the specific sequence of the citrate syn-
thase peptide is the major cause of this growth inhibition. If
the decrease in growth rate is actually caused by competi-
tion of the peptide with citrate synthase for an interacting
partner in a metabolon, then this should be overcome by
the overexpression of either citrate synthase or its interac-
tion partner.Indeed,when either citrate synthase or malate
dehydrogenase was overexpressed in yeast expressing the
GFP–citrate synthase peptide fusion protein, the growth
rate on acetate was restored. However, the overexpression
of aconitase did not overcome this growth inhibition.These
observations support the hypothesis that malate dehydro-
genase and citrate synthase must interact for optimal citric
acid cycle function and identify the interaction site for
malate dehydrogenase on citrate synthase as the 13-
residue Pro 354–Pro 366 peptide.
e. A Bacterial Isocitrate Dehydrogenase Is
Regulated by Phosphorylation
Escherichia coli isocitrate dehydrogenase is a dimer of
identical 416-residue subunits that is inactivated by phos-
phorylation of its Ser 113, an active site residue.In contrast,
most other enzymes that are known to be subject to cova-
lent modification/demodification, for example, glycogen
phosphorylase (Section 18-3), are phosphorylated at al-
losteric sites. In the case of isocitrate dehydrogenase, phos-
phorylation renders the enzyme unable to bind its sub-
strate, isocitrate.
Comparison of the X-ray structures, determined by
Daniel Koshland and Robert Stroud, of isocitrate dehydro-
genase alone, in its phosphorylated form, and with bound
isocitrate reveals only small conformational differences,
suggesting that electrostatic repulsions between the an-
ionic isocitrate and Ser phosphate groups prevent the en-
zyme from binding substrate. Evidently, phosphorylation
can regulate enzyme activity by directly interfering with ac-
tive site ligand binding as well as by inducing a conforma-
tional change from an allosteric site.
5 THE AMPHIBOLIC NATURE
OF THE CITRIC ACID CYCLE
Ordinarily one thinks of a metabolic pathway as being ei-
ther catabolic with the release (and conservation) of free
energy, or anabolic with a requirement for free energy. The
citric acid cycle is, of course, catabolic because it involves
degradation and is a major free energy conservation sys-
tem in most organisms. Cycle intermediates are only re-
quired in catalytic amounts to maintain the degradative func-
tion of the cycle. However, several biosynthetic pathways
utilize citric acid cycle intermediates as starting materials
Section 21-5. The Amphibolic Nature of the Citric Acid Cycle 817
JWCL281_c21_789-822.qxd 3/17/10 11:34 AM Page 817
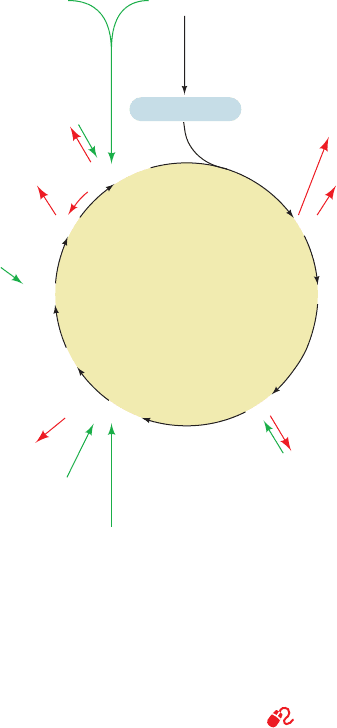
(anabolism). The citric acid cycle is therefore amphibolic
(both anabolic and catabolic).
All of the biosynthetic pathways that utilize citric acid
cycle intermediates also require free energy. Consequently,
the catabolic function of the cycle cannot be interrupted;
cycle intermediates that have been siphoned off must be re-
placed. Although the mechanistic aspects of the enzymes
involved in the pathways that utilize and replenish citric
acid cycle intermediates are discussed in subsequent chap-
ters, it is useful to briefly mention these metabolic inter-
connections here (Fig. 21-26).
a. Pathways That Utilize Citric Acid
Cycle Intermediates
Reactions that utilize and therefore drain citric acid
cycle intermediates are called cataplerotic reactions (empty-
ing; Greek: cata, down ⫹ plerotikos, to fill). These reactions
serve not only to synthesize important products but also to
avoid the inappropriate buildup of citric acid cycle inter-
mediates in the mitochondrion, for example, when there is
a high rate of breakdown of amino acids to citric acid cycle
intermediates (Section 26-3). Cataplerotic reactions occur
in the following pathways:
1. Glucose biosynthesis (gluconeogenesis; Section 23-1),
which occurs in the cytosol, utilizes oxaloacetate as its start-
ing material. Oxaloacetate is not transported across the mi-
tochondrial membrane, but malate is. Malate that has been
transported across the mitochondrial membrane is con-
verted to oxaloacetate in the cytosol for gluconeogenesis.
2. Lipid biosynthesis, which includes fatty acid biosyn-
thesis (Section 25-4) and cholesterol biosynthesis (Section
25-6A), is a cytosolic process that requires acetyl-CoA.
Acetyl-CoA is generated in the mitochondrion and is not
transported across the inner mitochondrial membrane.
Cytosolic acetyl-CoA is therefore generated by the break-
down of citrate, which can cross the inner mitochondrial
membrane, in a reaction catalyzed by ATP-citrate lyase
(Section 25-4D):
3. Amino acid biosynthesis utilizes citric acid cycle in-
termediates in two ways. ␣-Ketoglutarate is converted to
glutamate in a reductive amination reaction involving
either NAD
⫹
or NADP
⫹
catalyzed by glutamate dehydro-
genase (Section 26-1):
␣-Ketoglutarate and oxaloacetate are also used to synthe-
size glutamate and aspartate in transamination reactions
(Section 26-1):
and
4. Porphyrin biosynthesis (Section 26-4A) utilizes
succinyl-CoA as a starting material.
5. Complete oxidation of amino acids requires that the
citric acid cycle intermediates to which the amino acids are
degraded be converted first to PEP [in a reaction catalyzed
by phosphoenolpyruvate carboxykinase (PEPCK; Section
23-1): Oxaloacetate ⫹ GTP 12 PEP ⫹ GDP], then to pyru-
vate by pyruvate kinase (Section 17-2J), and finally to
acetyl-CoA by pyruvate dehydrogenase (Section 21-2A).
b. Reactions That Replenish Citric
Acid Cycle Intermediates
Reactions that replenish citric acid cycle intermediates
are called anaplerotic reactions (filling up, Greek: ana, up).
The main reaction of this type is catalyzed by pyruvate car-
boxylase, which produces oxaloacetate (Section 23-1Aa):
oxaloacetate ⫹ ADP ⫹ P
i
Pyruvate ⫹ CO
2
⫹ ATP ⫹ H
2
O Δ
Oxaloacetate ⫹ alanine Δ aspartate ⫹ pyruvate
␣-Ketoglutarate ⫹ alanine Δ glutamate ⫹ pyruvate
glutamate ⫹ NAD(P)
⫹
⫹ H
2
O
␣-Ketoglutarate ⫹ NAD(P)H ⫹ NH
⫹
4
Δ
ADP ⫹ P
i
⫹ oxaloacetate ⫹ acetyl-CoA
ATP ⫹ citrate ⫹ CoA Δ
818 Chapter 21. Citric Acid Cycle
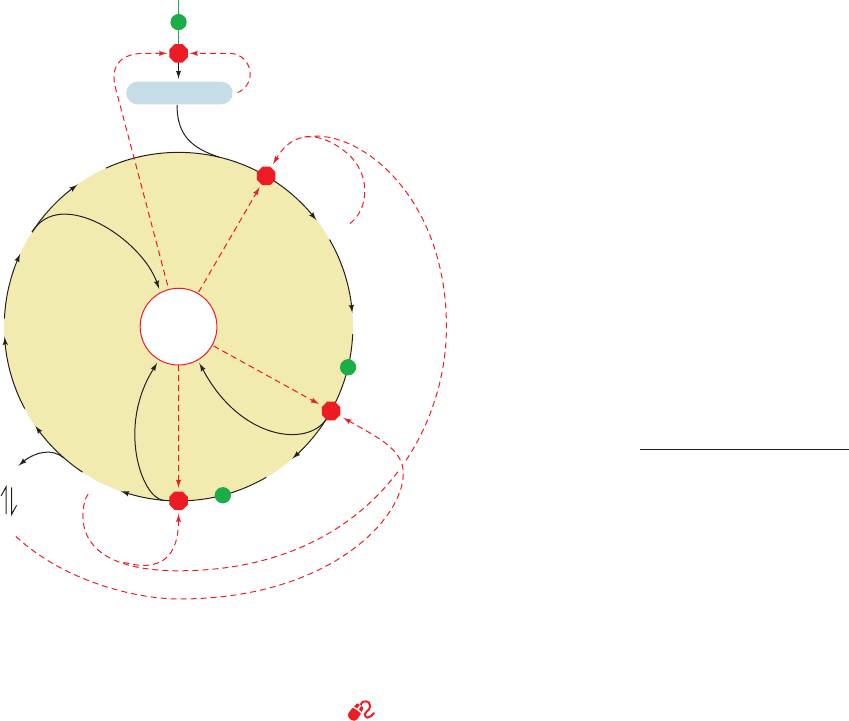
Figure 21-26 Amphibolic functions of the citric acid cycle.
This diagram indicates the positions at which intermediates are
cataplerotically drawn off for use in anabolic pathways (red
arrows) and the points where anaplerotic reactions replenish
depleted cycle intermediates (green arrows). Reactions involving
amino acid transamination and deamination are reversible, so
their direction varies with metabolic demand.
See the
Animated Figures
Pyruvate
2
CO
Oxaloacetate
Malate
Fumarate
Succinate
Succinyl-CoA
α-Ketoglutarate
Isocitrate
Citrate
Acetyl-CoA
Fatty
acids
Cholesterol
Amino
acids
Amino
acids
Glucose
Porphyrins
Odd-chain
fatty acids
Isoleucine
Methionine
Valine
A
sparate
Phenylalanine
Tyrosine
JWCL281_c21_789-822.qxd 3/17/10 11:34 AM Page 818

This enzyme “senses” the need for more citric acid cycle in-
termediates through its activator, acetyl-CoA. Any de-
crease in the rate of the cycle caused by insufficient ox-
aloacetate or other cycle intermediates results in an
increased level of acetyl-CoA because of its underutiliza-
tion.This activates pyruvate carboxylase,which replenishes
oxaloacetate, increasing the rate of the cycle. Of course, if
the citric acid cycle is inhibited at some other step, by high
NADH concentration, for example, increased oxaloacetate
concentration will not activate the cycle. The excess ox-
aloacetate instead equilibrates with malate, which is trans-
ported out of the mitochondria for use in gluconeogenesis.
Degradative pathways generate citric acid cycle
intermediates:
1. Oxidation of odd-chain fatty acids (Section 25-2E)
leads to the production of succinyl-CoA.
2. Breakdown of the amino acids isoleucine, methion-
ine, and valine (Section 26-3E) also leads to the production
of succinyl-CoA.
3. Transamination and deamination of amino acids lead
to the production of ␣-ketoglutarate and oxaloacetate.
These reactions are reversible and, depending on meta-
bolic demand, serve to remove or replenish these citric acid
cycle intermediates.
The citric acid cycle is truly at the center of metabolism
(see Fig. 16-1). Its reduced products, NADH and FADH
2
,
are reoxidized by the electron-transport chain during ox-
idative phosphorylation and the free energy released is
coupled to the biosynthesis of ATP. Citric acid cycle inter-
mediates are utilized in the biosynthesis of many vital cel-
lular constituents. In the next few chapters we shall explore
the interrelationships of these pathways in more detail.
Chapter Summary 819
1 Cycle Overview The citric acid cycle, the common
mode of oxidative metabolism in most organisms, is mediated
by eight enzymes that collectively convert 1 acetyl-CoA to 2
CO
2
molecules so as to yield 3 NADHs, 1 FADH
2
, and 1 GTP
(or ATP). The NADH and FADH
2
are oxidized by O
2
in the
electron-transport chain with the concomitant synthesis of
around 11 more ATPs, yielding a total of about 12 ATPs for
one turn of the citric acid cycle.
2 Metabolic Sources of Acetyl-Coenzyme A Pyruvate,
the end product of glycolysis under aerobic conditions, is con-
verted to acetyl-CoA by the pyruvate dehydrogenase multi-
enzyme complex (PDC), a cubic or dodecahedral cluster of
three enzymes: pyruvate dehydrogenase (E1), dihydrolipoyl
transacetylase (E2), and dihydrolipoyl dehydrogenase (E3).
The pyruvate dehydrogenase subunit catalyzes the conversion
of pyruvate to CO
2
and a hydroxyethyl-TPP intermediate.The
latter is channeled to dihydrolipoyl transacetylase, which oxi-
dizes the hydroxyethyl group to acetate and transfers it to
CoA to form acetyl-CoA. The lipoamide prosthetic group,
which is reduced to the dihydro form in the process, is reoxi-
dized by dihydrolipoyl dehydrogenase in a reaction involving
bound FAD that reduces NAD
⫹
to NADH. Dihydrolipoyl
transacetylase is inactivated by the formation of a covalent
adduct between lipoamide and As(III) compounds. Pyruvate
dehydrogenase’s two active sites are connected by an acidic tun-
nel through which they alternately supply and abstract protons
from one another and thereby keep their reactions out of phase.
Dihydrolipoyl dehydrogenase, which closely resembles glu-
tathione reductase, catalyzes a two-stage reaction. In the first
stage, dihydrolipoamide reduces the enzyme’s redox-active
disulfide group, yielding the reaction’s first product,
lipoamide. In the second stage, NAD
⫹
reoxidizes the reduced
enzyme through the intermediacy of the enzyme’s FAD pros-
thetic group, thereby closing the catalytic cycle and yielding
the enzyme’s second product, NADH. The activity of the
pyruvate dehydrogenase complex varies with the [NADH]/
[NAD
⫹
] and [acetyl-CoA]/[CoA] ratios. In eukaryotes, the
pyruvate dehydrogenase subunit is also inactivated by phos-
phorylation of a specific Ser residue and is reactivated by its
removal. These modifications are mediated, respectively, by
pyruvate dehydrogenase kinase and pyruvate dehydrogenase
phosphatase, which are components of the multienzyme
complex and respond to the levels of metabolic intermediates
such as NADH and acetyl-CoA.
3 Enzymes of the Citric Acid Cycle Citrate is formed by
the condensation of acetyl-CoA and oxaloacetate by citrate
synthase. The citrate is dehydrated to cis-aconitate and then
rehydrated to isocitrate in a stereospecific reaction catalyzed
by aconitase. This enzyme is specifically inhibited by (2R,3R)-
2-fluorocitrate, which is enzymatically synthesized from fluo-
roacetate and oxaloacetate. Isocitrate is oxidatively decar-
boxylated to ␣-ketoglutarate by isocitrate dehydrogenase,
which produces NADH and CO
2
.The ␣-ketoglutarate, in turn,
is oxidatively decarboxylated by ␣-ketoglutarate dehydroge-
nase, a multienzyme complex homologous to the PDC.This re-
action generates the second NADH and CO
2
. The resulting
succinyl-CoA is converted to succinate with the generation of
GTP (ATP in plants and bacteria) by succinyl-CoA syn-
thetase. The succinate is stereospecifically dehydrogenated to
fumarate by succinate dehydrogenase in a reaction that gener-
ates FADH
2
. The final two reactions of the citric acid cycle,
which are catalyzed by fumarase and malate dehydrogenase,
in turn hydrate fumarate to (S)-malate and oxidize this alco-
hol to its corresponding ketone, oxaloacetate, with concomi-
tant production of the pathway’s third and final NADH. The
citric acid cycle appears to have evolved through the piecing
together of enzymatic reactions that originally carried out
other metabolic functions.
4 Regulation of the Citric Acid Cycle The enzymes of
the citric acid cycle act as a functional unit that keeps pace
with the metabolic demands of the cell. The flux-controlling
enzymes appear to be citrate synthase, isocitrate dehydroge-
nase, and ␣-ketoglutarate dehydrogenase. Their activities are
controlled by substrate availability, product inhibition, inhibi-
tion by cycle intermediates, and activation by Ca
2⫹
. The en-
zymes of the citric acid cycle may be organized, in vivo, into a
metabolon for channeling the products of one enzyme to the
next enzyme in the cycle.
CHAPTER SUMMARY
JWCL281_c21_789-822.qxd 6/5/10 9:53 AM Page 819

820 Chapter 21. Citric Acid Cycle
History
Holmes, F.L., Hans Krebs: Vol. 1: The Formation of a Scientific
Life, 1900–1933; and Vol. 2: Architect of Intermediary Metabo-
lism, 1933–1937, Oxford University Press (1991 and 1993).
[The biography of the discoverer of the citric acid cycle
through the time of its discovery.]
Kornberg, H.L., Tricarboxylic acid cycles, BioEssays 7, 236–238
(1987). [A historical synopsis of the intellectual background
leading to the discovery of the citric acid cycle.]
Krebs, H.A., The history of the tricarboxylic acid cycle, Perspect.
Biol. Med. 14, 154–170 (1970).
Pyruvate Dehydrogenase Multienzyme Complex
Frank, R.A.W., Pratap, J.V., Pei, X.Y., Perham, R.N., and Luisi,
B.F., The molecular origins of specificity in the assembly of a
multienzyme complex, Structure 13, 1119–1130 (2005). [The X-
ray structure of B. stearothermophilus E1 ⴢ TPP ⴢ PSBD.]
Frank, R.A.W.,Titman, C.M., Pratap, J.V., Luisi, B.F., and Perham,
R.N., A molecular switch and proton wire synchronize the ac-
tive sites in thiamine enzymes, Science 306, 872–876 (2004).
Izard,T., Ævarsson,A.,Allen, M.D.,Westphal,A.H., Perham, R.N.,
de Kok, A., and Hol, W.G.J., Principles of quasi-equivalence
and euclidean geometry govern the assembly of cubic and do-
decahedral cores of pyruvate dehydrogenase, Proc. Natl.Acad.
Sci. 96, 1240–1245 (1999).
Karplus, P.A.and Schulz, G.E., Refined structure of glutathione re-
ductase at 1.54 Å resolution, J. Mol. Biol. 195, 701–729 (1987).
Karplus,P.A.and Schulz,G.E., Substrate binding and catalysis by glu-
tathione reductase derived from refined enzyme:substrate crystal
structures at 2Å resolution, J. Mol. Biol. 210, 163–180 (1989).
Lengyel, J.S., Stott, K.M., Wu, X., Brooks, B.R., Balbo,A., Schuck,
P., Perham, R.N., Subramaniam, S., and Milne, J.L.S., Extended
polypeptide linkers establish the spatial architecture of a pyru-
vate dehydrogenase multienzyme complex, Structure 16,
93–103 (2008).
Mande, S.S., Sarfaty, S.,Allen, M.D., Perham, R.N., and Hol,W.G.J.,
Protein–protein interactions in the pyruvate dehydrogenase
complex: dihydrolipoamide dehydrogenase complexed with
the binding domain of dihydrolipoamide acetyltransferase,
Structure 4, 277–286 (1996).
Mattevi, A., Obmolova, G., Sokatch, J.R., Betzel, C., and Hol,
W.G.J., The refined crystal structure of Pseudomonas putida
lipoamide dehydrogenase complexed with NAD
⫹
at 2.45 Å
resolution, Proteins 13, 336–351 (1992); and Mattevi, A.,
Schierbeek, A.J., and Hol, W.G.J., Refined crystal structure of
lipoamide dehydrogenase from Azotobacter vinelandii at 2.2 Å
resolution. A comparison with the structure of glutathione re-
ductase, J. Mol. Biol. 220, 975–994 (1991).
Milne, J.L.S.,Wu, X., Borgnia, M.J., Lengyel, J.S., Brooks, B.R., Shi,
D., Perham, R.N., and Subramaniam, S., Molecular structure of
a 9-MDa icosahedral pyruvate dehydrogenase subcomplex
containing the E2 and E3 enzymes using cryoelectron mi-
croscopy, J. Biol. Chem. 281, 4364–4370 (2006).
Patel, M.S. and Korotchkina, L.G., The biochemistry of the pyru-
vate dehydrogenase complex, Biochem. Mol. Biol. Educ. 31,
5–15 (2003).
Perham, R.N., Swinging arms and swinging domains in multifunc-
tional enzymes: Catalytic machines for multistep reactions,
Annu. Rev. Biochem. 69, 961–1004 (2000). [An authoritative
review on multienzyme complexes.]
Reed, L.J.,A trail of research from lipoic acid to ␣-keto acid dehy-
drogenase complexes, J. Biol. Chem. 276, 38329–38336 (2001).
[A scientific memoir.]
Roche, T.E., Baker, J.C.,Yan, X., Hiromasa, Y., Gong, X., Peng, T.,
Dong,J.,Turkan,A., and Kasten, S.E., Distinct regulatory prop-
erties of pyruvate dehydrogenase kinase and phosphatase iso-
forms, Prog. Nucleic Acid Res. Mol. Biol. 70, 33–75 (2001).
Williams, C.H., Jr., Lipoamide dehydrogenase, glutathione reduc-
tase, thioredoxin reductase, and mercuric ion reductase—A
family of flavoenzyme transhydrogenases, in Müller, F. (Ed.),
Chemistry and Biochemistry of Flavoenzymes, Vol. III,
pp. 121–211, CRC Press (2000).
Zhou, Z.H., McCarthy, D.B., O’Connor, C.M., Reed, L.J., and
Stoops, J.K., The remarkable structural and functional organi-
zation of the eukaryotic pyruvate dehydrogenase complexes,
Proc. Natl. Acad. Sci. 98, 14802–14807 (2001).
Enzymes of the Citric Acid Cycle
Beinert, H., Kennedy, M.C., and Stout, D.C., Aconitase as iron-
sulfur protein, enzyme, and iron-regulatory protein, Chem.
Rev. 96, 2335–2374 (1996).
Bolduc, J.M., Dyer, D.H., Scott, W.G., Singer, P., Sweet, R.M.,
Koshland, D.E., Jr., and Stoddard, B.L., Mutagenesis and Laue
structures of enzyme intermediates: Isocitrate dehydrogenase,
Science 268, 1312–1318 (1995).
Cleland, W.W. and Kreevoy, M.M., Low-barrier hydrogen bonds
and enzymic catalysis, Science 264, 1887–1890 (1994).
Frey, P.A. and Hegeman, A.D., Enzymatic Reaction Mechanisms,
Oxford (2007). [Contains discussions of the mechanisms of
various citric acid cycle enzymes.]
Huynen, M.A., Dandekar,T., and Bork, P.,Variation and evolution
of the citric-acid cycle: a genomic perspective, Trends Micro-
biol. 7, 281–291 (1999). [Discusses how genomic studies can al-
low reconstruction of metabolic pathways, even when some
enzymes appear to be missing.]
Karpusas, M., Branchaud, B., and Remington, S.J., Proposed
mechanism for the condensation reaction of citrate synthase:
1.9-Å structure of the ternary complex with oxaloacetate and
carboxymethyl coenzyme A, Biochemistry 29, 2213–2219
(1990).
Kurz, L.C., Nakra, T., Stein, R., Plungkhen, W., Riley, M., Hsu, F.,
and Drysdale, G.R., Effects of changes in three catalytic
residues on the relative stabilities of some of the intermediates
and transition states in the citrate synthase reaction, Biochem-
istry 37, 9724–9737 (1998).
Lauble, H., Kennedy, M.C., Beinert, H., and Stout, D.C., Crystal
structures of aconitase with isocitrate and nitroisocitrate
bound, Biochemistry 31, 2735–2748 (1992).
Mulholland, A.J., Lyne, P.D., and Karplus, M., Ab initio QM/MM
study of the citrate synthase mechanism. A low-barrier hydro-
gen bond is not involved, J. Am. Chem. Soc. 122, 534–535
(2000).
REFERENCES
5 The Amphibolic Nature of the Citric Acid Cycle Sev-
eral anabolic pathways cataplerotically utilize citric acid cycle
intermediates as starting materials. These essential substances
are replaced by anaplerotic reactions of which the major one
is synthesis of oxaloacetate from pyruvate and CO
2
by pyru-
vate carboxylase.
JWCL281_c21_789-822.qxd 6/5/10 9:53 AM Page 820
