Voet D., Voet Ju.G. Biochemistry
Подождите немного. Документ загружается.

(b)
as adapters that position the reactive E2–ubiquitin
thioester bond for the direct transfer of the ubiquitin to
the substrate protein Lys side chain.
In order for a target protein to be efficiently degraded, it
must be linked to a chain of at least four tandemly linked
ubiquitin molecules in which Lys 48 of each ubiquitin forms
an isopeptide bond with the C-terminal carboxyl group of
the succeeding ubiquitin (Fig. 32-78). These polyubiquitin
(polyUb) chains, which can reach lengths of 50 or more
ubiquitin molecules, are generated by the E3s, although
how they switch from transferring a ubiquitin to the target
protein to processively synthesizing a polyubiquitin chain
is unknown.
c. Ubiquitinated Proteins Are Hydrolyzed
in the Proteasome
A ubiquitinated protein is proteolytically degraded to
short peptides in an ATP-dependent process mediated by a
large (2000 kD, 26S) multisubunit protein complex named
the 26S proteasome (sometimes spelled “proteosome”) that
electron micrographic studies reveal has the shape of a bi-
capped hollow barrel (Fig. 32-79). Proteolysis occurs inside
the barrel, which permits this process to be extensive and
processive, while preventing nonspecific proteolytic dam-
age to other cellular components. PolyUb chains are the
signals that target a protein to the proteasome; the identity
of the target protein has little effect on the efficiency with
which it is degraded by the proteasome. Nevertheless, the
proteasome does not degrade ubiquitin molecules; they are
returned to the cell. The size and functional complexity of
this entire proteolytic system, which occurs in the nucleus
as well as the cytosol, rivals that of the ribosome (Section
32-3) and the spliceosome (Section 31-4A) and hence is
Section 32-6. Protein Degradation 1411
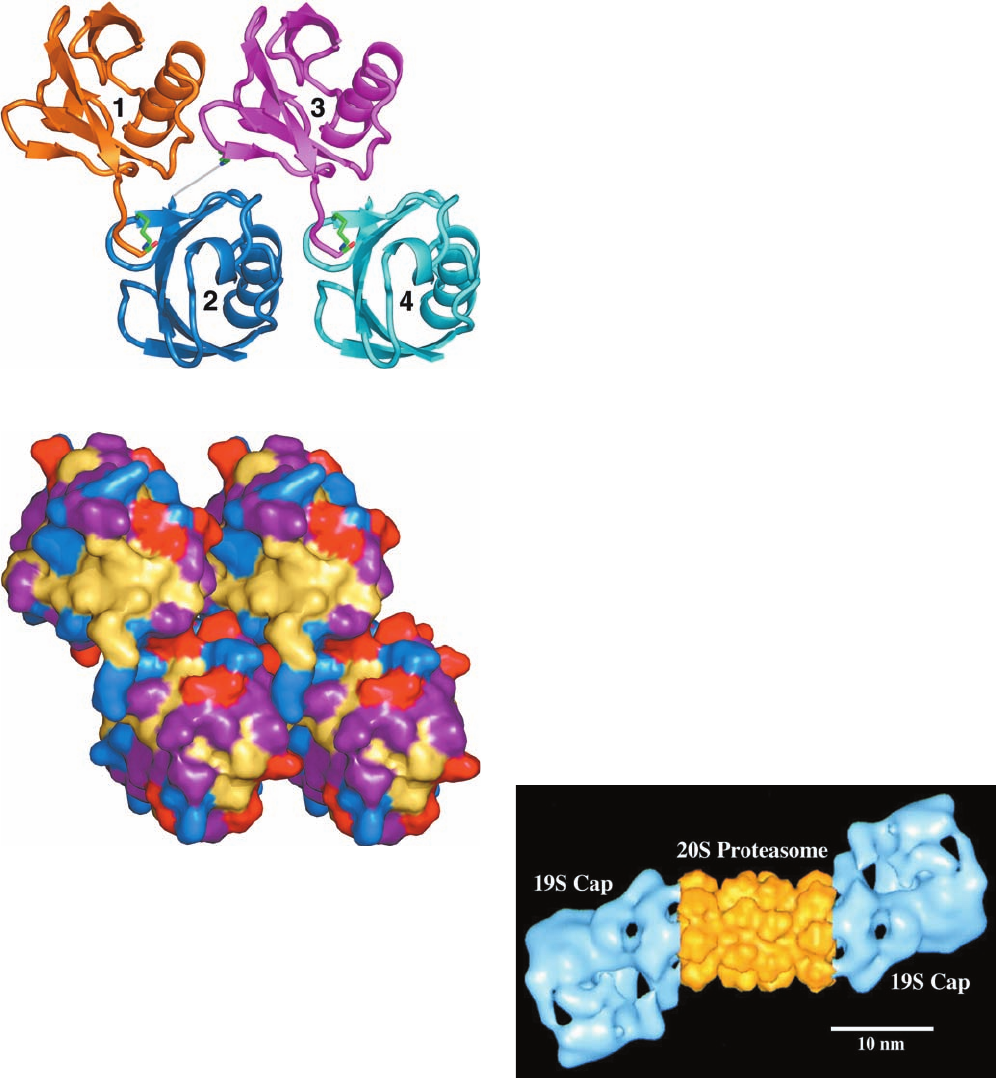
Figure 32-78 X-ray structure of human tetraubiquitin. (a) A
ribbon drawing in which the isopeptide bonds connecting
successive ubiquitin molecules, together with the Lys side chains
making them, are drawn in stick form with C green, N blue, and
O red. However, since the C-terminal three residues of ubiquitin
2 are disordered, the isopeptide bond connecting it to ubiquitin 3
is represented by a thin gray bond (this isopeptide bond
nevertheless exists, as was demonstrated by SDS–PAGE of
dissolved crystals).The monomer units in a multiubiquitin chain
of any length are likely to be arranged with the repeating
symmetry of the tetraubiquitin structure, although the weak
interactions between adjacent ubiquitin units suggests that this
chain is conformationally flexible. (b) A surface diagram, viewed
as in Part a, in which basic residues (Arg, Lys, His) are blue,
acidic residues (Asp, Glu) are red, uncharged polar residues
(Gly, Ser, Thr, Asn, Gln) are purple, and hydrophobic residues
(Ile, Leu,Val,Ala, Met, Phe,Tyr, Pro) are tan (ubiquitin lacks Cys
and Trp residues). Note the unusually large solvent-exposed
surface occupied by the hydrophobic residues. [Based on an
X-ray structure by William Cook, University of Alabama at
Birmingham, and Cecile Pickart, Johns Hopkins University.
PDBid 1TBE.]
Figure 32-79 Electron microscopy–based image of the
Drosophila melanogaster 26S proteasome. The complex is
around 450 190 Å.The central portion of this 2-fold symmetric
multiprotein complex (yellow), the 20S proteasome, consists of
four stacked 7-membered rings of subunits that form a hollow
barrel in which the proteolysis of ubiquitin-linked proteins
occurs.The 19S caps (cyan), which may attach to one or both
ends of the 20S proteasome, control the access of condemned
proteins to the 20S proteasome (see below). [Courtesy of
Wolfgang Baumeister, Max Planck Institute of Biochemistry,
Martinsreid, Germany.]
(a)
JWCL281_c32_1338-1428.qxd 9/7/10 2:23 PM Page 1411
indicative of the importance of properly managing protein
degradation. Indeed, ⬃5% of the proteins expressed by
yeast participate in protein degradation. We discuss the
structure and function of the 26S proteasome below.
d. Many E3s Have Elaborate Modular Structures
The proto-oncogene product c-Cbl (906 residues) is a
single-subunit, RING domain–containing E3 that functions
to ubiquitinate certain activated receptor tyrosine kinases
(RTKs; Section 19-3A), thereby terminating their signaling.
Nikola Pavletich determined the X-ray structure of the N-
terminal half of c-Cbl (residues 47–447) in its ternary com-
plex with the E2 protein UbcH7 (which consists of little
more than the ⬃150-residue E2 catalytic core) and a 9-
residue peptide containing the ubiquitination signal from a
nonreceptor tyrosine kinase (NRTK) named ZAP-70
(Fig. 19-44). The structure (Fig. 32-80) reveals that UbcH7
and c-Cbl’s RING domain and SH2-containing tyrosine ki-
nase–binding (TKB) domains interact with one another
across multiple interfaces to form a compact and apparently
rigid structure.The RING domain consists of a 3-stranded
sheet, an helix, and two large loops that are held together
by two tetrahedrally coordinated Zn
2
ions. UbcH7 adopts
the characteristic / fold of other E2s of known structure
(e.g., Fig. 32-77).The ZAP-70 peptide is bound on the oppo-
site side of the TKB domain from the UbcH7 active site Cys
residue (Cys 86) and is ⬃60 Å distant from it.
SCF complexes are multisubunit RING E3s that consist
of Cul1 (a member of the cullin family; 776 residues), Rbx1
(which contains the complex’s RING domain; 108
residues), Skp1 (163 residues), and a member of the F-box
protein family (⬃430 to 1000 residues; SCF for
Skp1–cullin–F-box protein). Rbx1 and Cul1 form the com-
plex’s catalytic core that binds E2; F-box proteins consist
of an ⬃40-residue F-box that binds Skp1 followed by
protein–protein interaction modules such as leucine-rich
repeats (LRRs) or WD40 repeats (Section 19-2C) that bind
substrate protein; and Skp1 functions as an adapter that
links the F-box to Cul1. Cells contain numerous different
F-box proteins (at least 38 in humans) that presumably
permit the specific ubiquitination of a diverse variety of
protein substrates (see below).
Pavletich has also determined the X-ray structures of
two segments of the SCF
Skp2
complex (where the super-
script identifies the complex’s F-box protein, here Skp2,
436 residues). The structure of the Skp1–Skp2 complex
(Fig. 32-81) reveals that it has the shape of a sickle with the
Skp1 and the 3-helix F-box of Skp2 forming the handle and
its 10 LRRs (⬃26 residues each) forming the curved blade.
The structure of the Cul1–Rbx1–Skp1–F-box
Skp2
quater-
nary complex (Fig. 32-82) shows that Cul1 is an elongated
protein that consists of a long stalk formed by three repeats
of a novel five-helix motif known as a cullin repeat fol-
lowed by a globular domain that binds Rbx1. Apparently
Cul1 acts like a rigid scaffold that organizes the Skp1–F-
box
Skp2
complex and Rbx1 so as to hold them over 100 Å
1412 Chapter 32. Translation
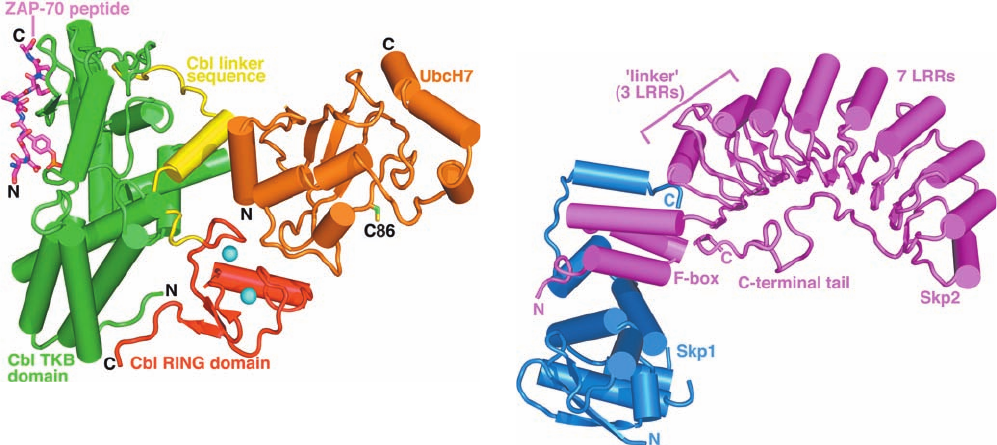
Figure 32-80 X-ray structure of the human c-Cbl–UbcH7–ZAP-
70 peptide ternary complex drawn in tube-and-arrow form.
UbcH7, an E2 that consists almost entirely of the E2 catalytic
core, is colored orange with the side chain of its active site Cys 86
shown in stick form with C green and S yellow. c-Cbl (residues
47–447 of the 903-residue protein), a monomeric RING E3, is
colored according to domain with its TKB domain green, its
RING domain red, and the linker joining them yellow. The
RING domain’s two bound Zn
2
ions are represented by cyan
spheres.The 9-residue ubiquitination site of the RTK ZAP-70,
whose fourth residue is phospho-Tyr, is drawn in stick form with
C magenta, N blue, and O red, and P orange. [Based on an X-ray
structure by Nikola Pavletich, Memorial Sloan-Kettering Cancer
Center, New York, New York. PDBid 1FBV.]
Figure 32-81 X-ray structure of the human Skp1–Skp2
complex. Skp1 and Skp2 are drawn in tube-and-arrow form in
blue and magenta. Skp2 consists of an N-terminal F-box that
forms three helices, followed by 3 noncanonical so-called linker
leucine-rich repeats (LRRs) that are contiguous with 7 LRRs
that were predicted from their amino acid sequences for a total
of 10 LRRs.After the tenth LRR, Skp2’s ⬃30-residue C-terminal
tail extends back past the first LRR by packing under the con-
cave surface of the LRR domain. [Based on an X-ray structure
by Nikola Pavletich, Memorial Sloan-Kettering Cancer Center,
New York, New York. PDBid 1FQV.]
JWCL281_c32_1338-1428.qxd 9/7/10 2:23 PM Page 1412

apart.The Rbx1 RING domain contains a 20-residue insert
that forms the binding site for a third tetrahedrally lig-
anded Zn
2
ion.
The apparent rigidity of the foregoing three structures
has enabled Pavletich to construct a model of the intact
SCF
Skp2
–E2 complex by superimposing Skp1–Skp2 on
Cul1–Rbx1–Skp1–F-box
Skp2
and docking the E2 UbcH7
onto the Rbx1 RING domain based on the c-Cbl–UbcH7
structure (Fig. 32-83). The model indicates that E2 and the
LRR-containing domain of Skp2 are on the same side of
the SCF complex but separated by a distance of ⬃50 Å.
This suggests that Cul1’s long stalk functions to separate
the complex’s substrate-binding and catalytic sites so that
substrates with different sizes and various distances be-
tween their ubiquitinated Lys residues and their ubiquiti-
nation signals can be accommodated.
e. The Ubiquitin System Has Both Housekeeping
and Regulatory Functions
Until the mid-1990s, it appeared that the ubiquitin sys-
tem functioned mainly in a “housekeeping” capacity to
maintain the proper balance among metabolic proteins
and to eliminate damaged proteins. Indeed, as Alexander
Varshavsky discovered, the half-lives of many cytoplasmic
proteins vary with the identities of their N-terminal residues
(Table 32-12). Thus, in a selection of 208 cytoplasmic pro-
teins known to be long lived, all have a “stabilizing”
residue, Met, Ser, Ala, Thr, Val, or Gly, at their N-termini.
This so-called N-end rule is true for both eukaryotes and
prokaryotes, which suggests the system that selects pro-
teins for degradation is conserved in eukaryotes and
prokaryotes, even though prokaryotes lack ubiquitin. The
N-end rule results from the actions of the single-subunit,
RING E3 named E3 (⬃1950 residues; also known as
Ubr1) whose ubiquitination signals are the destabilizing
N-terminal residues in Table 32-12.
Similarly, it has long been known that proteins with seg-
ments rich in Pro (P), Glu (E), Ser (S), and Thr (T), the so-
called PEST proteins, are rapidly degraded.This is because
these PEST elements often contain phosphorylation sites
that target their proteins for ubiquitination.
Section 32-6. Protein Degradation 1413
Table 32-12 Half-Lives of Cytoplasmic Enzymes as a
Function of Their N-Terminal Residues
N-Terminal Residue Half-Life
Stabilizing
Met 20 h
Ser
Ala
Thr
Va l
Gly
Destabilizing
Ile ⬃30 min
Glu
Ty r ⬃10 min
Gln
Highly Destabilizing
Phe ⬃3 min
Leu
Asp
Lys
Arg ⬃2 min
Source: Bachmair,A., Finley, D., and Varshavsky,A., Science 234,
180 (1986).
Figure 32-82 X-ray structure of the human
Cul1–Rbx1–Skp1–F-box
Skp2
quaternary complex. Cul1, Rbx1,
Skp1, and the Skp2 F-box are drawn in tube-and-arrow form and
respectively colored green, red, blue, and magenta.The three
cullin repeats of Cul1 are indicated.The three Zn
2
ions bound
to Rbx1 are represented by cyan spheres. [Based on an X-ray
structure by Nikola Pavletich, Memorial Sloan-Kettering Cancer
Center, New York, New York. PDBid 1LDK.]
Figure 32-83 Model of the SCF
Skp2
–E2 complex. This model,
which is based on the X-ray structures in Figs. 32-80, 32-81, and
32-82, is colored and viewed as in Fig. 32-82. E2 is yellow with its
active site Cys residue, to which ubiquitin would be covalently
linked, drawn in space-filling form in cyan.The Zn
2
ions
associated with the Rbx1 RING domain are represented by
yellow spheres.The gray arrow indicates the 50-Å gap between
the tip of the Skp2 LRR domain and the E2 active site. [Courtesy
of Nikola Pavletich, Memorial Sloan-Kettering Cancer Center,
New York, New York.]
JWCL281_c32_1338-1428.qxd 9/7/10 2:23 PM Page 1413
It is now clear, however, that the ubiquitin system is far
more sophisticated than a simple garbage disposal system.
Thus, the known E3s each respond to certain ubiquitina-
tion signals that often occur on a quite limited range of
target proteins, many of which have regulatory functions.
For example, the ubiquitination system has an essential
function in cell cycle progression. The cell cycle, as we have
seen in Section 30-4Aa and will further discuss in Section
34-4D, is regulated by a series of proteins known as cyclins.
A given cyclin, which is expressed immediately preceding
and/or during a specific phase of the cell cycle, binds to a
corresponding cyclin-dependent protein kinase (Cdk),
which then phosphorylates its target proteins so as to acti-
vate them to carry out the processes of that phase of the
cell cycle. Moreover, many cyclins also inhibit the transi-
tion to the subsequent phase of the cell cycle (e.g., DNA
replication or mitosis). Consequently, for a cell to progress
from one phase of the cell cycle to the next, the cyclin(s)
governing that phase must be eliminated. This occurs via
the specific ubiquitination of the cyclin, thereby condemn-
ing it to be destroyed by the proteasome.The E3s respon-
sible for this process are the SCF complexes containing
F-box proteins targeted to a corresponding cyclin and a
multisubunit complex known as the anaphase-promoting
complex (APC; alternatively the cyclosome; Section 34-
4Da).APC, an ⬃1500-kD RING domain–containing par-
ticle that in yeast consists of 11 subunits, specifically
ubiquitinates proteins that contain the 9-residue consen-
sus sequence RTALGDIGN, the so-called destruction
box, near their N-termini.
The transcription factor NF-B, which plays a central
role in immune and inflammatory responses (Section 34-
3Bs), is maintained in an inactive state in the cytosol
through its binding to the inhibitor IB in a way that oc-
cludes the short internal basic sequence that directs NF-
B’s import into the nucleus (its nuclear localization signal;
NLS). However, the stimulation of cell-surface receptors
by proinflammatory cytokines such as tumor necrosis
factor- (TNF; Section 19-3Db) and interleukin-1 (IL-1;
Section 19-3Eb) initiate a signal transduction pathway
(Section 19-3D) that phosphorylates IB bound to NF-
B at both Ser residues in the sequence DSGLDS. This
phosphorylated sequence is the ubiquitination signal for
the SCF complex containing the F-box protein -TrCP
(605 residues), which mediates the ubiquitination of the
phosphorylated IB.The consequent destruction of IB
exposes the NLS of NF-B, which is then translocated to
the nucleus where it activates the transcription of its target
genes (Section 34-3Bs).
Some viruses usurp the ubiquitin system. Oncogenic
forms of human papillomavirus (HPV), the cause of
nearly all cervical cancers (a leading cause of death of
women in developing countries), encode the ⬃150-residue
E6 protein, which combines with the 875-residue cellular
protein named E6-associated protein (E6AP; the first E3
known to contain a HECT domain) to ubiquitinate p53,
thereby marking it for destruction. This latter protein is a
transcription factor that monitors genome integrity and
hence is important in preventing malignant transforma-
tion and the proliferation of cancer cells (Section 34-4Ca),
that is, it is a tumor suppressor (a protein whose loss of
function is a cause of cancer). Consequently, HPV pro-
vokes the uncontrolled growth of the cells it infects and
hence its own proliferation. E6AP normally functions to
ubiquitinate certain members of the Src family of protein
tyrosine kinases (Section 19-3Ba), including Src itself. The
deletion of the segment of chromosome 15 that contains
the E6AP gene causes Angelman syndrome, which as we
have seen (Section 30-7d) is characterized by severe men-
tal retardation and is exclusively maternally inherited due
to genomic imprinting.
The foregoing are only a few examples of the numerous
cellular processes that are regulated by the ubiquitin-medi-
ated proteolysis system. Not surprisingly, therefore, many
pathological conditions in humans, including inflammatory,
neurodegenerative, and muscle-wasting diseases, are at-
tributable to malfunctioning ubiquitination systems.
f. The 20S Proteasome Catalyzes Proteolysis Inside
a Hollow Barrel
The 26S proteasome (Fig. 32-79) is an ⬃2100-kD multi-
subunit protein that catalyzes the ATP-dependent hydroly-
sis of ubiquitin-linked proteins. This yields oligopeptides
with lengths of 4 to 25 residues and averaging 7 to 9
residues that are subsequently degraded to their compo-
nent amino acids by cytosolic exopeptidases. The 26S pro-
teasome consists of a 20S proteasome (⬃670 kD), the bar-
rel-shaped catalytic core of the 26S proteasome, and its 19S
caps (⬃700 kD; also known as PA700 and the 19S regula-
tor), which associate with the ends of the 20S proteasome
and stimulate its activity (PA for proteasome activator).
The 20S proteasome only hydrolyzes unfolded proteins in
an ATP-independent manner; the 19S caps function to
identify and unfold the ubiquitinated protein substrates.
The 20S proteasome occurs in the nuclei and cytosol of
all eukaryotic cells and in all archaebacteria yet examined.
However,the only eubacteria in which it occurs are those of
the class Actinobacteria, which suggests that they obtained
it via horizontal gene transfer from some other organism.
The 20S proteasome of Thermoplasma acidophilum (an
archaebacterium) consists of 14 copies each of and sub-
units (233 and 203 residues) that electron microscopy stud-
ies revealed form a 150-Å long and 110-Å-diameter barrel
in which the subunits are arranged in four stacked rings (as
is evident in the central portion of the 26S proteasome seen
in Fig. 32-79).The and subunits are 26% identical in se-
quence except for an ⬃35-residue N-terminal tail of the
subunit, which the subunit lacks. Eukaryotic 20S protea-
somes are more complex in that they consist of 7 different
-like and 7 different -like subunits versus only one of
each type for the T. acidophilum 20S proteasome.
The X-ray structure of the T. acidophilum 20S protea-
some, determined by Baumeister and Robert Huber, re-
veals that its two inner rings each consist of 7 subunits
and its two outer rings each consist of 7 subunits, all
arranged with D
7
symmetry (Fig. 32-84). Thus the overall
1414 Chapter 32. Translation
JWCL281_c32_1338-1428.qxd 8/4/10 4:45 PM Page 1414
structure of the 20S proteasome superficially resembles
that of the unrelated molecular chaperone GroEL (Section
9-2Ca). The structures of the and subunits are remark-
ably similar (Fig. 32-84a) except, of course, for the sub-
unit’s N-terminal tail (blue in Fig. 32-84a), which extends
radially inward to contact the N-terminal tail of a neigh-
boring subunit. This accounts for the observation that
subunits alone spontaneously assemble into 7-membered
rings (a capacity that is abolished by the deletion of their
N-terminal 35 residues), whereas subunits alone remain
monomeric.
The central cavity of the T. acidophilum 20S proteasome
consists of three large chambers (Fig.32-84c): Two are located
at the interfaces between adjoining rings of and subunits,
with the third, larger chamber centrally located between the
two rings of subunits. Unfolded polypeptide substrates en-
ter the central chamber of the barrel (where the protea-
some’s active sites are located; see below) through ⬃13-Å-
diameter axially located apertures in the rings that are
lined with hydrophobic residues. This allows only unfolded
proteins to enter the central chamber, thereby protecting
properly folded proteins from indiscriminant degradation
by this omnivorous protein-dismantling machine.
The X-ray structure of the yeast 20S proteasome, deter-
mined by Huber, demonstrates that its outer and inner
rings respectively consist of seven different -type subunits
and seven different -type subunits, all of which are
uniquely arranged (Fig. 32-85). The -like subunits have
folds that are similar to one another as well as to that of the
T. acidophilum 20S proteasome and likewise for the -like
subunits. Consequently, this 28-subunit, 6182-residue pro-
tein complex has exact 2-fold rotational symmetry relating
its two pairs of rings but only pseudo-7-fold rotational sym-
metry relating the subunits within each ring. The narrow
axial apertures in the rings through which unfolded
polypeptides enter the hydrolytic chamber (Fig. 32-85d)
are occluded in the closed state (Fig. 32-85c) by a plug
formed by the interdigitation of its subunits’ N-terminal
tails.This indicates that the 19S caps of the 26S proteasome,
which have been shown to activate the 20S proteasome,
Section 32-6. Protein Degradation 1415
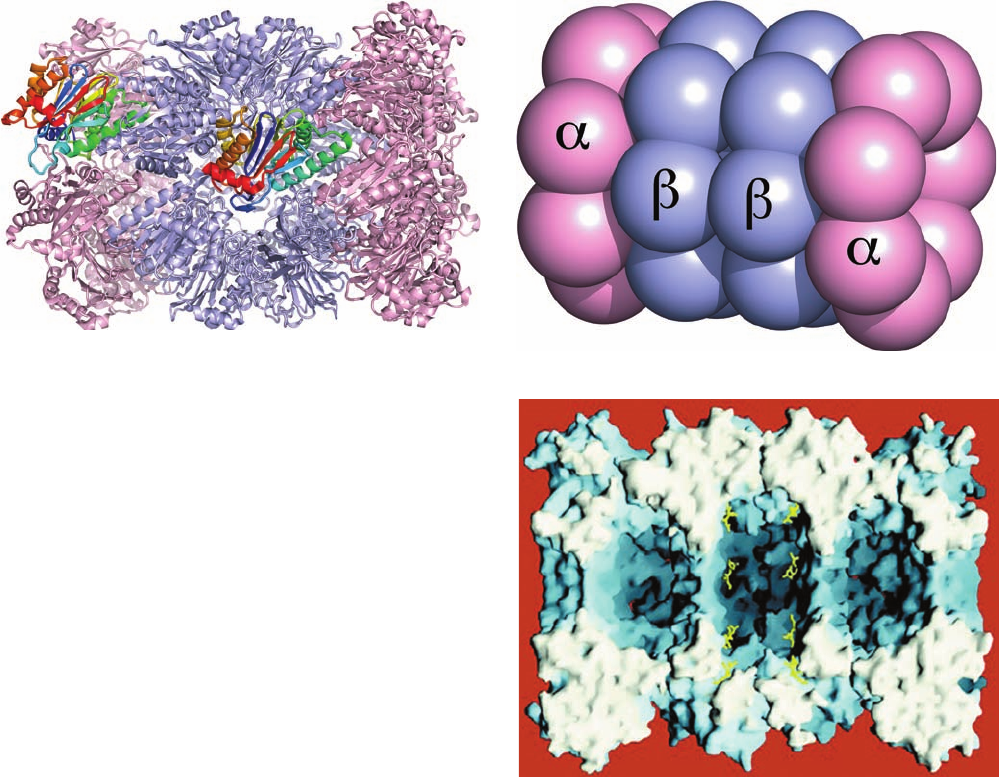
Figure 32-84 X-ray structure of the T. acidophilum 20S
proteasome. (a) Ribbon diagram, viewed with its 7-fold axis
tipped toward the viewer by 20°, in which the subunits are pink
and the subunits are light blue except for one subunit (left)
and a similarly oriented subunit (middle), which are both
colored in rainbow order from their N-termini (blue) to their
C-termini (red). Note their close structural resemblance. (b)
Diagram, viewed as in Part a, in which the subunits are
represented by equal-sized spheres with subunits pink and
subunits light blue. (c) Surface diagram, viewed along a 2-fold
axis, with the subunits nearest the viewer removed to expose the
proteasome’s triple-chambered internal cavity, which is
maximally ⬃100 Å long and ⬃60 Å in diameter.The active sites
on the subunits are marked by the bound inhibitor, LLnL,
which is drawn in stick form in yellow. [Part a based on an X-ray
structure by and Part c courtesy of Robert Huber, Max-Planck-
Institut für Biochemie, Martinsried, Germany. PDBid 1PMA.]
(b)
(c)
(a)
JWCL281_c32_1338-1428.qxd 9/7/10 2:23 PM Page 1415
1416 Chapter 32. Translation
(a)
(c) (d)
(b)
JWCL281_c32_1338-1428.qxd 9/7/10 2:23 PM Page 1416

control the access to it by inducing conformational changes
in its rings (see below).The X-ray structure of the bovine
20S proteasome, determined by Tomitake Tsukihara, re-
veals that its arrangement of seven -type and seven
-type subunits is similar to that in yeast.
g. The Proteasome Catalyzes Peptide Hydrolysis via
a Novel Mechanism
The X-ray structure of the T. acidophilum 20S protea-
some in complex with the aldehyde inhibitor acetyl-Leu-
Leu-norleucinal (LLnL)
reveals that its active sites are on the inner surfaces of its
rings of subunits, with the aldehyde function of the LLnL
close to the side chain of the highly conserved Thr 1.
Deletion of this Thr or its mutation to Ala yields properly
assembled 20S proteasomes that are completely inactive.
Evidently, 20S proteasomes catalyze peptide hydrolysis by
a novel mechanism in which the hydroxyl group of its Thr
1 is the attacking nucleophile. This, as yet, poorly under-
stood mechanism, in which the amino group at the N-
terminus and possibly a bound water molecule act to nucle-
ophilically activate the hydroxyl side chain, is now known
to be employed by other hydrolases (e.g., glutamate syn-
thase; Section 26-5Aa), which are collectively known as the
N-terminal nucleophile (Ntn) family of hydrolases. The T.
acidophilum subunits preferably cleave polypeptides af-
ter hydrophobic residues. However, in the yeast and the
bovine 20S proteasomes, only subunits 1, 2, and 5 are
catalytically active. Their respective preferences for cleav-
age after acidic (caspase-like), basic (trypsin-like), and hy-
drophobic (chymotrypsin-like) residues are explained by
Acetyl-Leu-Leu-norleucinal (LLnL)
CH
3
CH
3
O
O
C Leu Leu NH CH CH
(CH
2
)
3
the respective basic, acidic, and nonpolar characters of
their pockets that bind the side chain of the residue preced-
ing the scissile peptide bond, although this specificity is rel-
atively low. The functions of the four different catalytically
inactive subunits are unknown, although mutagenically
modifying an inactive subunit can abolish the catalytic
activity of an active subunit.
h. The 19S Caps Control the Access of Ubiquitinated
Proteins to the 20S Proteasome
The 20S proteasome probably does not exist alone in
vivo; it is most often in complex with two 19S caps that
function to recognize ubiquitinated proteins, unfold them,
and feed them into the 20S proteasome in an ATP-depend-
ent manner (it may also associate with other regulatory
complexes; see below). The 19S cap, which consists of ⬃18
different subunits, is poorly characterized due in large part
to its low intrinsic stability. Its so-called base complex con-
sists of 9 different subunits, 6 of which are ATPases that
form a ring that abuts the ring of the 20S proteasome
(Fig. 32-79). Each of these ATPases contains an ⬃230-
residue ATPase module that is a member of the AAA
family (Section 30-2Ca). Cecile Pickart demonstrated via
cross-linking experiments that one of these ATPases,
named S6 (alternatively Rpt5), contacts the polyUb signal
that targets a condemned protein to the 26S proteasome.
This suggests that the recognition of the polyUb chain as
well as substrate protein unfolding are ATP-driven
processes. Moreover, the ring of ATPases must function to
open (gate) the otherwise closed axial aperture of the 20S
proteasome so as to permit the entry of the unfolded sub-
strate protein.
Eight additional subunits form the so-called lid com-
plex, the portion of the 19S cap that is more distal to (dis-
tant from) the 20S proteasome. The functions of the lid
subunits are largely unknown, although a truncated 26S
proteasome that lacks the lid subunits is unable to degrade
polyubiquitinated substrates. Several other subunits may
be transiently associated with the 19S cap and/or with the
20S proteasome.
i. Deubiquitinating Enzymes Have Several Functions
The enzymes that hydrolytically cleave the isopeptide
bonds linking successive ubiquitin units in polyUb are
known as deubiquitinating enzymes (DUBs). Cells contain
a surprisingly large number of DUBs (at least 17 in yeast
and ⬃100 in humans). Nearly all known DUBs are cysteine
proteases, enzymes whose catalytic mechanism resembles
that of serine proteases (Section 15-3C) but whose attack-
ing nucleophile is Cys¬S
rather than Ser¬OH.
DUBs may release entire polyUb chains from a con-
demned protein or sequentially release ubiquitin units
from the chain terminus. It has been proposed that this lat-
ter process functions as a clock to time the protein degra-
dation process. If a polyUb chain is trimmed to less than
four ubiquitin units before degradation begins, then its at-
tached protein is likely to escape destruction. This would
spare proteins that had been inappropriately tagged with
only short polyUb chains.
Section 32-6. Protein Degradation 1417
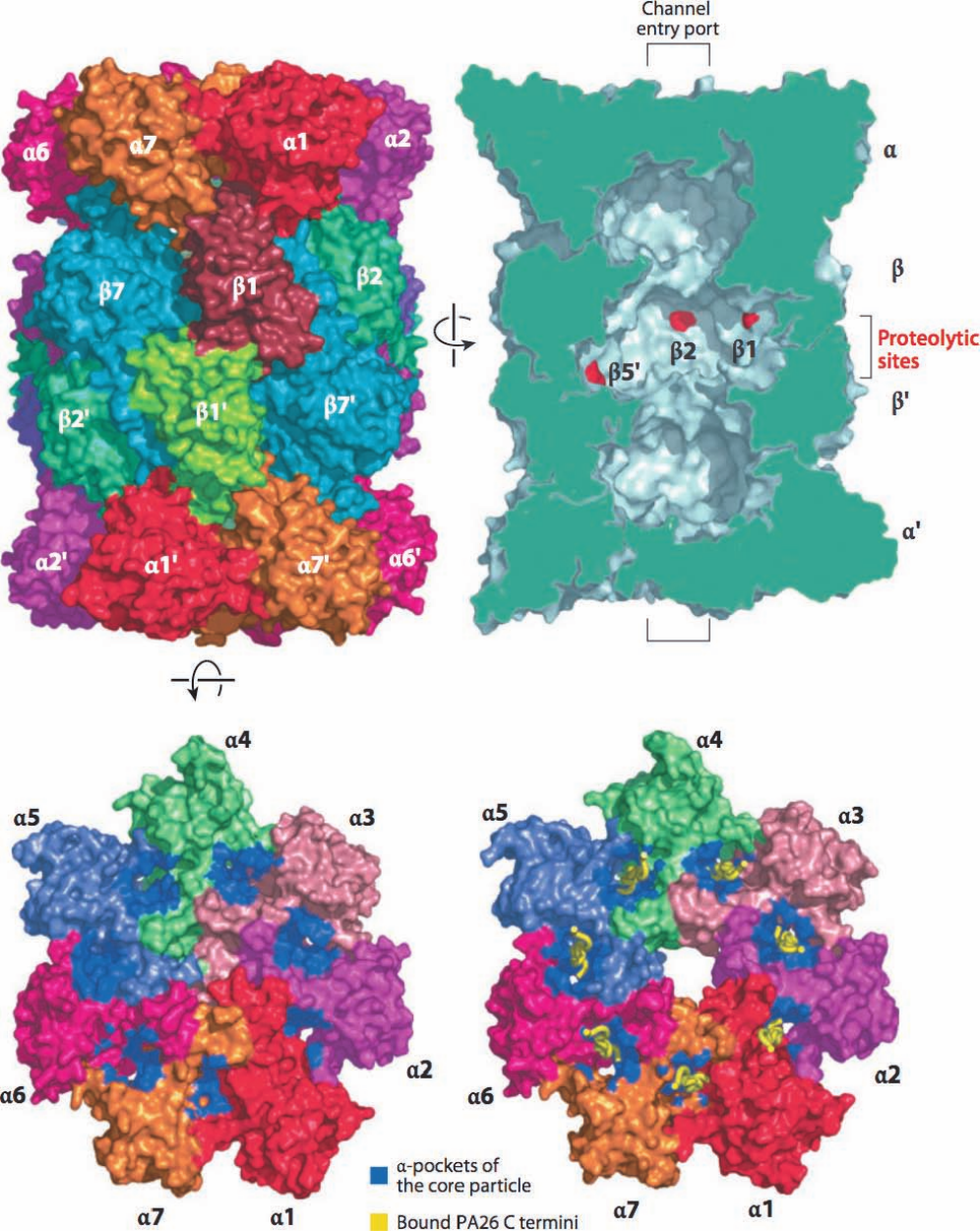
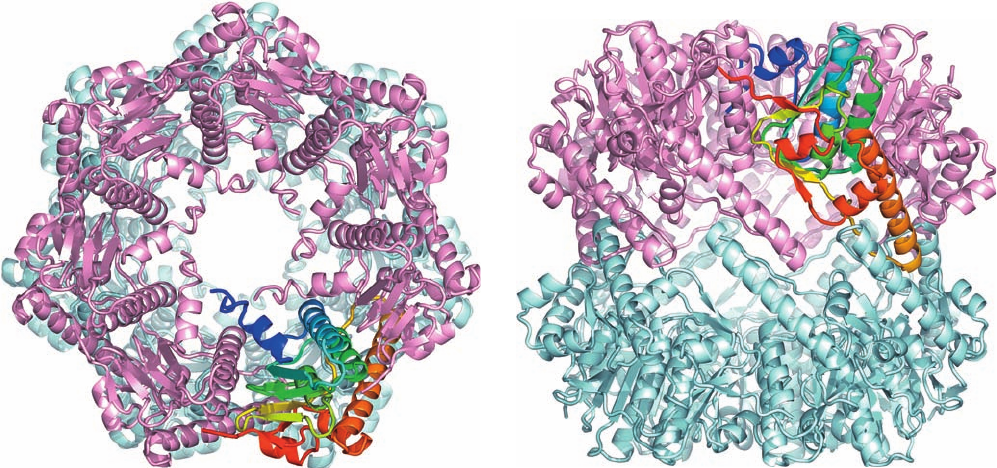
Figure 32-85 (Opposite) X-ray structure of the yeast 20S
proteasome. (a) Surface diagram viewed along the 28-mer’s
2-fold axis. Each pair of identical subunits has the same color
except for 1 and the symmetry related 1¿, which are colored
differently. (b) Cutaway surface diagram sliced along the
complex’s pseudo-7-fold axis. The slice surface is green and the
active sites of the 1, 2, and 5¿ subunits are marked in red. The
brackets indicate the approximate positions of the channel entry
ports as seen in the open state. (c) End view of the 20S
proteasome showing its -ring in its closed state represented as
in Part a.The pockets where PA26 binds (see below) are
highlighted in blue. (d) The open state represented as in Part c
and with the C-termini of its bound PA26 subunits shown in
worm form in yellow. [Courtesy of Daniel Finley, Harvard
Medical School. Based on X-ray structures by Robert Huber,
Max-Planck-Institut für Biochemie, Martinsried, Germany
(closed state), and Christopher Hill, University of Utah (open
state). PDBids 1RYP and 1FNT.]
JWCL281_c32_1338-1428.qxd 8/4/10 4:45 PM Page 1417
(a)
The mammalian 19S lid subunit known as POH1
(Rpn11 for the 65% identical yeast subunit) appears to be
responsible for the deubiquitination of target proteins
prior to their degradation; its inactivation prevents target
protein degradation. Curiously, this DUB is a Zn
2
-de-
pendent protease (as is carboxypeptidase A; Fig. 15-42)
rather than a cysteine protease.
Certain DUBs function to dismember polyUb chains that
have been released from substrate proteins by sequentially
removing ubiquitin units from the end of the chain that is
nearest to the substrate protein (that with a free C-termi-
nus). Consequently, these DUBs cannot remove ubiquitin
units from polyUb chains that are still attached to substrate
proteins, thereby preventing their premature removal.
Cells express ubiquitin as polyproteins containing sev-
eral ubiquitin units (Section 32-5Ac) or with ubiquitin fused
to certain ribosomal subunits (there is no gene that encodes
a single ubiquitin unit). These polyproteins are rapidly
processed by certain DUBs to yield free ubiquitin.
j. The 11S Activator Forms a Heptameric Barrel That
Opens the 20S Proteasome
Higher eukaryotes contain an 11S activator (alterna-
tively, 11S regulator) that functions to open the channel
into the 20S proteasome in an ATP-independent manner
so as to permit the entrance of polypeptides (but not
folded proteins). The mammalian 11S activator, which
functions in the generation of peptides for presentation to
the immune system (Section 35-2E), is named REG (alter-
natively PA28). It is a heteroheptameric complex of two
⬃245-residue subunits, REG and REG, that exhibit
⬃50% sequence identity except for a highly variable inter-
nal 18-residue segment that is thought to confer subunit-
specific properties. Indeed REG alone forms a heptamer
whose biochemical properties are similar to that of REG
(although both subunits must be present in vivo).
The trypanosome Trypanosoma brucei, which lacks 19S
caps, expresses a homoheptameric 11S activator named
PA26 that is only 14% identical to human REG. Neverthe-
less, the various 11S activators activate 20S proteasomes
from widely divergent species.Thus,rat 20S proteasome is ac-
tivated by PA26 and the yeast 20S proteasome is activated by
human REG despite the fact that yeast lacks 11S activators.
1418 Chapter 32. Translation
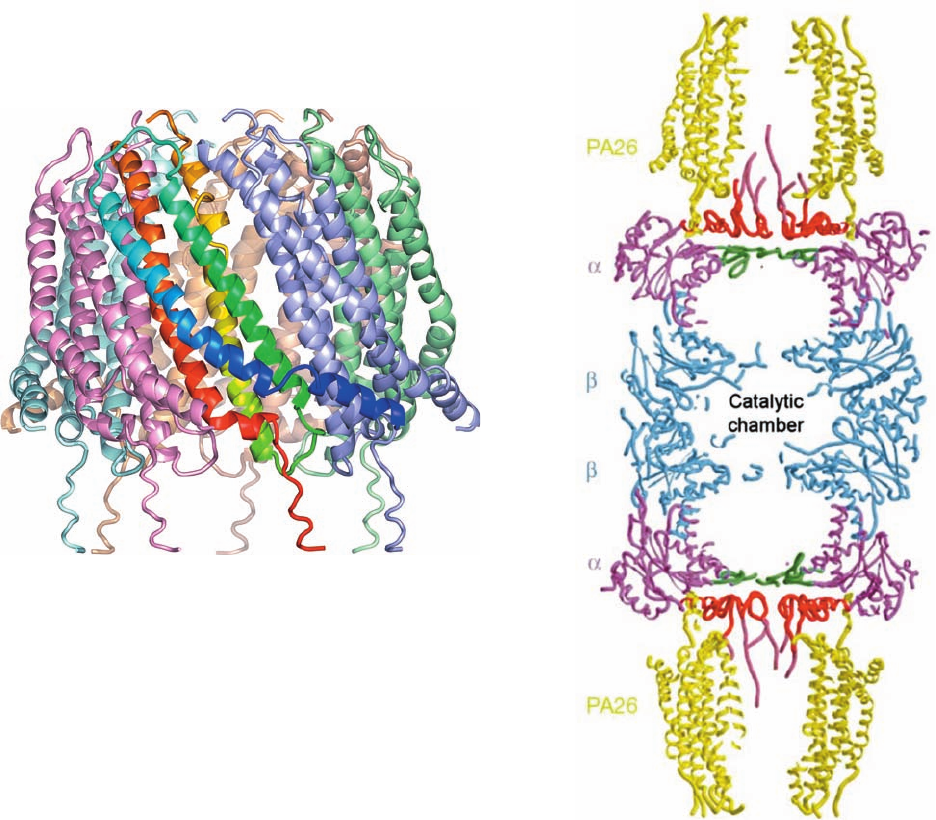
Figure 32-86 X-ray structure of T. brucei PA26 in complex
with the yeast 20S proteasome. (a) The PA26 heptamer in ribbon
form viewed with its 7-fold axis vertical. Each of its subunits are
differently colored with that closest to the viewer colored in
rainbow order from its N-terminus (blue) to its C-terminus (red).
(b) Cutaway diagram of the entire complex drawn in worm form
and viewed with its 7-fold axis vertical.The PA26 is yellow, the
and subunits of the 20S proteasome are magenta and blue, its
-annulus is green, and its N-terminal segments that are ordered
and partially disordered are red and pink. [Part a based on an
X-ray structure by and Part b courtesy of Christopher Hill,
University of Utah. PDBid 1FNT.]
(b)
JWCL281_c32_1338-1428.qxd 8/19/10 10:06 PM Page 1418
The X-ray structure of PA26 in complex with the yeast
20S proteasome, determined by Christopher Hill, reveals
that each PA26 monomer consists of an up–down–up–down
4-helix bundle. These monomers form a 7-fold symmetric
heptameric barrel that is 90 Å in diameter, 70 Å long, and
has a 33-Å-diameter central pore (Fig. 32-86a) and which
closely resembles the previously determined X-ray struc-
ture of human REG.Two PA26 barrels associate coaxially
with the 20S proteasome, one at each end (Fig. 32-86b).The
conformation of the 20S proteasome in this complex, for
the most part, is closely similar to that of the 20S protea-
some alone (Fig. 32-85). However, the C-terminal tails of
the PA26 subunits insert into pockets on the 20S protea-
some’s subunits in a way that induces conformational
changes in its N-terminal tails that clear the 20S protea-
some’s otherwise blocked central aperture (Fig. 32-85c,d),
thus permitting unfolded polypeptides to enter the protea-
some’s central chamber.
k. Bacteria Contain a Variety of Self-
Compartmentalized Proteases
Nearly all eubacteria lack 20S proteasomes. Neverthe-
less they have ATP-dependent proteolytic assemblies that
share the same barrel-shaped architecture and carry out
similar functions. For example, in E. coli, two proteins
known as Lon and Clp mediate up to 80% of the bac-
terium’s protein degradation,with additional contributions
from at least three other proteins including heat shock lo-
cus UV (HslUV). Thus, all cells appear to contain proteases
whose active sites are only available from the inner cavity of
a hollow particle to which access is controlled. These so-
called self-compartmentalized proteases appear to have
arisen early in the history of cellular life, before the advent
of eukaryotic membrane-bound organelles such as the
lysosome, which similarly carry out degradative processes
in a way that protects the cell contents from indiscriminant
destruction.
Clp protease consists of two components, the proteolyti-
cally active ClpP and one of several ATPases, which in E.
coli are ClpA and ClpX. The X-ray structure of ClpP, deter-
mined by John Flanagan, reveals that it oligomerizes to
form an ⬃90-Å-long and -wide hollow barrel that consists
of two back-to-back 7-fold symmetric rings of 193-residue
subunits (Fig.32-87) and thereby has the same D
7
symmetry
as does the 20S proteasome. Nevertheless, the ClpP subunit
has a novel fold that is different from that of the 20S protea-
some’s homologous and subunits. The ClpP active site,
which is only exposed on the inside of the barrel, contains a
catalytic triad composed of Ser 97, His 122, and Asp 171,
and hence is a serine protease (Section 15-3Ab).
HslUV protease appears to be a hybrid of Clp and the
26S proteasome. Its HslV subunits in Haemophilus influen-
zae (174 residues) are 18% identical to the subunits of
the T. acidophilum 20S proteasome, whereas its regulatory
HslU caps (444 residues) have ATPase activity and are ho-
mologous to E. coli ClpX. The X-ray structure of H. in-
fluenzae HslUV, determined by David McKay, indi-
cates that HslV forms a dimer of hexameric rather than
Section 32-6. Protein Degradation 1419
Figure 32-87 X-ray structure of E. coli ClpP. (a) View of the
heptameric complex along its 7-fold axis, drawn in ribbon form in
which the lower ring is pale cyan and the upper ring is pink with
one subunit colored in rainbow order from its N-terminus (blue)
(a)
(b)
to its C-terminus (red). (b) View along the complex’s 2-fold axis
(rotated 90° about a horizontal axis with respect to Part a).
[Based on an X-ray structure by John Flanagan, Brookhaven
National Laboratory, Upton, New York. PDBid 1TYF.]
JWCL281_c32_1338-1428.qxd 8/19/10 10:06 PM Page 1419
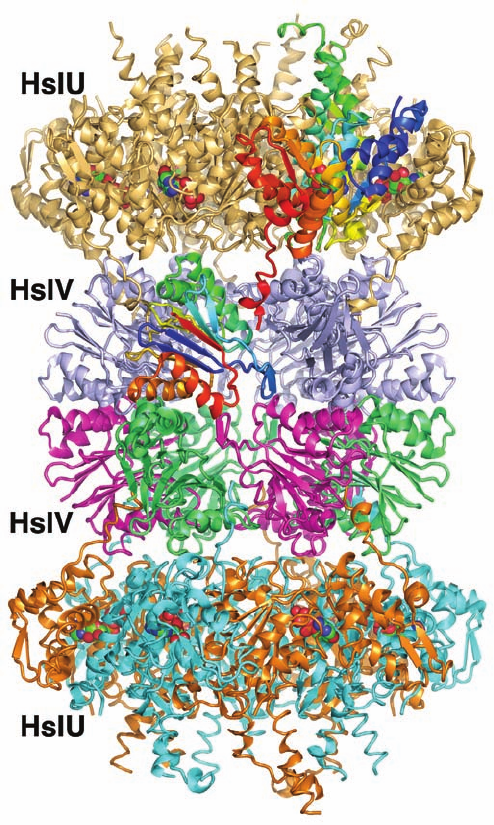
heptameric rings (Fig. 32-88).A hexameric ring of HslU sub-
units binds to both ends of the HslV dodecamer to form a
24-subunit assembly with D
6
symmetry, rather than the D
7
symmetry of the 26S proteasome. Nevertheless, both the
fold and the intersubunit contacts of the HslV subunits are
closely similar to those of the 20S proteasome subunits.
In addition, both have N-terminal Thr residues.Thus, HslV
can be regarded as the eubacterial homolog of archaebac-
terial and eukaryotic 20S proteasomes.
Thermoplasma acidophilum contains another large pro-
teolytic complex, which is unrelated to the proteasome.The
X-ray structure of this protease (Fig. 32-89), determined by
Huber, indicates that it forms a 730-kD toroidal hexameric
ring with D
3
symmetry that has a peculiar triangular shape
reminiscent of a tricorn (a hat whose brim is turned up on
three sides) and hence was named tricorn protease. Cryo-
EM studies indicate that 20 of these tricorn hexamers asso-
ciate to form a 14,600-kD hollow icosahedron (Fig. 32-89c;
an icosahedron is shown in Fig. 8-65c), making it by far the
largest homooligomeric enzyme complex known (it is even
larger than some virus particles, many of which also have
icosahedral symmetry; Section 33-2Aa).
l. Ubiquitination Has Multiple Proteasome-
Independent Functions
Proteins may be monoubiquitinated or polyubiquiti-
nated or even monoubiquitinated on more than one Lys
residue. Moreover, ubiquitin has seven Lys residues so that
seven types of polyubiquitin chains are possible.
Many types of ubiquitination mediate processes other
than directing their associated proteins to the proteasome.
This occurs through mechanisms reminiscent of protein
phosphorylation, but instead of being recognized by spe-
cialized phosphoprotein-binding motifs such as SH2 (Sec-
tion 19-3Cb),ubiquitinated proteins are recognized by con-
served ubiquitin-binding domains (UBDs). Moreover,
whereas protein phosphorylation is reversible through the
action of protein phosphatases (Section 19-3F), ubiquitina-
tion is reversible through the agency of DUBs.
Ubiquitination participates in regulating such diverse
cellular processes as endocytosis, protein trafficking,
DNA repair, intracellular signaling, and transcription.
For example, during S phase of the cell cycle, the mo-
noubiquitination of PCNA (the sliding clamp associated
with the eukaryotic DNA replication fork; Section 30-
4Ba) at its Lys 164 recruits an error-prone translesion
DNA polymerase to the replication fork at a DNA dam-
age site, whereas the polyubiquitination of the same site
with a Lys 63–linked chain recruits DNA polymerases
that mediate error-free lesion repair. In another example,
in the signal transduction pathway that activates NF-B
(Section 32-6Be), IB is phosphorylated by IB kinase
(IKK), which has a regulatory subunit named NEMO (for
NF-B essential modulator). The binding of cytokines
such as TNF and IL-1 to their transmembrane receptors
in this pathway activates the receptor to Lys 63–linked
polyubiquitinate NEMO, which in turn activates IKK to
phosphorylate IB. This, as we saw, induces the forma-
tion of the Lys 48–linked polyubiquitination signal that
leads to the destruction of IB and hence the transloca-
tion of NF-B to the nucleus. Yersinia pestis, the bac-
terium that causes bubonic plague (Section 19-3Fc), pro-
duces a virulence factor named YopJ, which functions as
a DUB that prevents the activation of NF-B (which is an
1420 Chapter 32. Translation
Figure 32-88 X-ray structure of H. influenzae HslVU in
complex with ATP. The 821-kD complex is drawn in ribbon form
viewed along a 2-fold axis with its 6-fold axis vertical.The D
6
symmetric dodecamer of HslV subunits is coaxially bound at
both ends by C
6
symmetric HslU hexamers to yield a complex
with overall D
6
symmetry. The subunits of the lower HslU
hexamer are alternately orange and cyan, whereas those of the
upper HslU hexamer are pale orange except for one subunit,
which is colored in rainbow order from its N-terminus (blue) to
its C-terminus (red).The subunits of the lower HslV hexamer are
alternately green and magenta, whereas those of the upper
hexamer are pale blue except for one subunit, which is colored in
rainbow order.The ATPs, which are bound at the interfaces
between neighboring HslU subunits, are drawn in space-filling
form with C green, N blue, O red, and P orange. [Based on an
X-ray structure by David McKay, Stanford University School of
Medicine. PDBid 1G3I.]
JWCL281_c32_1338-1428.qxd 9/7/10 2:23 PM Page 1420
