Voet D., Voet Ju.G. Biochemistry
Подождите немного. Документ загружается.

by bisulfite treatment (Section 30-7b), which reestablishes
the “missing” base pair as U ⴢ A. Similarly, Sec–tRNA
Sec
,
which is also not bound by EF-Tu (but rather by SELB;
Section 32-2De), has 8 bp in its acceptor stem vs 7 bp in
those of other elongator tRNAs. However, initiator tRNAs
from several sources have fully base paired acceptor stems,
and the U1 ⴢ A72 base pair of tRNA
Gln
is opened up on
binding to GlnRS (Section 32-2Cc).
c. EF-Tu Undergoes a Major Conformational Change
on Hydrolyzing GTP
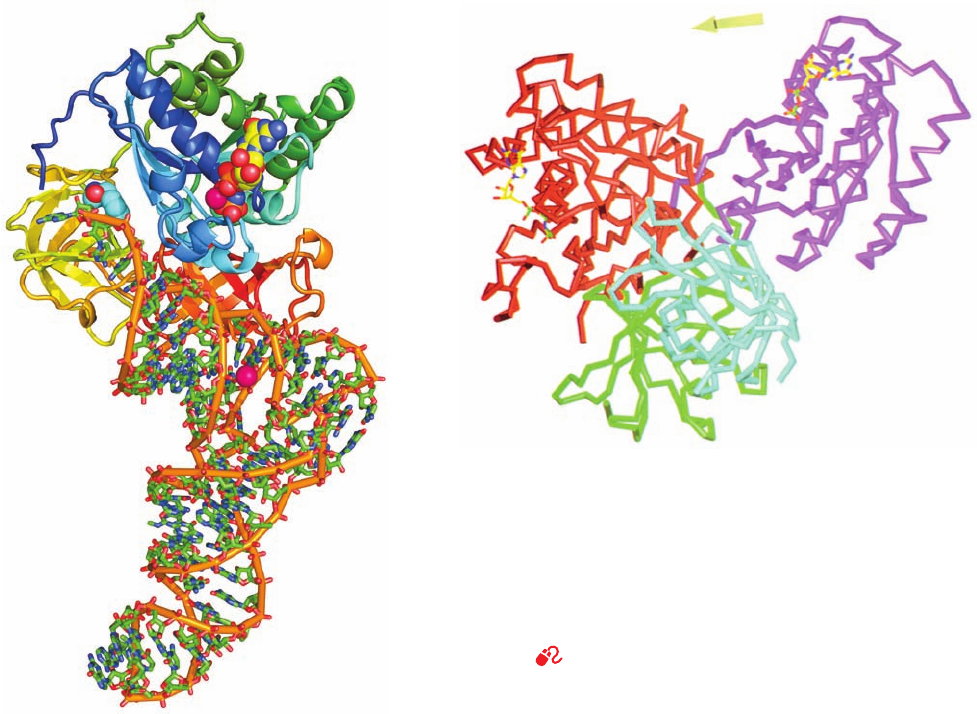
Morten Kjeldgaard and Nyborg determined the X-ray
structures of T. aquaticus EF-Tu (405 residues) in com-
plex with GMPPNP and the 70% identical E. coli EF-Tu
(393 residues) in complex with GDP (Fig. 32-48).The con-
formation of EF-Tu in its complex with only GMPPNP
closely resembles that in its ternary complex with
Phe–tRNA
Phe
and GMPPNP (Fig. 32-47). However, com-
parison of the GMPPNP and GDP complexes indicates
that, on hydrolyzing its bound GTP, EF-Tu undergoes a
major structural reorganization. Its greatest local confor-
mational changes occur in the Switch I and Switch II re-
gions of domain 1, which in all G-proteins signal the state
of the bound nucleotide to interacting partners (Section
19-2Cb; here domains 2 and 3): Switch I converts from a 
hairpin to a short ␣ helix and the ␣ helix of Switch II shifts
toward the C-terminus by 4 residues. As a consequence,
this latter helix reorients by 42°, which results in domain 1
rigidly changing its orientation with respect to domains 2
and 3 by a dramatic 91° rotation.The tRNA binding site is
thereby eliminated.
d. EF-Ts Disrupts the Binding of GDP to EF-Tu
EF-Tu has a 100-fold higher affinity for GDP than GTP.
Hence, replacement of the EF-Tu–bound GDP by GTP
must be facilitated by the interaction of EF-Tu with EF-Ts
(Fig. 32-46, top). The X-ray structure of the EF-Tu ⴢ EF-Ts
complex, determined by Stephen Cusack and Reuben
Section 32-3. Ribosomes and Polypeptide Synthesis 1381
Figure 32-47 X-ray structure of the ternary complex of yeast
Phe–tRNA
Phe
, Thermus aquaticus EF-Tu, and GMPPNP. The
EF-Tu is drawn in ribbon form colored in rainbow order from its
N-terminus (blue) to its C-terminus (red).The tRNA is shown in
stick form colored according to atom type with C green, N blue,
O red, and P orange and with orange rods linking successive P
atoms.The tRNA’s appended aminoacyl–Phe residue and the
GMPPNP that is bound to the EF-Tu are drawn in space-filling
form with C atoms cyan and yellow, respectively. Two bound
Mg
2⫹
ions are represented by magenta spheres. [Based on an
X-ray structure by Jens Nyborg, University of Aarhus, Århus,
Denmark. PDBid 1TTT.]
Figure 32-48 Comparison of the X-ray structures of EF-Tu in
its complexes with GDP and GMPPNP. The protein is
represented by its C
␣
backbone with domain 1, its GTP-binding
domain, purple in the GDP complex and red in the GMPPNP
complex. Domain 2 and domain 3, which have the same
orientation in both complexes, are green and cyan.The bound
GDP and GMPPNP are shown in stick form with C yellow, N
blue, O red, and P green. [Courtesy of Morten Kjeldgaard and
Jens Nyborg, University of Aarhus, Århus, Denmark. PDBid
1EFT.]
See Interactive Exercise 45
JWCL281_c32_1338-1428.qxd 10/19/10 8:24 AM Page 1381
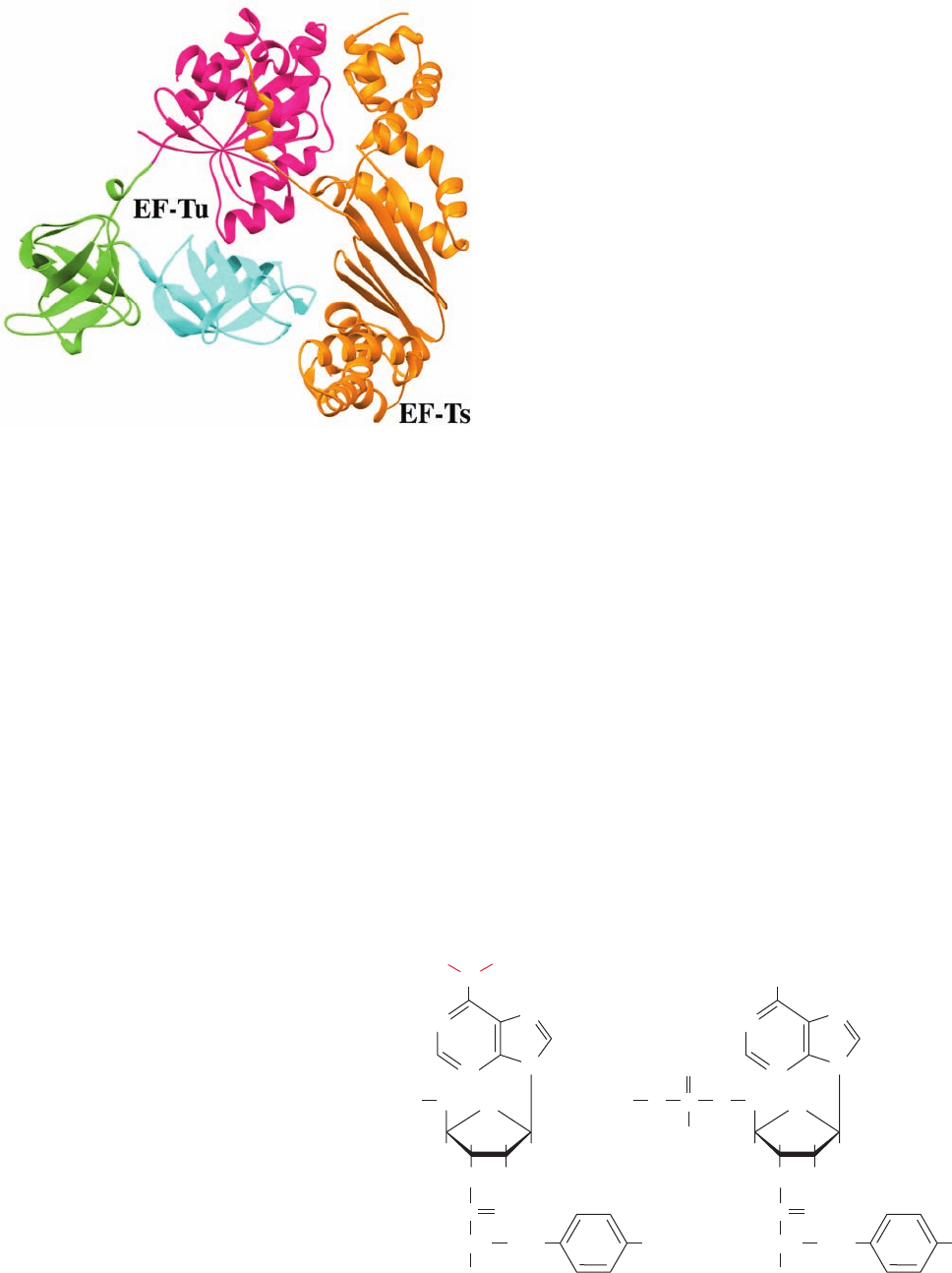
Leberman, reveals that the EF-Tu has a conformation re-
sembling that of its GDP complex (Fig. 32-49) but with
its domains 2 and 3 swung away from domain 1 by ⬃18°.
EF-Ts is an elongated molecule that binds along the right side
of EF-Tu as shown in Fig. 32-49, where it contacts EF-Tu’s
domains 1 and 3. The intrusive interactions of EF-Ts side
chains with the GDP binding pocket on EF-Tu domain 1
disrupts the Mg
2
ion binding site. This reduces the affinity
of EF-Tu for GDP, thereby facilitating its exchange for
GTP (after EF-Ts has dissociated), which has a 10-fold
higher concentration in the cell than does GDP (the GEF-
containing segment of Sos similarly interferes with Mg
2
binding and hence guanine nucleotide binding by Ras; Sec-
tion 19-3Cf). EF-Tu’s subsequent binding of a charged
elongator tRNA increases its affinity for GTP.
e. Transpeptidation
In the transpeptidation stage of the elongation cycle
(Fig. 32-46), the peptide bond is formed through the nucle-
ophilic displacement of the P-site tRNA by the amino
group of the 3¿-linked aminoacyl–tRNA in the A site (Fig
32-39). The nascent polypeptide chain is thereby length-
ened at its C-terminus by one residue and transferred to
the A-site tRNA. The reaction occurs without the need of
activating cofactors such as ATP because the ester linkage
between the nascent polypeptide and the P-site tRNA is a
“high-energy” bond. The peptidyl transferase center that
catalyzes peptide bond formation is located entirely on the
large subunit as is demonstrated by the observation that in
high concentrations of organic solvents such as ethanol, the
large subunit alone catalyzes peptide bond formation. The
organic solvent apparently distorts the large subunit in a
way that mimics the effect of small subunit binding.
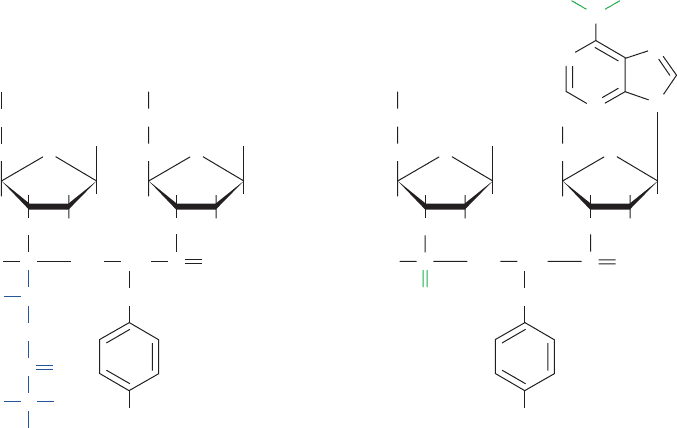
f. Puromycin Is an Aminoacyl–tRNA Analog
The ribosomal elongation cycle was originally character-
ized through the use of the antibiotic puromycin (Fig.32-50).
This product of Streptomyces alboniger, which resembles the
3¿ end of Tyr–tRNA, causes the premature termination of
polypeptide chain synthesis. Puromycin, in competition with
the mRNA-specified aminoacyl–tRNA but without the
need of elongation factors, binds to the ribosomal A site
which, in turn, catalyzes a normal transpeptidation reaction
to form peptidyl–puromycin. Yet, the ribosome cannot
catalyze the transpeptidation reaction in the next elonga-
tion cycle because puromycin’s “amino acid residue” is
linked to its “tRNA” via an amide rather than an ester bond.
Polypeptide synthesis is therefore aborted and the pep-
tidyl–puromycin is released.
In the absence of the elongation factor EF-G (see be-
low), an active ribosome cannot bind puromycin because
its A site is at least partially occupied by a peptidyl–tRNA.
A newly initiated ribosome, however, violates this rule; it
1382 Chapter 32. Translation
Figure 32-50 Puromycin. This antibiotic (left)
resembles the 3¿-terminus of tyrosyl–tRNA (right).
Figure 32-49 X-ray structure of the E. coli EF-Tu EF-Ts
complex. Domains 1, 2, and 3 of EF-Tu are magenta, green, and
cyan, respectively, and EF-Ts is orange. [Based on an X-ray
structure by Stephen Cusack and Reuben Leberman, EMBL,
Grenoble Cedex, France. PDBid 1EFU.]
CH
3
CH
3
N
N
N
N
CH
2
HH
HH
OH
O
O
H
HN
OC
CH
CH
2
O
N
H
3
C
NH
3
+
Puromycin
N
N
N
N
CH
2
tRNA
HH
HH
O OH
O
CH
2
NH
2
NH
3
+
OC
CH
OH
Tyrosyl–tRNA
O
P
O
O
–
O
JWCL281_c32_1338-1428.qxd 8/4/10 4:45 PM Page 1382
...
O
O
–
O
–
O
C
C
CH
O
H
H
C
O
H
H
A
O OH
OH
CH
NH
NH CH
NH
CH
2
CH
2
O
O
H
H
H
H
A
O OH
CH
2
OCH
3
CH
2
(a) (b)
tRNA (P site) tRNA (A site)
O
O
P
O
H
H
C
H
H
A
O
O
H
NH CH
CH
2
O
HO
H
H
H
H
N
N
N
N
N
NH OH
CH
2
CC
R
n
R
n–1
Peptidyl transferase
tetrahedral intermediate
H
3
C
CH
3
CCdA-p-Puro
catalyzes fMet–puromycin formation. These observations
demonstrated the functional existence of the ribosomal P
and A sites and established that binds directly
to the P site, whereas other aminoacyl–tRNAs must first en-
ter the A site.
g. Transpeptidation: The Ribosome Is a Ribozyme
What is the nature of the peptidyl transferase center,
that is, does it consist of RNA, protein, or both? Since all
proteins, including those associated with ribosomes, are ri-
bosomally synthesized, the primordial ribosome must have
preceded the primordial proteins and hence consisted
entirely of RNA. Despite this (in hindsight) obvious evolu-
tionary argument, the idea that rRNA functions catalyti-
cally was not seriously entertained until after it had been
discovered that RNA can, in fact, act as a catalyst (Section
31-4Ae). Several other observations further indicate that
the ribosome is a ribozyme:
1. The absence from the 50S subunit of any one of its
proteins but L2, L3, and L4 does not abolish its peptidyl
transferase function.
2. rRNAs are more highly conserved throughout evolu-
tion than are ribosomal proteins.
3. Most mutations that confer resistance to antibiotics
that inhibit protein synthesis occur in genes encoding
rRNAs rather than ribosomal proteins.
Nevertheless, the unambiguous demonstration that rRNA
functions catalytically in polypeptide synthesis proved to be
fMet–tRNA
Met
f
surprisingly elusive. Noller succeeded in showing that the T.
thermophilus large ribosomal subunit from which ⬃95% of
the protein had been removed by treatment with SDS and
proteinase K followed by phenol extraction (which dena-
tures proteins; Section 6-6A) maintained 80% of its pep-
tidyl transferase activity in a model reaction. Moreover, this
activity was abolished by RNase treatment. However, since
the remaining protein was due to several intact ribosomal
proteins (which are presumably sequestered within the 23S
RNA), it could be argued that these proteins are essential
for ribosomal catalytic function,a reasonable expectation in
light of the 3.5 billion years over which ribosomal proteins
and RNAs have coevolved.
Steitz and Moore unequivocally determined the nature
of the peptidyl transferase center through its identification
in the X-ray structure of the 50S subunit. Peptide bond for-
mation presumably resembles the reverse of peptide bond
hydrolysis such as that catalyzed by serine proteases (Sec-
tion 15-3C). The ribosomal reaction’s tetrahedral interme-
diate (Fig. 32-51a) is mimicked by a compound synthesized
by Michael Yarus that consists of the trinucleotide CCdA
linked to puromycin via a phosphoramidite group (Fig. 32-
51b).This compound, which is named CCdA-p-Puro, binds
tightly to the ribosome so as to inhibit its peptidyl trans-
ferse activity. The X-ray structure of the 50S subunit in
complex with CCdA-p-Puro reveals that the inhibitor
binds to domain V of the 23S RNA (Fig. 32-27b) at the en-
trance to the ⬃100-Å-long polypeptide exit tunnel that
runs through to the back of the subunit (Figs. 32-30 and 32-
34c). There, the inhibitor is completely enveloped in RNA
Section 32-3. Ribosomes and Polypeptide Synthesis 1383
Figure 32-51 Ribosomal tetrahedral intermediate and its
analog. (a) The chemical structure of the tetrahedral intermediate
(red C) in ribosomally mediated peptide bond formation in
which the A-site aminoacyl residue is Tyr. (b) CCdA-p-Puro, the
transition state analog of the tetrahedral intermediate in Part a
produced by linking the 3¿-OH group of CCdA to the amino
group of puromycin’s O-methyltyrosine residue via a phosphoryl
group.
JWCL281_c32_1338-1428.qxd 8/4/10 4:45 PM Page 1383
with no protein side chain approaching closer than ⬃18 Å to
the inhibitor’s phosphoramidite group and with the nearest
Mg
2
ion 8.5 Å away—both too far away to participate in
catalysis. Moreover, all the nucleotides that contact the
CCdA-p-Puro are 95% conserved among all three king-
doms of life. Clearly, the ribosomal transpeptidase reaction
is catalyzed by RNA.
Despite the foregoing, the X-ray structure of the T.
thermophilus ribosome in complex with tRNA and
mRNA (Fig. 32-34) reveals that the N-terminal tail of L27
interacts with the phosphate group of residue C75 of the
A-site tRNA. Moreover, L16 interacts with the elbow re-
gion of the A-site tRNA via interactions between con-
served Arg residues and phosphate and 2¿-OH groups in
the elbow region of the tRNA. These observations con-
firm previous mutational studies indicating that L16 and
L27 help stabilize the binding of the A-site tRNA. How-
ever, some organisms lack L27, which indicates that its
binding to A-site tRNA is not part of an evolutionarily
conserved mechanism.
h. Peptide Bond Formation Does Not Occur via
Acid–Base Catalysis
The ribosomal peptidyl transferase reaction occurs
⬃10
7
-fold faster than the uncatalyzed reaction. How does
the ribosome catalyze this reaction? Peptide bond forma-
tion is naively expected to proceed via the nucleophilic at-
tack of the amino group on the carbonyl group of an ester
to form a tetrahedral intermediate that collapses to an
amide and an alcohol (Fig. 32-39). However, in the physio-
logical pH range, the attacking amino group is predomi-
nantly in its ammonium form (RNH
3
), and hence lacks
the lone pair necessary to undertake a nucleophilic attack.
This suggests that the peptidyl transferase reaction is cat-
alyzed in part by a general base that abstracts a proton
from the ammonium group to generate the required free
amino group (RNH
2
).
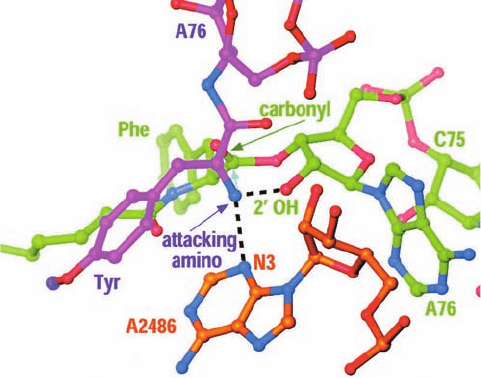
Inspection of the peptidyl transferase center in H.
marismortui reveals that the only basic group within 5 Å of
the inferred position of the attacking amino group is atom
N3 of the invariant rRNA base A2486 (A2451 in E. coli). It
is ⬃3 Å from and hence hydrogen bonded to the attacking
amino group (Fig.32-52).This further suggests that the pro-
tonated A2486-N3 electrostatically stabilizes the oxyanion
of the tetrahedral reaction intermediate and then donates
the proton to the leaving group of the P-site tRNA to yield
a 3¿-OH group (general acid catalysis). However, in order
for A2486-N3 to act as a general base in abstracting the
proton from an ammonium group (whose pK is ⬃10), it
must have a pK of at least 7 (recall that proton transfers be-
tween hydrogen-bonded groups occur at physiologically
significant rates only when the pK of the proton donor is
no more than 2 or 3 pH units greater than that of the pro-
ton acceptor; Section 15-3Dd).Yet, the pK of N3 in AMP is
3.5. Moreover, several lines of evidence indicate that
A2486 does not function as an acid–base catalyst including
(1) the model displayed in Fig. 32-52 indicates that the
tetrahedral intermediate’s oxyanion would point away
from and hence could not be stabilized by protonated
A2486-N3; and (2) mutagenic replacement of A2486 by
any other base does not greatly alter the ribosome’s cat-
alytic rate.
i. The Ribosome Is an Entropy Trap
If the peptidyl transfer reaction does not involve
acid–base catalysis, what is the origin of the ribosome’s
catalytic power? Marina Rodnina and Richard Wolfenden
noted that the uncatalyzed reaction of esters with amines
to form amides occurs quite facilely in aqueous solution.
They therefore measured the rates of both uncatalyzed
peptide bond formation by model compounds and pep-
tidyl transfer by the ribosome at several different temper-
atures. This provided values of and , the re-
action’s change in the enthalpy and entropy of activation
by the ribosome relative to the uncatalyzed reaction. Here
¢¢H
‡
cat
T ¢¢S
‡
cat
¢¢G
‡
cat
¢G
‡
cat
(uncat) ¢G
‡
(cat),
¢¢S
‡
cat
¢¢H
‡
cat
1384 Chapter 32. Translation
Figure 32-52 Model of the substrate complex of the 50S
ribosomal subunit. Atoms are colored according to type with the
A-site substrate C and P purple, P-site substrate C and P green,
23S rRNA C and P orange, N blue, and O red.The attacking
amino group of the A-site aminoacyl residue is held in position
for nucleophilic attack (cyan arrow) on the carbonyl C of the
P-site aminoacyl ester through hydrogen bonds (dashed black
lines) to A2486-N3 and the 2¿-O of the P-site A76. [Courtesy of
Peter Moore and Thomas Steitz, Yale University.]
where is the change in the reaction’s free energy ¢¢G
‡
cat
of activation by the ribosome, G
‡
(cat), relative to that of
the uncatalyzed reaction, G
‡
(uncat) (Section 14-1C).
The measured value of is 19 kJ mol
1
, a quan-
tity that would be positive, not negative, if the ribosomal
reaction had a significant component of chemical catalysis
such as acid–base catalysis and/or the formation of new
hydrogen bonds. In contrast, the value of is 52 kJ
mol
1
, which indicates that the Michaelis complex in the ri-
bosomal reaction is significantly more ordered relative to
the transition state than is the uncatalyzed reaction. This
value of largely accounts for the observed ⬃10
7
-
fold rate enhancement of the ribosomal reaction relative to
T ¢¢S
‡
cat
T ¢¢S
‡
cat
¢¢H
‡
cat
JWCL281_c32_1338-1428.qxd 8/4/10 4:45 PM Page 1384
the uncatalyzed reaction (the rate enhancement by the ri-
bosome is given by ; Section 14-1Cd). Evidently, the
ribosome enhances the rate of peptide bond formation by
properly positioning and orienting its substrates and/or ex-
cluding water from the preorganized electrostatic environ-
ment of the active site (a form of reactant ordering) rather
than by chemical catalysis.
The X-ray structures of the large ribosomal subunit in
complex with aminoacyl–tRNA and peptidyl–tRNA indi-
cate that the ribosome uses an induced fit mechanism, as
occurs in enzymes such as hexokinase (Section 17-2Aa).
Conformational changes in the 23S rRNA, presumably
triggered by proper binding of the aminoacyl–tRNA in the
A site, orient the ester group of the peptidyl–tRNA for nu-
cleophilic attack. The hydrogen bond between the 2¿-OH
of the P-site A76 and the attacking amino group (Fig. 32-
52) is crucial in doing so. In fact, replacing this 2¿-OH group
with H or F reduces the reaction rate by at least 10
6
. This
suggests that the peptidyl transferase reaction occurs via
the substrate-assisted proton shuttle mechanism dia-
grammed in Fig. 32-53. Although the peptidyl transferase
reaction is relatively sluggish compared to the reactions
catalyzed by many protein enzymes, it is sufficiently fast to
keep up with the other ribosomal processes (which collec-
tively link together 10–20 residues/s).Apparently, the ribo-
some’s peptidyl transferase function is a molecular fossil
from the RNA world.
In the absence of a tRNA in the A site, the ester bond
linking the peptidyl group to the P-site tRNA is shielded by
U2585 of the 23S RNA from nucleophilic attack by water,
which would otherwise release the peptidyl group from the
ribosome.
e
¢¢G
‡
cat
>RT
j. Translocation: The Ribosome Moves to the
Next Codon
In the translocation stage of the elongation cycle, the now
uncharged P-site tRNA (at first but subsequently an
elongator tRNA) is transferred to the E site (not shown in
Fig. 32-46), its former occupant having been previously ex-
pelled (see below). Simultaneously, the peptidyl–tRNA in the
A site, together with its bound mRNA, is moved to the P site.
This prepares the ribosome for the next elongation cycle.
The maintenance of the peptidyl–tRNA’s codon–anticodon
association is no longer necessary for amino acid specifica-
tion. Rather, it acts as a place-keeper that permits the ribo-
some to precisely step off the three nucleotides along the
mRNA required to preserve the reading frame. Indeed, the
observation that frameshift suppressor tRNAs induce a
four-nucleotide translocation (Section 32-2Ea) indicates
that mRNA movement is directly coupled to tRNA move-
ment.An Mg
2
-stabilized kink in the mRNA between the A
and P codons apparently helps prevent slippage.
k. EF-G Structurally Mimics the
EF-Ts tRNA Complex
The translocation process requires the participation of
elongation factor EF-G (also called EF2), which binds to
the ribosome together with GTP and is only released on
hydrolysis of the GTP to GDP P
i
(Fig. 32-46). EF-G re-
lease is a prerequisite for beginning the next elongation cy-
cle because EF-G and EF-Tu bind to the same site of the ri-
bosome and hence their binding is mutually exclusive.
The X-ray structure of T. thermophilus EF-G GMPPNP,
determined by Anders Liljas and Derek Logan, reveals a
tadpole-shaped monomeric protein that consists of five
tRNA
Met
f
Section 32-3. Ribosomes and Polypeptide Synthesis 1385
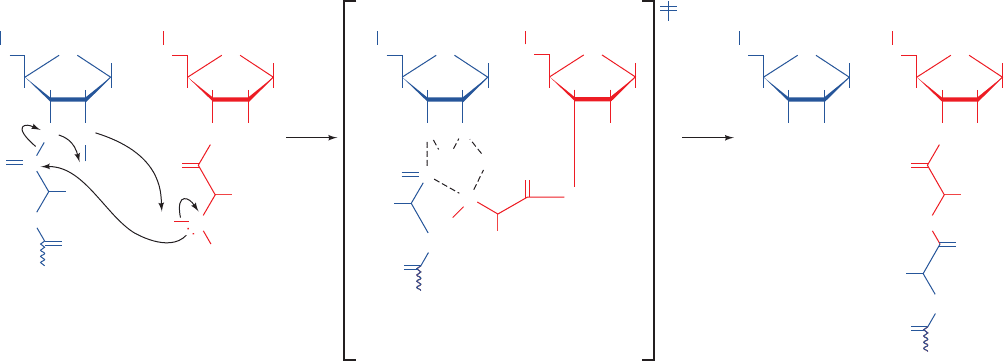
Figure 32-53 The mechanism of ribosome-catalyzed peptidyl
transfer. The nucleophilic attack of the -amino group of the
aminoacyl–tRNA (red) on the carbonyl C of the peptidyl–tRNA
(blue) occurs in concert with a proton shuttle that involves the
O3¿ and 2¿-OH of the P-site A76 together with the -amino
A siteP siteA siteP siteA siteP site
HH
HH
OOH
O
A
O
R
n+1
R
n
HN
NH
HH
HH
OH
O
O
A
tRNA
OH
O
tRNA
O
O
HH
HH
O
O
O
A
tRNA
HH
HH
OOH
O
O
A
tRNA
O
O
R
n+1
R
n
N
H
H
C
O
O
HN
H
HH
HH
O
O
O
A
tRNA
O
O
R
n+1
R
n
C
O
H
H
N
H
O
NH
HH
HH
O
OH
O
O
A
tRNA
group of the aminoacyl–tRNA.The reaction proceeds through a
transition state (center; enclosed by square brackets) that contains
a six-membered ring of partially bonded atoms and which
collapses to the reaction products drawn on the right.
JWCL281_c32_1338-1428.qxd 9/7/10 2:23 PM Page 1385
domains (Fig. 32-54).The first two domains closely resemble
those in EF-Tu GMPPNP rather than those in EF-Tu
GDP (Fig. 32-48). This, it is argued, is because the two elon-
gation factors have reciprocal functions with EF-Tu GTP
facilitating the conversion of the ribosome from its post- to
its pre-translocational state and EF-G GTP promoting the
reverse transition. This idea is supported by the intriguing
observation that the Phe–tRNA
Phe
EF-Tu GMPPNP and
EF-G GMPPNP complexes are almost identical in appear-
ance: EF-G’s three C-terminal domains (yellow through red
in Fig. 32-54), which have no counterparts in EF-Tu, closely
resemble the EF-Tu–bound tRNA in shape, a remarkable
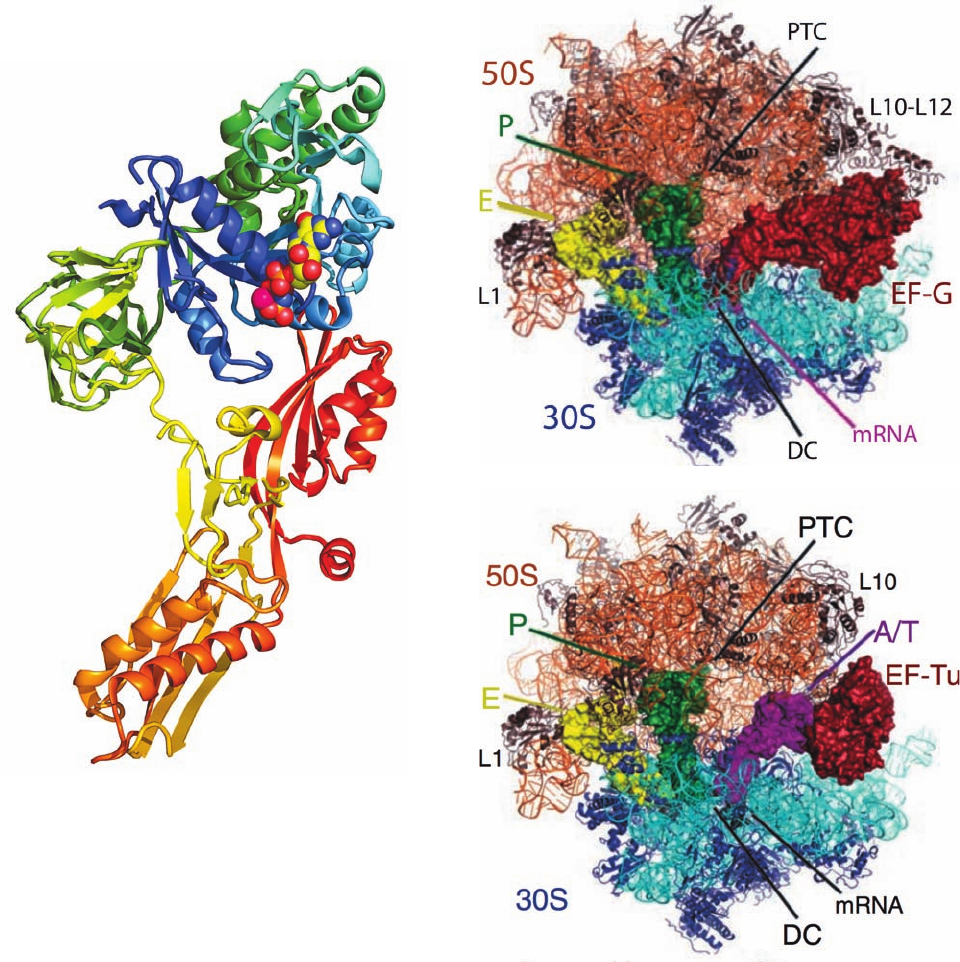
case of macromolecular mimicry. Indeed, EF-G occupies the
same ribosomal site as does EF-Tu (Fig. 32-55).
EF-G is unusual among G-proteins in that it has no cor-
responding GEF. However, its N-terminal guanine nu-
cleotide–binding domain contains a unique helical insert
(green in Fig. 32-54) that contacts the domain’s conserved
core at sites analogous to those in EF-Tu that interact with
EF-Ts. This suggests that this subdomain acts as an inter-
nal GEF.
1386 Chapter 32. Translation
Figure 32-55 X-ray structure of the T. thermophilus ribosome
in its complexes with (a) EF-G and (b) EF-Tu. The ribosome is
drawn in ribbon form with its 23S RNA orange, its 50S subunit
proteins brown, its 16S RNA cyan, and its 30S proteins blue. Its
bound tRNAs, mRNA, EF-G, and EF-Tu are represented by
their surface diagrams with E-site tRNA yellow, P-site tRNA
green,A-site tRNA magenta, mRNA black, and both EF-G and
EF-Tu red. In Part b, the A-site tRNA bound to EF-Tu is in the
A/T conformation (see below).The positions of the decoding
center (DC) and the peptidyl transferase center (PTC) are
indicated. [Courtesy of Martin Schmeing and Venki
Ramakrishnan, MRC Laboratory of Molecular Biology,
Cambridge, U.K. PDBids 2WRI, 2WRJ, 2WRN, and 2WRO.]
Figure 32-54 X-ray structure of EF-G from T. thermophilus in
complex with GMPPNP. The protein is drawn in ribbon form
colored in rainbow order from its N-terminus (blue) to its
C-terminus (red).The GMPPNP is drawn in space-filling form
colored according to atom type with C yellow, N blue, O red, and
P orange. An Mg
2
ion that is bound to the GMPPNP is
represented by a magenta sphere. Portions of the structure are
not visible. Note the remarkable resemblance in shape between
this structure and that of Phe–tRNA
Phe
EF-Tu GMPPNP (Fig.
32-47). [Based on an X-ray structure by Anders Liljas and Derek
Logan, Lund University, Lund, Sweden. PDBid 2BV3.]
(a)
(b)
JWCL281_c32_1338-1428.qxd 9/7/10 2:23 PM Page 1386
E
Transpeptidation
Accommodation
= tRNA = amino acid
residue
Site:
50S
30S
Binding state:
P
Post-translocational
state
1
3
A
E/E P/P
OH
EPA
A/AP/P
EP A
A/AP/P
OH
EPA
A/PP/E
P/P A/T
5
aa-tRNA
•
EF-Tu
•
GTP
EF-Tu
•
tRNA
EF-G
•
GTP
EF-G
•
GDP+ P
i
EF-G
2
4
EF-Tu
•
GDP + P
i
Pre-translocational
state
OH
OH
GTP
l. Translocation Occurs via Intermediate States
Chemical footprinting studies (Section 31-2Ab) by
Noller revealed that certain bases in the 16S rRNA are
protected by tRNAs bound in the ribosomal A and P sites
and that certain bases in the 23S rRNA are protected by
tRNAs in the A, P, and E sites.Almost all of these protected
bases are absolutely conserved in evolution and many of
them have been implicated in ribosomal function through
biochemical or genetic studies.
Variations in chemical footprinting patterns during the
elongation cycle together with the more recently determined
X-ray and cryo-EM structures indicate that the translocation
of tRNA occurs in several discrete steps (Fig. 32-56):
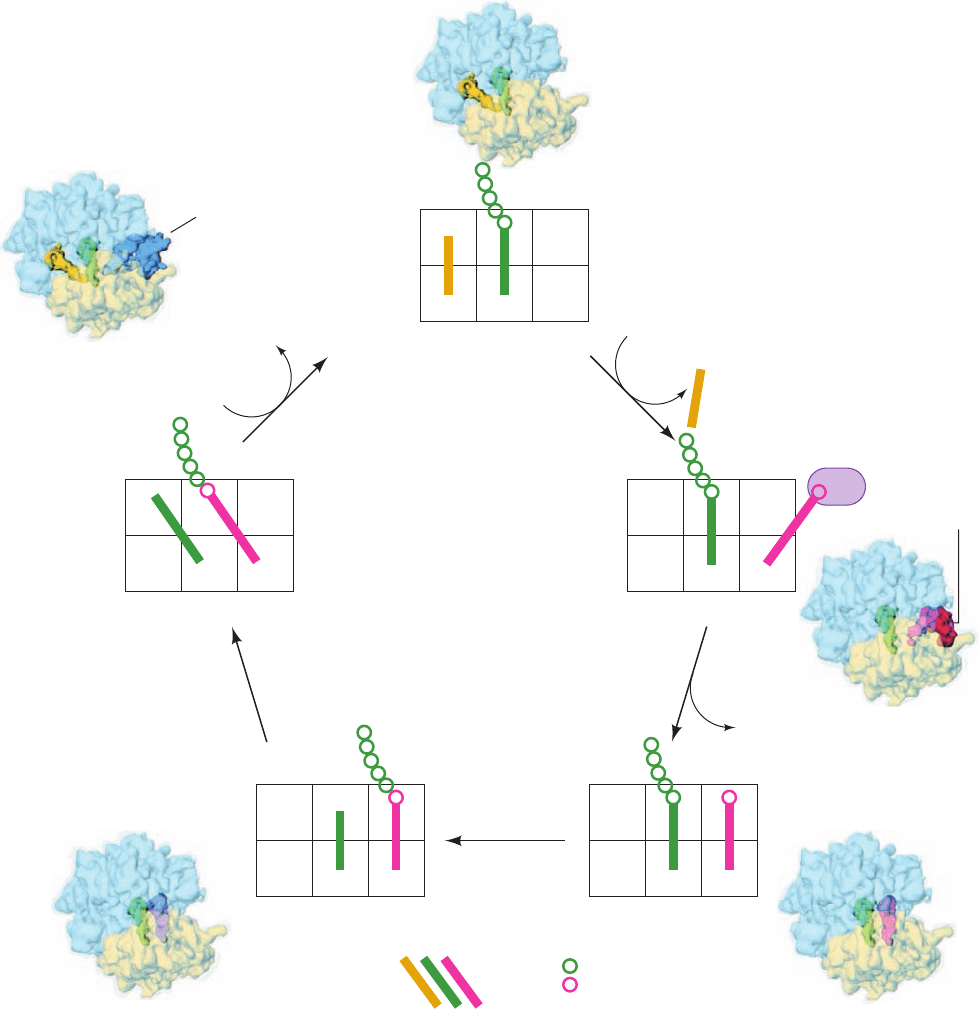
1. Let us start with the ribosome in its post-transloca-
tional state: a deacylated tRNA bound to the E subsites of
both the 30S and 50S subunits (the E/E binding state), a
peptidyl–tRNA bound in the P subsites of both subunits
Section 32-3. Ribosomes and Polypeptide Synthesis 1387
Figure 32-56 Ribosomal binding states in the elongation cycle.
Note how this scheme elaborates the classic elongation cycle
diagrammed in Fig. 32-46. The drawings are accompanied by
17-Å-resolution cryo-EM–based images of the E. coli 70S
ribosome in the corresponding binding states in which the 30S
subunit is transparent yellow, the 50S subunit is transparent blue,
and the tRNAs and elongation factors are colored as in the
drawing they accompany. [Cryo-EM images courtesy of Knud
Nierhaus, Max-Planck-Institut für Molekulare Genetik, Berlin,
Germany, and Joachim Frank,Wadsworth Center, State
University of New York at Albany.]
JWCL281_c32_1338-1428.qxd 8/19/10 10:06 PM Page 1387
(the P/P state), and the A site empty. An aminoacyl–tRNA
(aa–tRNA) in ternary complex with EF-Tu and GTP binds
to the A site accompanied by the release of the E-site
tRNA (but see below). This yields a complex in which the
incoming aa–tRNA is bound in the 30S subunit’s A subsite
via a codon–anticodon interaction (recall that the mRNA
is bound to the 30S subunit) but with the EF-Tu preventing
the entry of the tRNA’s aminoacyl end into the 50S
subunit’s A subsite, an arrangement termed the A/T state
(T for EF-Tu).
2. EF-Tu hydrolyzes its bound GTP to GDP P
i
and is
released from the ribosome. This permits the aa–tRNA to
fully bind to the A site (the A/A state), a process called ac-
commodation in which the 3¿ end of the tRNA swings
around by nearly 70 Å.
3. The peptidyl transferase reaction occurs, yielding the
pre-translocational state.
4. The acceptor end of the new peptidyl–tRNA shifts
from the A subsite of the 50S subunit to its P-subsite, while
the tRNA’s anticodon end remains associated with the A
subsite of the 30S subunit (yielding the A/P hybrid binding
state). The acceptor end of the newly deacylated tRNA si-
multaneously moves from the P subsite to the E subsite of
the 50S subunit while its anticodon end remains associated
with the P subsite of the 30S subunit (the P/E state).
5. The ribosomal binding of the EF-G GTP complex
and the subsequent GTP hydrolysis impel the anticodon
ends of these tRNAs, together with their bound mRNA, to
move relative to the small ribosomal subunit such that the
peptidyl–tRNA assumes the P/P state and the deacylated
tRNA assumes the E/E state (the post-translocational
state), thereby completing the elongation cycle.
The binding of tRNA to the A and E sites, as Nierhaus
has shown, exhibits negative allosteric cooperativity. In the
pre-translocational state, the E site binds the newly deacy-
lated tRNA with high affinity (the E site is sterically unable
to bind an aminoacyl–tRNA), whereas the empty A site
has low affinity for aminoacyl–tRNA. However, in the
post-translocational state, the ribosome has undergone a
conformational change that converts the A site to a high-
affinity state and the E site to a low-affinity state, which
consequently releases the deacylated tRNA when
aa–tRNA EF-Tu GTP binds to the A site.Thus, the E site
is not simply a passive holding site for spent tRNAs but
performs an essential function in the translation process.
The GTP hydrolysis by the elongation factors EF-Tu and
EF-G as well as the peptidyl transferase reaction appar-
ently function to reduce the activation barriers between
these conformational states.The unidirectional A S P S E
flow of tRNAs through the ribosome is thereby facilitated.
Certain aspects of the foregoing mechanism are not
fully resolved. For example, X-ray studies of the 70S ribo-
some in complex with three tRNAs (e.g., Fig. 32-34) sug-
gest that the E-site tRNA is not released from the ribo-
some until Step 2 of Fig. 32-56. However, Nierhaus and
Frank argue that these complexes were crystallized in the
presence of unphysiologically high tRNA concentrations.
Whatever the case, it is clear that the changes in binding
states result in large-scale tRNA movements, in some in-
stances 50 Å. Moreover, cryo-EM studies indicate that on
binding EF-G GDP(CH
2
)P (like GMPPNP but with a
CH
2
group rather than an NH group bridging its and
phosphates), the 30S subunit rotates with respect to the
50S subunit by 6° clockwise when viewed from the 30S sub-
unit’s solvent side, which results in a maximum displace-
ment of ⬃19 Å at the periphery of the ribosome.This rota-
tion is accompanied by many smaller conformational
changes in both subunits, particularly in the regions about
the entrance and exit to the mRNA channel. Clearly, we
are far from fully understanding how the ribosome works
at the molecular level.
m. The Eukaryotic Elongation Cycle Resembles That
of Prokaryotes
The eukaryotic elongation cycle closely resembles that of
prokaryotes. In eukaryotes, the functions of EF-Tu and EF-
Ts are respectively assumed by the eukaryotic elongation
factors eEF1A and eEF1B, with yeast eEF1B consisting of
two subunits: eEF1B, which catalyzes nucleotide ex-
change, and eEF1B, which has unknown function (in
higher eukaryotes, eEF1B contains a third subunit,
eEF1B, that possesses a nucleotide exchange activity sim-
ilar to that of eEF1B). Likewise, eEF2 functions in a man-
ner analogous to EF-G. However, the corresponding eu-
karyotic and prokaryotic elongation factors are not
interchangeable.
The X-ray structure of yeast eEF1A eEF1B (Fig. 32-57),
determined by Kjeldgaard and Nyborg, reveals that
eEF1A structurally resembles the homologous EF-Tu (Fig.
32-49), whereas eEF1B exhibits no resemblance to EF-
Ts, either in sequence or in structure. Nevertheless,
eEF1B functionally interacts with eEF1A much as EF-Ts
interacts with EF-Tu: Both GEFs associate with their cor-
responding G-protein so as to disrupt the Mg
2
binding site
associated with its bound guanine nucleotide.
E. Translational Accuracy
The genetic code is normally expressed with remarkable fi-
delity. We have already seen that transcription and tRNA
aminoacylation both proceed with high accuracy (Sections
31-2Ec and 32-2Ce).The accuracy of ribosomal mRNA de-
coding was estimated from the rate of misincorporation of
[
35
S]Cys into highly purified flagellin, an E. coli protein
(Section 35-3I) that normally lacks Cys. These measure-
ments indicated that the mistranslation rate is ⬃10
4
errors
per codon. This rate is greatly increased in the presence of
streptomycin, an antibiotic that increases the rate of ribo-
somal misreading (Section 32-3Ga). From the types of
reading errors that streptomycin is known to induce, it was
deduced that the mistranslation arose almost entirely from
the confusion of the Arg codons CGU and CGC for the
Cys codons UGU and UGC.The above error rate is there-
fore largely caused by mistakes in ribosomal decoding.
An aminoacyl–tRNA is selected by the ribosome only
according to its anticodon. Yet the binding energy loss
1388 Chapter 32. Translation
JWCL281_c32_1338-1428.qxd 8/19/10 10:06 PM Page 1388

arising from a single base mismatch in a codon–anticodon in-
teraction is estimated to be ⬃12 kJ mol
1
, which, accord-
ing to Eq. [32.1], cannot account for a ribosomal decoding
accuracy of less than ⬃10
2
errors per codon. Moreover,
the base pairing interaction between the UUU codon for
Phe and the GAA anticodon of tRNA
Phe
would be naively
expected to be less stable than the incorrect pairing be-
tween the UGC codon for Ser and the GCG anticodon of
tRNA
Arg
. This is because both interactions have one G U
base pair and the former correct interaction’s remaining
two A U base pairs are weaker than the latter incorrect
interaction’s remaining two G C base pairs. Evidently, the
ribosome has some sort of proofreading mechanism that
increases its overall decoding accuracy.
a. The Ribosome Monitors the Formation of a
Correct Codon–Anticodon Complex
As we have seen (Figs. 32-55b and 32-56), the amino-
acyl–tRNA EF-Tu GTP ternary complex initially binds to
the ribosome with the tRNA in the A/T binding state.The
tRNA only assumes the fully bound A/A state (accommo-
dation) after the GTP has been hydrolyzed and the EF-Tu
GDP complex has been released from the ribosome.These
two states presumably permit the ribosome to double-
check (proofread) the codon–anticodon complex that the
mRNA makes with the incoming tRNA.
The X-ray structure of the T. thermophilus 30S subunit
in complex with a U
6
hexanucleotide mRNA and a 17-nt
RNA consisting of the tRNA
Phe
anticodon stem–loop
(Fig. 32-11, although its nucleotides are unmodified), de-
termined by Ramakrishnan, revealed how an mRNA-
specified tRNA initially binds to the ribosome. The
codon–anticodon association is stabilized by its interac-
tions with three universally conserved ribosomal bases,
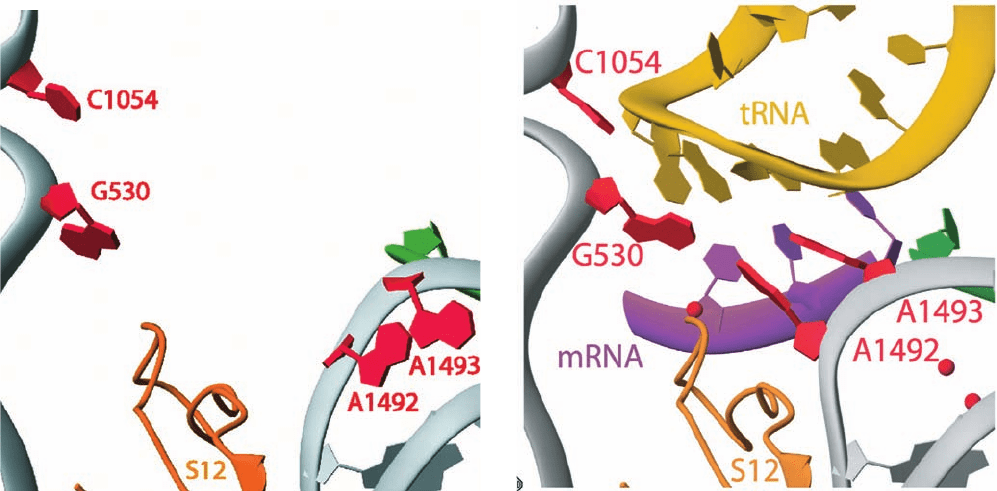
A1492, A1493, and G530 (Fig. 32-58):
1. The first codon–anticodon base pair, that between
mRNA U1 and tRNA A36, is stabilized by the binding of
the rRNA A1493 base in the base pair’s minor groove (Fig.
32-58a).
2. The second codon–anticodon base pair, that between
U2 and A35, is bolstered by A1492 and G530, which both
bind in this base pair’s minor groove (Fig. 32-58b).
3. The third codon–anticodon base pair (the wobble
pair; Section 32-2Db), that between U3 and G34, is rein-
forced through minor groove binding by G530 (Fig. 32-58c).
Section 32-3. Ribosomes and Polypeptide Synthesis 1389
Figure 32-58 Codon–anticodon interactions in the ribosome.
The (a) first, (b) second, and (c) third codon–anticodon base
pairs as seen in the X-ray structure of the T. thermophilus 30S
subunit in complex with U
6
(a model mRNA) and the 17-nt
anticodon stem–loop of tRNA
Phe
(whose anticodon is GAA).
The structures are drawn in ball-and-stick form embedded in
Figure 32-57 X-ray structure of yeast eEF1A eEF1B␣.
Domains 1, 2, and 3 of eEF1A are magenta, green, and cyan,
respectively, and eEF1B is orange. The complex is oriented so
as to emphasize the structural resemblance between eEF1A and
the similarly colored EF-Tu in its complex with EF-Ts (Fig.
32-49). Note the lack of resemblance between eEF1B and
EF-Ts. [Based on an X-ray structure by Morten Kjeldgaard and
Jens Nyborg, University of Aarhus, Århus, Denmark. PDBid 1F60.]
their semitransparent van der Waals surfaces. Codons are purple,
anticodons are yellow, and rRNA is brown or gray with non-C
atoms colored according to type (N blue, O red, and P green).
Protein C atoms are gray and Mg
2
ions are represented by
magenta spheres. [Courtesy of Venki Ramakrishnan, MRC
Laboratory of Molecular Biology, Cambridge, U.K. PDBid 1IBM.]
(a) (b) (c)
JWCL281_c32_1338-1428.qxd 8/19/10 10:06 PM Page 1389
This latter interaction appears to be less stringent than
those in the first and second codon–anticodon positions,
which is consistent with the need for the third codon–
anticodon pairing to tolerate non-Watson–Crick base pairs
(Section 32-2D).
Comparison of this structure with that of the 30S subunit
alone reveals that the foregoing rRNA nucleotides un-
dergo conformational changes on the formation of a
codon–anticodon complex (Fig. 32-59). In the absence of
tRNA, the bases of A1492 and A1493 stack in the interior
of an RNA loop but flip out of this loop to form the
codon–anticodon complex, whereas the G530 base
switches from the syn to the anti conformation (Section
29-2Aa).These interactions enable the ribosome to mon-
itor whether an incoming tRNA is cognate to the codon
in the A site; a non-Watson–Crick base pair could not
bind these ribosomal bases in the same way. Indeed, any
mutation of A1492 or A1493 is lethal because pyrim-
idines in these positions could not reach far enough to in-
teract with the codon–anticodon complex or G530 and
because a G in either position would be unable to form
the required hydrogen bonds and its N2 would be sub-
jected to steric collisions. An incorrect codon–anticodon
provides insufficient free energy to bind the tRNA to the
ribosome and it therefore dissociates from it, still in its
ternary complex with EF-Tu and GTP.
b.
GTP Hydrolysis by EF-Tu Is a Thermodynamic
Prerequisite to Ribosomal Proofreading
A proofreading step must be entirely independent of the
initial selection step. Only then can the overall probability
of error be equal to the product of the probabilities of er-
ror of the individual selection steps. We have seen that
DNA polymerases and aminoacyl–tRNA synthetases
maintain the independence of their two selection steps by
carrying them out at separate active sites (Sections 30-2Ac
and 32-2Ce). Yet the ribosome only recognizes the incom-
ing aminoacyl–tRNA according to its anticodon’s comple-
mentarity to the codon in the A site. Consequently, the ri-
bosome must somehow examine this codon–anticodon
interaction in two separate ways.
The formation of a correct codon–anticodon complex
induces EF-Tu to hydrolyze its bound GTP, although how
this occurs is unclear (note that EF-Tu’s GTPase domain is
bound in the 50S subunit which, together with the observa-
tion that GTP hydrolysis requires an intact tRNA, suggests
that the hydrolysis signal is at least in part transmitted
through the tRNA). The resulting conformational change
in EF-Tu (Fig. 32-48) swings its bound tRNA into the A/A
state (accommodation), a process that moves the 3¿ end of
the tRNA by nearly 70 Å. This, it is hypothesized, subjects
the codon–anticodon interaction to a strain that only a cor-
rect pairing can withstand. The codon–anticodon interac-
tion is thereby subjected to a second screening that only
1390 Chapter 32. Translation
Figure 32-59 Ribosomal decoding site. The X-ray structures
of T. thermophilus 30S subunit (a) alone and (b) in its complex
with U
6
and the 17-nt anticodon stem–loop of tRNA
Phe
.The
RNAs are drawn as ribbons with their nucleotides in paddle
form with tRNA gold, A-site mRNA purple, rRNA gray, and
(a) (b)
nucleotides that undergo conformational changes red. Protein
S12 is tan and Mg
2
ions are represented by red spheres. Compare
Part b with Fig. 32-58. [Courtesy of Venki Ramakrishnan, MRC
Laboratory of Molecular Biology, Cambridge, U.K. PDBids
(a) 1FJF and (b) 1IBM.]
JWCL281_c32_1338-1428.qxd 8/19/10 10:06 PM Page 1390
