Voet D., Voet Ju.G. Biochemistry
Подождите немного. Документ загружается.

important immune system activator). We shall see in Sec-
tion 34-3Bff that the monoubiquitination of certain his-
tones functions to regulate transcription.
m. Ubiquitinlike Modifiers Participate in a Variety of
Regulatory Processes
Eukaryotic cells express several proteins that have the
same -Grasp fold as ubiquitin and are similarly conju-
gated to other proteins, although their surface residues and
charge distributions differ significantly.These ubiquitinlike
modifiers (Ubls), which participate in a variety of funda-
mental cellular processes, each have a corresponding acti-
vating enzyme (E1), at least one conjugating enzyme (E2),
and one or more ligases (E3s), that function to link the Ubl
to its target protein(s) in a manner closely resembling that
of ubiquitin.
Two of the most extensively studied Ubls are SUMO
(small ubiquitin-related modifier; 18% identical to ubiqui-
tin) and RUB1 (related-to-ubiquitin 1; called NEDD8 in
vertebrates; 50% identical to ubiquitin), proteins that are
highly conserved from yeast to humans.The sumoylation of
PCNA at the same Lys residue at which it is ubiquitinated
promotes normal DNA replication during S phase. The
sumoylation of IB at the same residue (Lys 21) at which
it is ubiquitinated blocks its ubiquitination and subsequent
degradation and thereby prevents the translocation of NF-
B to the nucleus. Evidently, there is a complex regulatory
interplay between the ubiquitination and sumoylation of
both PCNA and IB. SUMO also modifies two mam-
malian glucose transporters, GLUT1 and GLUT4 (Section
20-2Eb), and in doing so, increases the availability of
GLUT4 but decreases that of GLUT1.
All known RUB1 targets are cullins, all of which are
subunits of SCF complexes, the multisubunit RING E3s
(Section 32-6Bd). In fact, -TrCP, the E3 that directs the
ubiquitination of IB, must be conjugated to RUB1 be-
fore it can do so, thereby adding further complexity to the
control of NF-B.
Section 32-6. Protein Degradation 1421
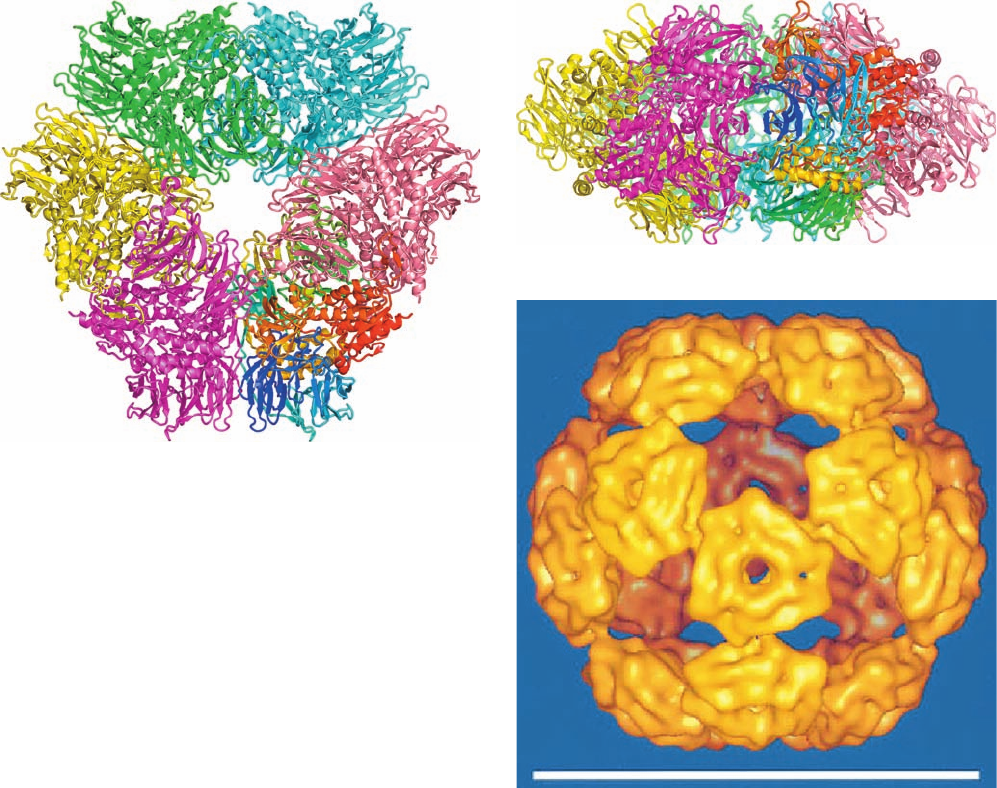
Figure 32-89 Structure of the tricorn protease. (a) The X-ray
structure of its D
3
symmetric hexamer drawn in ribbon form and
viewed along its 3-fold axis. Its subunits are differently colored
with that on the lower right colored in rainbow order from its
N-terminus (blue) to its C-terminus (red). (b) View along the
hexameric complex’s 2-fold axis (rotated 90° about a horizontal
axis with respect to Part a). (c) A cryo-EM–based image of the
icosahedral complex. Each of its component “plates” represents
a hexameric complex such as those drawn in Parts a and b. Note
how neighboring plates are related by both 3-fold axes (which
are coincident with those of the hexameric complexes) and
5-fold axes.The white scale bar is 50 nm long. [Parts a and b are
based on an X-ray structure by Robert Huber, Max-Planck-
Institut für Biochemie, Martinsried, Germany. PDBid 1K32. Part
c is courtesy of Wolfgang Baumeister, Max-Planck-Institut für
Biochemie, Martinsried, Germany.]
(a)
(b)
(c)
JWCL281_c32_1338-1428.qxd 9/7/10 2:24 PM Page 1421

1422 Chapter 32. Translation
1 The Genetic Code Point mutations are caused by either
base analogs that mispair during DNA replication or by sub-
stances that react with bases to form products that mispair. In-
sertion/deletion (frameshift) mutations arise from the associa-
tion of DNA with intercalating agents that distort the DNA
structure. The analysis of a series of frameshift mutations that
suppress one another established that the genetic code is an
unpunctuated triplet code. In a cell-free protein synthesizing sys-
tem, poly(U) directs the synthesis of poly(Phe), thereby demon-
strating that UUU is the codon specifying Phe.The genetic code
was elucidated through the use of polynucleotides of known
composition but random sequence, by the ability of defined
triplets to promote the ribosomal binding of tRNAs bearing spe-
cific amino acids, and through the use of synthetic mRNAs of
known alternating sequences.The latter investigations have also
demonstrated that the 5¿ end of mRNA corresponds to the N-
terminus of the polypeptide it specifies and have established the
sequences of the Stop codons. Degenerate codons differ mostly
in the identities of their third base. Small single-stranded DNA
phages such as X174 contain overlapping genes in different
reading frames.The genetic code used by mitochondria differs in
several codons from the “standard” genetic code.
2 Transfer RNA and Its Aminoacylation Transfer RNAs
consist of 54 to 100 nucleotides that can be arranged in the
cloverleaf secondary structure. As many as 10% of a tRNA’s
bases may be modified. Yeast tRNA
Phe
forms a narrow,
L-shaped, three-dimensional structure that resembles that of
other tRNAs. Most of the bases are involved in stacking and
base pairing associations including nine tertiary interactions
that appear to be essential for maintaining the molecule’s na-
tive conformation. Amino acids are appended to their cognate
tRNAs in a two-stage reaction catalyzed by the corresponding
aminoacyl–tRNA synthetase (aaRS). There are two classes of
aaRSs, each containing 10 members. Class I aaRSs have two
conserved sequence motifs that occur in the Rossmann fold
common to the catalytic domain of these enzymes. Class II
aaRSs have three conserved sequence motifs that occur in the
7-stranded antiparallel sheet–containing fold that forms the
core of their catalytic domains. In binding only their cognate
tRNAs, aaRSs recognize only an idiosyncratic but limited num-
ber of bases (identity elements) that are, most often, located at
the anticodon and in the acceptor stem. The great accuracy of
tRNA charging arises from the proofreading of the bound
amino acid by certain aminoacyl–tRNA synthetases via a dou-
ble-sieve mechanism and at the expense of ATP hydrolysis.
Many organisms and organelles lack a GlnRS and instead
synthesize Gln–tRNA
Gln
by the GluRS-catalyzed charging of
tRNA
Gln
with glutamate followed by its transamidation using
glutamine as the amido group source in a reaction mediated
by Glu–tRNA
Gln
amidotransferase (Glu-AdT). Ribosomes se-
lect tRNAs solely on the basis of their anticodons. Sets of de-
generate codons are read by a single tRNA through wobble
pairing. The UGA codon, which is normally the opal Stop
codon may, depending on its context in mRNA, specify a se-
lenoCys (Sec) residue, which is carried by a specific tRNA
(tRNA
Sec
), thereby forming a selenoprotein. Nonsense muta-
tions may be suppressed by tRNAs whose anticodons have
mutated to recognize a Stop codon.
3 Ribosomes and Polypeptide Synthesis The ribosome
consists of a small and a large subunit whose complex shapes
have been revealed by cryoelectron microscopy and X-ray
crystallography. The three RNAs and 52 proteins comprising
the E. coli ribosome self-assemble under proper conditions.
Both ribosomal subunits consist of an RNA core in which the
proteins are embedded, mainly as globular domains on the
back and sides of the particle, with long basic polypeptide seg-
ments that infiltrate between the RNA helices so as to neutral-
ize their anionic charges. Eukaryotic ribosomes are larger and
more complex than those of prokaryotes.
Ribosomal polypeptide synthesis proceeds by the addition
of amino acid residues to the C-terminal end of the nascent
polypeptide. The mRNAs are read in the 5¿S3¿ direction.
mRNAs are usually simultaneously translated by several ribo-
somes in the form of polysomes.The ribosome has three tRNA-
binding sites: the A site, which binds the incoming aminoacyl–
tRNA; the P site, which binds the peptidyl–tRNA;and the E site,
which transiently binds the outgoing deacylated tRNA. During
polypeptide synthesis, the nascent polypeptide is transferred to
the aminoacyl–tRNA, thereby lengthening the nascent
polypeptide by one residue. The newly deacylated tRNA is
translocated to the E site and the new peptidyl–tRNA, with its
associated codon, is translocated to the P site. In prokaryotes,
the initiation sites on mRNA are recognized through their
Shine–Dalgarno sequences and by their initiating codon.
Prokaryotic initiating codons specify . Initiation
involves the participation of three initiation factors that induce
the assembly of the ribosomal subunits with in
the P site and mRNA. Eukaryotic initiation is a far more com-
plicated process that requires the participation of at least 11 ini-
tiation factors. The system binds the mRNA’s 5¿ cap and scans
along the mRNA until it finds its AUG initiation codon, usually
the mRNA’s first AUG, through codon–anticodon interactions
with the initiating tRNA, .
Polypeptides are elongated in a three-part cycle, consisting
of aminoacyl–tRNA decoding, transpeptidation, and translo-
cation, that requires the participation of elongation factors
and is vectorially driven by GTP hydrolysis. EF-Tu, which
functions to escort aminoacyl–tRNAs into the ribosomal A
site, undergoes a major conformational change on hydrolyzing
its bound GTP. The X-ray structure of the 50S subunit clearly
shows that the ribosomal peptidyl transferase center is distant
from any protein and hence that the ribosome is a ribozyme.
Peptide bond formation is catalyzed by a substrate-assisted
mechanism in which the ribosome functions as an entropy
trap. Translocation is motivated through the EF-G–catalyzed
hydrolysis of GTP. EF-G GDP, which binds to the same ribo-
somal site as aminoacyl–tRNA EF-Tu GTP, is a macromol-
ecular mimic of this complex. Translocation occurs via inter-
mediate states, the A/P and P/E states, in which the newly
formed peptidyl–tRNA and the newly deacylated tRNA are
respectively bound to the A and P subsites of the 30S subunit
and to the P and E subsites of the 50S subunit, following which
EF-G hydrolyzes its bound GTP and shifts these tRNAs to the
P/P and E/E states. The ribosome initially selects an aminoa-
cyl–tRNA whose anticodon is cognate to its A-site–bound
codon through interactions involving three universally con-
Met–tRNA
Met
i
fMet–tRNA
Met
f
fMet–tRNA
Met
f
CHAPTER SUMMARY
JWCL281_c32_1338-1428.qxd 8/19/10 10:06 PM Page 1422
served 30S subunit bases while the tRNA is in the A/T binding
state.The codon–anticodon interaction is then proofread in an
independent process that follows the hydrolysis of the EF-
Tu–bound GTP and which occurs when the tRNA has shifted
to the A/A binding state, a process called accommodation.
Termination codons bind release factors that induce the hy-
drolysis of the peptidyl–tRNA bond. Eukaryotic elongation
and termination resemble those of prokaryotes. Ribosomal in-
hibitors, many of which are antibiotics, are medically important
and biochemically useful in elucidating ribosomal function.
Streptomycin causes mRNA misreading and inhibits prokary-
otic chain initiation, chloramphenicol inhibits prokaryotic pep-
tidyl transferase, paromomycin causes codon misreading,tetra-
cycline inhibits aminoacyl–tRNA binding to the prokaryotic
30S subunit, and diphtheria toxin ADP-ribosylates eEF2.
4 Control of Eukaryotic Translation Several mechanisms
of translational control have been elucidated in eukaryotes.
eIF2 kinases catalyze the phosphorylation of eIF2,which then
tightly binds eIF2B so as to prevent it from exchanging eIF2-
bound GDP for GTP and hence inhibits translational initiation.
These eIF2 kinases include heme-regulated inhibitor (HRI),
which functions to coordinate globin synthesis with heme avail-
ability; double-stranded RNA-activated protein kinase (PKR),
an interferon-induced protein that functions to inhibit viral pro-
liferation; and PKR-like endoplasmic reticulum kinase (PERK),
which functions to protect the cell from the irreversible damage
caused by the accumulation of unfolded proteins in the ER.
GCN2, in contrast, is an eIF2 kinase that, when amino acids are
scarce, stimulates the translation of the transcriptional activator
GCN4 by causing the 40S ribosomal subunit to scan across four
upstream open reading frames (uORFs) in the GCN4 mRNA,
thereby permitting the ribosome to initiate translation at the
GCN4 coding sequence. The phosphorylation of eIF4E (cap-
binding protein) by a MAP kinase cascade increases eIF4E’s
affinity for capped mRNA and thereby stimulates translational
initiation. The binding of 4E-BPs to eIF4E blocks its binding of
eIF4G and hence prevents initiation. However, the insulin-in-
duced phosphorylation of the 4E-BPs causes them to dissociate
from eIF4E. The mRNAs in certain animal oocytes are masked
by the binding of proteins, which prevents their translation.
Many oocyte mRNAs have short poly(A) tails that are pre-
ceded by a cytoplasmic polyadenylation element (CPE) that is
bound by CPE-binding protein (CPEB). CPEB binds maskin
which binds eIF4E, thereby inhibiting translational initiation.
However, when CPEB is phosphorylated, it recruits poly(A)
polymerase (PAP), which extends the mRNA’s poly(A) tail
such that it is bound by poly(A)-binding protein (PABP).
PABP then binds to eIF4G, which in turn displaces maskin
from eIF4E, thereby permitting the translation of the mRNA.
Antisense oligonucleotides can be used to inhibit the transla-
tion of their complementary mRNAs.Although the delivery of
antisense oligonucleotides to their sites of action has proved
to be a difficult problem, their use is starting to show some
medical and biotechnological successes.
5 Post-Translational Modification Proteins may be post-
translationally modified in a variety of ways. Proteolytic cleav-
ages, usually by specific peptidases, activate proproteins. The
signal peptides of preproteins are removed by signal peptidases.
Covalent modifications alter many types of side chains in a va-
riety of ways that modulate the catalytic activities of enzymes,
provide recognition markers, and stabilize protein structures.
Protein splicing occurs via the intein-catalyzed self-excision
between an N-extein and a C-extein accompanied by the liga-
tion of the N- and C-exteins via a peptide bond. Most inteins
contain a homing endonuclease that makes a double-strand
nick in a gene similar to that encoding the corresponding extein,
thereby triggering a recombinational double-strand DNA re-
pair process that copies the gene encoding the intein into the
break. Inteins therefore appear to be molecular parasites.
6 Protein Degradation Proteins in living cells are contin-
ually turning over. This controls the level of regulatory en-
zymes and disposes of abnormal proteins that would other-
wise interfere with cellular processes. Proteins are degraded
by lysosomes via a nonspecific process as well via a process
specific for KFERQ proteins that is stimulated during starva-
tion. A cytosolically based ATP-dependent system degrades
normal as well as abnormal proteins in a process that flags
these proteins by the covalent attachment of Lys 48–linked
polyubiquitin chains to their Lys residues. This process is me-
diated by three consecutively acting enzymes: ubiquitin-acti-
vating enzyme (E1), ubiquitin-conjugating enzyme (E2), and
ubiquitin–protein ligase (E3). Most cells have one species of
E1, several species of E2, and numerous species of E3, each of
which is served by one or a few E2s. The polyubiquitinated
protein is proteolytically degraded in the 26S proteasome.
E3s can have complicated modular structures, each having
different specificities for target proteins. SCF complexes, one
of whose several subunits contains a RING domain,are partic-
ularly elaborate. The RING E3 known as E3 functions to
ubiquitinate proteins that satisfy the N-end rule.The transcrip-
tion factor NF-B is activated through the ubiquitination and
subsequent destruction of its otherwise bound inhibitor IB
by the SCF -TrCP, which is activated through phosphoryla-
tion via a signal transduction cascade. Cyclins, which mediate
the cell cycle, are destroyed in a programmed manner through
ubiquitination by their cognate E3s, one of which is anaphase-
promoting complex (APC).
The 26S proteasome consists of a hollow protein barrel
formed by two rings of seven subunits flanking two rings of
seven subunits known as the 20S proteasome, which is
bound at each end by 19S caps that each consist of ⬃18 sub-
units. The active sites of the subunits, which are members of
the N-terminal nucleophile (Ntn) family of hydrolases, are in-
side the barrel. Ubiquitinated proteins are selected by the 19S
caps, which unfold them in an ATP-dependent matter and
then feed them into the 20S proteasome via its axial channel.
The polyubiquitin (polyUb) chains are excised from the con-
demned protein by proteasome-associated deubiquitinating en-
zymes (DUBs), while other DUBs dismember the polyUb
chains to their component ubiquitin units, thereby recycling
them. The 11S activator is a heptameric complex that, on bind-
ing to one end of a 20S proteasome, opens its axial channel in an
ATP-independent manner, thereby permitting polypeptides, but
not folded proteins, to enter the 20S proteasome. Eubacteria,
nearly all of which lack proteasomes, nevertheless express a va-
riety of self-compartmentalized proteases, including ClpP, heat
shock locus UV (HslUV), and tricorn protease, that function to
proteolytically dispose of their cellular proteins. Monoubiquiti-
nation and polyubiquitination with chains linked through ubiq-
uitin residues other than Lys 48 regulate a variety of cellular
processes. Ubiquitinlike modifiers (Ubls), such as SUMO and
RUB1, also participate in numerous regulatory processes.
Chapter Summary 1423
JWCL281_c32_1338-1428.qxd 9/7/10 2:24 PM Page 1423

1424 Chapter 32. Translation
General
Watson, J.D., Baker, T.A., Bell, S.P., Gann, A., Levine, M., and
Losick, R., Molecular Biology of the Gene (6th ed.), Chap. 14
and 15, Cold Spring Harbor Laboratory Press (2008).
The Genetic Code
Attardi, G., Animal mitochondrial DNA: an extreme example of
genetic economy, Int. Rev. Cytol. 93, 93–145 (1985).
Benzer, S., The fine structure of the gene, Sci. Am. 206(1), 70–84
(1962).
Crick, F.H.C.,The genetic code, Sci.Am. 207(4), 66–74 (1962) [The
structure of the code as determined by phage genetics]; and
The genetic code: III, Sci.Am. 215(4),55–62 (1966).[A descrip-
tion of the nature of the code after its elucidation was almost
complete.]
Crick, F.H.C., Burnett, L., Brenner, S., and Watts-Tobin, R.J., Gen-
eral nature of the genetic code for proteins, Nature 192,
1227–1232 (1961).
Fox,T.D., Natural variation in the genetic code, Annu. Rev. Genet.
21, 67–91 (1987).
Judson, J.F., The Eighth Day of Creation, Expanded Edition, Part
II, Cold Spring Harbor Laboratory Press (1996). [A fascinating
historical narrative on the elucidation of the genetic code.]
Khorana, H.G., Nucleic acid synthesis in the study of the genetic
code, Nobel Lectures in Molecular Biology, 1933–1975, pp.
303–331, Elsevier (1977).
Knight, R.D., Freeland, S.J., and Landweber, L.F., Selection, his-
tory and chemistry: the three faces of the genetic code, Trends
Biochem. Sci. 24, 241–247 (1999).
Nirenberg, M., The genetic code, Nobel Lectures in Molecular Bi-
ology, 1933–1975, pp. 335–360, Elsevier (1977).
Nirenberg, M., Historical review: deciphering the genetic code—a
personal account, Trends Biochem. Sci. 29, 46–54 (2004).
Nirenberg, M. and Leder, P., RNA code words and protein synthe-
sis, Science 145, 1399–1407 (1964). [The determination of the
genetic code by the ribosomal binding of tRNAs using specific
trinucleotides.]
Nirenberg, M.W. and Matthaei, J.H., The dependence of cell-free
protein synthesis in E. coli upon naturally occurring or syn-
thetic polyribonucleotides, Proc. Natl. Acad. Sci. 47, 1588–1602
(1961). [The landmark paper reporting the finding that
poly(U) stimulates the synthesis of poly(Phe).]
Singer, B. and Kusmierek, J.T., Chemical mutagenesis, Annu. Rev.
Biochem. 51, 655–693 (1982).
The Genetic Code, Cold Spring Harbor Symp. Quant. Biol. 31
(1966). [A collection of papers describing the establishment of
the genetic code. See especially the articles by Crick, Niren-
berg, and Khorana.]
Yanofsky, C., Establishing the triplet nature of the genetic code,
Cell 128, 815–818 (2007).
Yarus, M., Caporaso, J.G., and Knight, R., Origins of the genetic
code: the escaped triplet theory, Annu. Rev. Biochem. 74,
179–198 (2005).
Transfer RNA and Its Aminoacylation
Alexander, R.W. and Schimmel, P., Domain–domain communica-
tion in aminoacyl–tRNA synthetases, Prog. Nucleic Acid Res.
Mol. Biol. 69, 317–349 (2001).
Ambrogelly,A., Palioura, S., and Söll,D., Natural expansion of the
genetic code, Nature Chem. Biol. 3, 29–35 (2007).
Björk, G.R., Ericson, J.U., Gustafsson, C.E.D., Hagervall, T.G.,
Jösson,Y.H., and Wikström, P.M.,Transfer RNA modification,
Annu. Rev. Biochem. 56, 263–287 (1987).
Böck,A., Forschhammer, K., Heider, J., and Baron, C., Selenopro-
tein synthesis: an expansion of the genetic code, Trends
Biochem. Sci. 16, 463–467 (1991).
Crick, F.H.C., Codon–anticodon pairing: the wobble hypothesis,
J. Mol. Biol. 19, 548–555 (1966).
Fukai, S., Nureki, O., Sekine, S., Shimada, A., Tao, J., Vassylyev,
D.G., and Yokoyama, S., Structural basis for double-sieve dis-
crimination of
L-valine from L-isoleucine and L-threonine by
the complex of tRNA
Va l
and valyl–tRNA synthetase, Cell 103,
793–803 (2000).
Ibba, M. and Söll, D., Aminoacyl–tRNA synthesis, Annu. Rev.
Biochem. 69, 617–650 (2000).
Ibba, M.A., Francklyn, C., and Cusack, S. (Eds.), The Aminoacyl-
tRNA Synthetases, Landes Bioscience/Eurekah.com (2005).
[Contains reviews on each of the twenty aminoacyl–tRNA
synthetases.]
Jacquin-Becker, C.,Ahel, I.,Ambrogelly,A., Ruan, B., Söll, D., and
Stathopoulos, C., Cysteinyl–tRNA formation and prolyl–tRNA
synthetase, FEBS Lett. 514, 34–36 (2002).
Kim, S.H., Suddath, F.L., Quigley, G.J., McPherson,A., Sussman, J.L.,
Wang, A.M.J., Seeman, N.C., and Rich, A., Three-dimensional
tertiary structure of yeast phenylalanine transfer RNA, Science
185, 435–440 (1974); and Robertus, J.D., Ladner, J.E., Finch, J.T.,
Rhodes, D., Brown, R.S., Clark, B.F.C., and Klug,A., Structure of
yeast phenylalanine tRNA at 3 Å resolution, Nature 250,
546–551 (1974). [The landmark papers describing the high-reso-
lution structure of a tRNA.]
Krzycki, J.A., The direct genetic encoding of pyrrolysine, Curr. Opin.
Microbiol. 8, 706–712 (2005); and Nozawa, K., O’Donoghue, P.,
Gundllapalli, S., Araiso, Y., Ishitani, R., Umehara, T., Söll, D., and
Nureki, O., Pyrolysyl–tRNA synthetase–tRNA
Pyl
structure reveals
the molecular basis of orthogonality, Nature 457, 1163–1167 (2009).
Ling,J., Reynolds, N., and Ibba, M.,Aminoacyl–tRNA synthesis and
translational control, Annu. Rev. Microbiol. 63, 61–78 (2009).
Nureki, O., Vassylyev, D.G., Tateno, M., Shimada, A., Nakama, T.,
Fukai, S., Konno, M., Hendrickson, T.L., Schimmel, P., and
Yokoyama, S., Enzyme structure with two catalytic sites for
double-sieve selection of substrate, Science 280, 578–582
(1998). [The X-ray structures of IleRS in complexes with
isoleucine and valine.]
Rould, M.A., Perona, J.J., and Steitz,T.A., Structural basis of anti-
codon loop recognition by glutaminyl–tRNA synthetase,
Nature 352, 213–218 (1991).
Ruff, M., Krishnaswamy, S., Boeglin, M., Poterszman, A.,
Mitschler, A., Podjarny, A., Rees, B., Thierry, J.C., and Moras,
D., Class II aminoacyl transfer RNA synthetases: Crystal
structure of yeast aspartyl–tRNA synthetase complexed with
tRNA
Asp
, Science 252, 1682–1689 (1991).
Schimmel, P. and Beebe, K., Aminoacyl tRNA synthetases: from
the RNA to the theater of proteins. In Gesteland, R.F., Cech,
T.R., and Atkins, J.F. (Eds.), The RNA World (3rd ed.), pp.
227–255, Cold Spring Harbor Laboratory Press (2006).
Silvian, L.F.,Wang, J., and Steitz,T.A., Insights into editing from an
Ile-tRNA synthetase structure with tRNA
Ile
and mupirocin,
Science 285, 1074–1077 (1999).
Söll, D. and RajBhandary, U.L. (Eds.),tRNA:Structure, Biosynthe-
sis, and Function, ASM Press (1995).
Stadtman, T.C., Selenocysteine, Annu. Rev. Biochem. 65, 83–100
(1996).
REFERENCES
JWCL281_c32_1338-1428.qxd 8/4/10 4:46 PM Page 1424
Ribosomes and Polypeptide Synthesis
Aitken, C.E., Petrov,A., and Puglisi, J.D., Single ribosome dynam-
ics and the mechanism of translation, Annu. Rev. Biophys. 39,
491–513 (2010).
Ban, N., Nissen, P., Hansen, J., Moore, P.B., and Steitz,T., The com-
plete atomic structure of the large ribosomal subunit at 2.4 Å
resolution, Science 289, 905–920 (2000).
Bashan, A. and Yonath, A., Correlating ribosome function with
high-resolution structures,Trends Microbiol. 16, 326–335 (2008).
Bell, C.E. and Eisenberg, D.E., Crystal structure of diphtheria
toxin bound to nicotinamide adenine dinucleotide, Biochem-
istry 35, 1137–1149 (1996).
Brandt, F., Etchells, S.A., Ortiz, J.O., Elcock,A.H., Hartl, F.U., and
Baumeister, W., The native 3D organization of bacterial
polysomes, Cell 136, 261–271 (2009).
Brodersen, D.E., Clemons, W.M., Jr., Carter, A.P., Morgan-
Warren, R.J., Wimberly, B.T., and Ramakrishnan, V.,The struc-
tural basis for the action of the antibiotics tetracycline,
pactamycin, and hygromycin B on the 30S ribosomal subunit,
Cell 103, 1143–1154 (2000).
Czworkowski, J.,Wang, J., Steitz, J.A., and Moore, P.B., The crystal
structures of elongation factor G complexed with GDP, at 2.7
Å resolution; and Ævarsson, A., Brazhnikov, E., Garber, M.,
Zheltonosova, J., Chirgadze, Yu., Al-Karadaghi, S., Svensson,
L.A., and Liljas,A.,Three-dimensional structure of the riboso-
mal translocase: elongation factor G from Thermus ther-
mophilus, EMBO J. 13, 3661–3668 and 3669–3677 (1994).
Dintzis, H.M., Assembly of the peptide chains of hemoglobin,
Proc. Natl. Acad Sci. 47, 247–261 (1961); and The wandering
pathway to determining N to C synthesis of proteins: Some
recollections concerning protein structure and biosynthesis,
Biochem. Mol. Biol. Educ. 34, 241–246 (2006).[The determina-
tion of the direction of polypeptide biosynthesis.]
Dunkle, J.A. and Cate, J.H.D., Ribosome structure and dynamics
during translocation and termination, Annu. Rev. Biophys. 39,
227–244 (2010).
Frank, J., Single-particle imaging of macromolecules by cryo-
electron microscopy, Annu. Rev. Biophys. Biomol. Struct. 31,
303–319 (2002).
Frank, J. and Agrawal, R.K., A ratchet-like inter-subunit reorgan-
ization of the ribosome during translocation, Nature 406,
318–322 (2000).
Frank, J. and Gonzalez, R.L., Jr., Structure and dynamics of a pro-
cessive Brownian motor: the translating ribosome, Annu. Rev.
Biochem. 79, 381–412 (2010).
Gao, H., et al., RF3 induces ribosomal conformational changes
responsible for dissociation of class I release factors, Cell 129,
929–941 (2007).
Gingras, A.-C., Raught, B., and Sonnberg, N., eIF4 initiation fac-
tors: effectors of mRNA recruitment to ribosomes and regula-
tors of translation, Annu. Rev. Biochem. 68, 913–963 (1999).
Held, W.A., Ballou, B., Mizushima, S., and Nomura, M., Assembly
mapping of 30S ribosomal proteins from Escherichia coli, J.
Biol. Chem. 249, 3103–3111 (1974).
Holbrook, S.R., Structural principles from large RNAs, Annu.
Rev. Biophys. 37, 445–464 (2008).
Jackson, R.J., Hellen, C.U.T., and Pestova, T.V., The mechanism of
eukaryotic translation and principles of its regulation, Nature
Rev. Mol. Cell Biol. 10, 113–127 (2010).
Kawashima, T., Berthet-Colominas, C., Wulff, M., Cusack, S., and
Leberman, R.,The structure of the Escherichia coli EF-Tu EF-
Ts complex at 2.5 Å resolution, Nature 379, 511–518 (1996).
Kiel, M.C., Kaji, H., and Kaji, A., Ribosome recycling, Biochem.
Mol. Biol. Educ. 35, 40-44 (2007); and Hirokawa, G., Demesh-
kina, N., Iwakura, N., Kaji, H., and Kaji, A., The ribosome-
recycling step: consensus or controversy? Trends Biochem. Sci.
31, 143–149 (2006).
Kjeldgaard, M. and Nyborg, J., Refined structure of elongation fac-
tor EF-Tu from Escherichia coli, J. Mol. Biol. 223, 721–742
(1992); and Kjeldgaard, M., Nissen, P.,Thirup, S., and Nyborg, J.,
The crystal structure of elongation factor EF-Tu from Thermus
aquaticus in the GTP conformation, Structure 1, 35–50 (1993).
Korostelev, A. and Noller, H.F., The ribosome in focus: new struc-
tures bring new insights, Trends Biochem. Sci. 32, 434–441
(2007); and Korostelev, A., Trakhanov, S., Laurberg, M., and
Noller, H.F., Crystal structure of the 70S ribosome–tRNA
complex reveals functional interactions and rearrangements,
Cell 126, 1065–1077 (2006).
Laurberg, M., Asahara, H., Korostelev, A., Zhu, J., Trakhanov, S.,
and Noller,H.F.,Structural basis for translation termination on
the 70S ribosome, Nature 454, 852–857 (2008); Korostelev, A.,
Asahara, H., Lancaster, L., Laurberg, M., Hirschi, A., Zhu, J.,
Trakhanov, S., Scott,W.G., and Noller, H.F., Crystal structure of
a translation termination complex formed with release factor
RF2, Proc. Natl.Acad. Sci. 105, 19684–19689 (2008); andWeixl-
baumer, A., Jin, H., Neubauer, C., Voorhees, R.M., Petry, S.,
Kelley, A.C., and Ramakrishnan, V., Insights into translational
termination from the structure of RF2 bound to the ribosome,
Science 322, 953–956 (2008).
Marcotrigiano, J., Gingras, A.-C., Sonenberg, N., and Burley, S.K.,
Cocrystal structure of the messenger RNA 5¿ cap-binding pro-
tein (eIF4E) bound to 7-methyl-GDP, Cell 89, 951–961 (1997).
Moazed, D. and Noller, H.F., Intermediate states in the movement
of transfer RNA in the ribosome, Nature 342, 142–148 (1989).
Moore, P.B. and Steitz,T.A.,The involvement of RNA in ribosome
function, Nature 418, 229–235 (2002); and The structural basis
of large ribosomal subunit function, Annu. Rev. Biochem. 72,
813–850 (2003).
Munro, J.B., Sanbonmatsu, K.Y., Spahn, C.M.T., and Blanchard,
S.C., Navigating the ribosome’s metastable energy landscape,
Trends Biochem. Sci. 31, 390–399 (2009).
Nissen, P., Kjeldgaard, M.,Thirup, S., Polekhina, G., Reshetnikova,
L., Clark, B.F.C., and Nyborg, J., Crystal structure of the ter-
nary complex of Phe–tRNA
Phe
, EF-Tu, and a GTP analog, Sci-
ence 270, 1464–1472 (1995).
Noller, H.F., Hoffarth, V., and Zimniak, L., Unusual resistance
of peptidyl transferase to protein extraction procedures, Sci-
ence 256, 1416–1419 (1992); and Noller, H.F., Peptidyl trans-
ferase: protein, ribonucleoprotein, or RNA? J. Bacteriol. 175,
5297–5300 (1993).
Ogle, J.M., Brodersen, D.E., Clemons,W.M., Jr.,Tarry, M.J., Carter,
A.P., and Ramakrishnan, V., Recognition of cognate transfer
RNA by the 30S ribosomal subunit, Science 292, 897–902
(2001); and Ogle, J.M., Carter, A.P., and Ramakrishnan, V., In-
sights into the decoding mechanism from recent ribosome
structures, Trends Biochem. Sci. 28, 259–266 (2003).
Pioletti, M., et al., Crystal structure of complexes of the small ribo-
somal subunit with tetracycline, edeine and IF3, EMBO J. 20,
1829–1839 (2001).
Rodnina, M.V., Beringer, M., and Wintermeyer, W., How the ri-
bosome makes peptide bonds, Trends Biochem. Sci. 32,
20–26 (2007).
Schluenzen, F., Tocilj, A., Zarivach, R., Harms, J., Gluehmann, M.,
Janell, D., Bashan,A., Bartels, H.,Agmon, I., Franceschi, F., and
Yonath,A., Structure of functionally activated small ribosomal
subunit at 3.3 Å resolution, Cell 102, 615–623 (2000).
Schmeing, T.M., and Ramakrishnan, V., What recent ribosome
structures have revealed about the mechanism of translation,
References 1425
JWCL281_c32_1338-1428.qxd 8/19/10 10:06 PM Page 1425
Nature 461, 1234–1242 (2009); Simonovic, M. and Steitz, T.A.,
A structural view of the mechanism of the ribosome-catalyzed
peptide bond mechanism, Biochim. Biophys. Acta 1789,
612–623 (2009); and Zimmerman, E. and Yonath,A., Biological
implications of the ribosome’s stunning stereochemistry,
ChemBioChem 10, 63–72 (2009). [Authoritative reviews.]
Schmeing,T.M.,Voorhees, R.M., Kelley,A.C., Gao,Y.-G., Murphy,
F.V., IV,Weir, J.R., and Ramakrishnan,V.,The crystal structure
of the ribosome bound to EF-Tu and aminoacyl–tRNA; and
Gao, Y.-G., Selmer, M., Dunham, M., Weixlbaumer, A., Kelley,
A.C., and Ramakrishnan, V., The structure of the ribosome
with elongation factor G trapped in the post-translocational
state, Science 326, 688–693 and 694–699 (2009).
Selmer, M., Dunham, C.M., Murphy, F.V., IV, Weixlbaumer, A.,
Petry, S., Kelley, A.C., Weir, J.R., and Ramakrishnan,V., Struc-
ture of the 70S ribosome complexed with mRNA and tRNA,
Science 313, 1935–1942 (2006); and Voorhees, R.M., Weixl-
baumer,A., Loakes, D., Kelley,A.C., and Ramakrishnan,V., In-
sights into substrate stabilization from snapshots of the pep-
tidyl transferase center of the intact 70S ribosome, Nature
Struct. Mol. Biol. 16, 528–533 (2009).
Sievers, A., Beringer, M., Rodnina, M.V., and Wolfenden, R., The
ribosome as an entropy trap. Proc. Natl. Acad. Sci. 101,
7897–7901 (2004).
Sonenberg, N., eIF4E, the mRNA cap-binding protein: from basic
discovery to translational research, Biochem. Cell Biol. 86,
178–183 (2008).
Sonenberg, N. and Dever, T.E., Eukaryotic translation initiation
factors and regulators, Curr. Opin. Struct. Biol. 13, 56–63 (2003).
Spahn, C.M.T., Beckmann, R., Eswar, N., Penczek, P.A., Sali, A.,
Blobel, G., and Frank, J., Structure of the 80S ribosome from
Saccharomyces cerevisiae—tRNA-ribosome and subunit-
subunit interactions, Cell 107, 373–386 (2001).
Steitz, J.A. and Jakes, K., How ribosomes select initiator regions in
mRNA: base pair formation between the 3¿ terminus of 16S
RNA and the mRNA during initiation of protein synthesis in
Escherichia coli, Proc. Natl. Acad. Sci. 72, 4734–4738 (1975).
Sykes, M.T. and Williamson, J.R., A complex assembly landscape
for the 30S ribosomal subunit, Annu. Rev. Biophys. 38, 197–215
(2009); and Woodson, S.A., RNA folding and ribosome assem-
bly, Curr. Opin. Struct. Biol. 12, 667–673 (2008).
Vestergaard, B., Van, L.B., Andersen, G.R., Nyborg, J., Bucking-
ham, R.H., and Kjeldgaard, M., Bacterial polypeptide release
factor RF2 is structurally distinct from eukaryotic eRF1, Mol.
Cell 8, 1375–1382 (2001).
Weixlbaumer, A., Petry, S., Dunham, C.M., Selmer, M., Kelley,
A.C., and Ramakrishnan, V., Crystal structure of the ribosome
recycling factor bound to the ribosome, Nature Struct. Mol.
Biol. 14, 733–737 (2007).
Wimberly, B.T., Broderson, D.E., Clemons, W.M., Jr., Morgan-
Warren, R., von Rhein, C., Hartsch, T., and Ramakrishnan, V.,
Structure of the 30S ribosomal subunit, Nature 407, 327–339
(2000); and Broderson, D.E., Clemons, W.M., Jr., Carter, A.P.,
Wimberly, B.T., and Ramakrishnan, V., Crystal structure of the
30S ribosomal subunit from Thermus thermophilus: Structure
of the proteins and their interactions with 16S RNA, J. Mol.
Biol. 316, 725–768 (2002).
Yonath,A.,The search and its outcome:High resolution structures
of ribosomal particles from mesophilic, thermophilic, and
halophilic bacteria at various functional states, Annu. Rev. Bio-
phys. Biomol. Struct. 31, 257–273 (2002).
Yonath, A.,Antibiotics targeting ribosomes: resistance, selectivity,
synergism, and cellular regulation, Annu. Rev. Biochem. 74,
649–679 (2005).
Youngman, E.M., McDonald, M.E., and Green, R., Peptide re-
lease on the ribosome: mechanism and implications for trans-
lational control, Annu. Rev. Microbiol. 62, 33–373 (2008).
Yusupova, G.Z., Yusupov, M.M., Cate, J.D.H., and Noller, H.F.,
The path of messenger RNA through the ribosome, Cell 106,
233–241 (2001).
Control of Eukaryotic Translation
Branch, A.D., A good antisense molecule is hard to find, Trends
Biochem. Sci. 23, 45–50 (1998).
Calkhoven, C.F., Müller, C., and Leutz, A., Translational control of
gene expression and disease, Trends Mol.Med. 8, 577–583 (2002).
Chen, J.-J., and London, I.M., Regulation of protein synthesis by
heme-regulated eIF-2 kinase, Trends Biochem. Sci. 20,
105–108 (1995).
Clemens, M.J., PKR—A protein kinase regulated by double-
stranded RNA, Int. J. Biochem. Cell Biol. 29, 945–949 (1997).
Dever, T.E., Gene-specific regulation by general translation fac-
tors, Cell 108, 545–556 (2002).
Gray, N.K. and Wickens, M., Control of translation initiation in an-
imals, Annu. Rev. Cell Dev. Biol. 14, 399–458 (1998).
Lawrence, J.C., Jr. and Abraham, R.T., PHAS/4E-BPs as regula-
tors of mRNA translation and cell proliferation, Trends
Biochem. Sci. 22, 345–349 (1997).
Lebedeva, I. and Stein, C.A., Antisense oligonucleotides: promise
and reality, Annu. Rev. Pharmacol.Toxicol. 41, 403–419 (2001).
Matthews, M.B., Sonnenberg, N., and Hershey, J.W.B. (Eds.),
Translational Control in Biology and Medicine, Cold Spring
Harbor Laboratory Press (2007).
Mendez, R. and Richter, J.D., Translational control by CPEB: a
means to the end, Nature Rev. Mol. Cell Biol. 2, 521–529 (2001).
Phillips, M.I. (Ed.), Antisense Technology: Part A. General Meth-
ods, Methods of Delivery, and RNA Studies; and Part B. Appli-
cations, Methods Enzymol. 313 and 314 (2000).
Sen, G.C. and Lengyel, P., The interferon system, J. Biol. Chem.
267, 5017–5020 (1992).
Sheehy, R.E., Kramer, M., and Hiatt, W.R., Reduction of poly-
galacturonase activity in tomato fruit by antisense RNA, Proc.
Natl.Acad. Sci. 85, 8805–8809 (1988).
Sonenberg, N. and Hinnebusch, A.G., Regulation of translation
initiation in eukaryotes: mechanisms and biological targets,
Cell 136, 731–745 (2009).
Tafuri, S.R. and Wolffe, A.P., Dual roles for transcription and
translation factors in the RNA storage particles of Xenopus
oocytes, Trends Cell. Biol. 3, 94–98 (1993).
Tamm, I., Dörken, B., and Hartmann, G., Antisense therapy in on-
cology: new hope for an old idea? Lancet 358, 489–497 (2001).
Post-Translational Modification
Gogarten, J.P., Senejani, A.G., Zhaxybayeva, O., Olendzenski, L.,
and Hilario, E., Inteins: structure, function, and evolution,
Annu. Rev. Microbiol. 56, 263–287 (2002).
Harding, J.J., and Crabbe, M.J.C. (Eds.), Post-Translational Modi-
fications of Proteins, CRC Press (1992).
Klabunde, T., Sharma, S., Telenti, A., Jacobs, W.R., Jr., and Sac-
chetini, J.C., Crystal structure of Gyr A protein from Mycobac-
terium xenopi reveals structural basis of splicing, Nature Struct.
Biol. 5, 31–36 (1998).
Liu, X.-Q., Protein-splicing intein: genetic mobility, origin, and
evolution,Annu. Rev. Genet. 34, 61–76 (2000).
Saleh, L. and Perler, F.B., Protein splicing in cis and in trans,Chem.
Rec. 6, 183–193 (2006).
Wold, F., In vivo chemical modification of proteins, Annu. Rev.
Biochem. 50, 783–814 (1981).
1426 Chapter 32. Translation
JWCL281_c32_1338-1428.qxd 9/7/10 2:24 PM Page 1426

Problems 1427
1. What is the product of the reaction of guanine with nitrous
acid? Is the reaction mutagenic? Explain.
2. What is the polypeptide specified by the following DNA
antisense strand? Assume translation starts at the first initiation
codon.
5¿-TCTGACTATTGAGCTCTCTGGCACATAGCA-3¿
*3. The fingerprint of a protein from a phenotypically rever-
tant mutant of bacteriophage T4 indicates the presence of an al-
tered tryptic peptide with respect to the wild type. The wild-type
and mutant peptides have the following sequences:
Mutant
Cys-Glu-Thr- Met-Ser- His-Ser- Tyr-Arg
Wild type
Cys-Glu-Asp-His- Val-Pro-Gln-Tyr-Arg
PROBLEMS
Wold, F. and Moldave, K. (Eds.), Posttranslational Modifications,
Parts A and B, Methods Enzymol. 106 and 107 (1984). [Con-
tains extensive descriptions of the amino acid “zoo.”]
Protein Degradation
Bochtler, M., Ditzel, L., Groll, M., Hartmann, C., and Huber, R.,
The proteasome, Annu. Rev. Biophys. Biomol. Struct. 28,
295–317 (1999).
Brandstetter,H., Kim,J.-S., Groll, M., and Huber,R.,Crystal struc-
ture of the tricorn protease reveals a protein disassembly line,
Nature 414, 466–470 (2001); Walz, J., Tamura, T., Tamura, N.,
Grimm, R., Baumeister, W., and Koster, A.J., Tricorn protease
exists as an icosahedral supermolecule in vivo, Mol. Cell 1,
59–65 (1997); and Walz, J., Koster, A.J., Tamura, T., and
Baumeister, W., Capsids of tricorn protease studied by cryo-
microscopy, J. Struct. Biol. 128, 65–68 (1999).
Cook,W.J., Jeffrey, L.C.,Kasperek,E.,and Pickart,C.M., Structure
of tetraubiquitin shows how multiubiquitin chains can be
formed, J. Mol. Biol. 236, 601–609 (1994).
Cook,W.J., Jeffrey, L.C., Sullivan, M.L., and Vierstra, R.D., Three-
dimensional structure of a ubiquitin-conjugating enzyme (E2),
J. Biol. Chem. 267, 15116–15121 (1992).
DeMartino, G.N. and Gillette,T.G., Proteasomes: machines for all
reasons, Cell 129, 659–662 (2007).
Deshaies, R.J. and Joazeiro, C.A.P., RING domain E3 ubiquitin
ligases, Annu. Rev. Biochem. 78, 399–434 (2009).
Dye, B.T. and Schulman, B.A., Structural mechanisms underlying
posttranslational modification by ubiquitin-like proteins,
Annu. Rev. Biophys. Biomol. Struct. 36, 131–150 (2007).
Finley, D., Recognition and processing of ubiquitin–protein conju-
gates by the proteasome, Annu.Rev.Biochem. 78, 477–513 (2009).
Glickman, M.H. and Ciechanover, A., The ubiquitin-proteasome
proteolytic pathway: destruction for the sake of construction,
Physiol. Rev. 82, 373–428 (2002).
Hershko, A. and Ciechanover, A., The ubiquitin system, Annu.
Rev. Biochem. 67, 425–479 (1998).
Jentsch, S. and Pyrowalakis, G. Ubiquitin and its kin: how close
are the family ties, Trends Cell Biol. 10, 335–342 (2003). [Dis-
cusses Ubls.]
Liu, F. and Walters, K.J., Multitasking with ubiquitin through mul-
tivalent interactions, Trends Biochem. Sci. 35, 352–360 (2010).
Löwe, J., Stock, D., Jap, B., Zwicki, P., Baumeister, W., and Huber,
R., Crystal structure of the 20S proteasome from the archeon T.
acidophilum at 3.4 Å resolution, Science 268, 533–539 (1995);
and Groll, M., Ditzel, L., Löwe, J., Stock, D., Bochtler, M., Bar-
tunik, H.D., and Huber, R., Structure of 20S proteasome from
yeast at 2.4 Å resolution, Nature 386, 463–471 (1997).
Manchado, E., Eguren, M., and Malumbres, M.,The anaphase-pro-
moting complex/cyclosome (APC/C): cell-cycle-dependent and
-independent functions, Biochem. Soc.Trans. 38, 65–71 (2010).
Mukhopadhyay, D. and Riezman, H., Proteosome-independent
functions of ubiquitin in endocytosis and signaling, Science
315, 201–205 (2007).
Navon, A. and Ciechanover, A., The 26S proteosome: from basic
mechanism to drug targeting, J. Biol. Chem. 284, 33713–33718
(2009).
Page, A.M. and Hieter, P.,The anaphase-promoting complex: new
subunits and regulators, Annu. Rev. Biochem. 68, 583–609
(1999).
Pickart, C.M., Back to the future with ubiquitin, Cell 116, 181–190
(2004); and Mechanisms underlying ubiquitination, Annu. Rev.
Biochem. 70, 503–533 (2001).
Schwartz, A.L. and Ciechanover, A., Targeting proteins for de-
struction by the ubiquitin system: implication for human
pathobiology, Annu. Rev. Pharmacol.Toxicol. 49, 73–96 (2009).
Senahdi,V.-J., Bugg, C.E.,Wilkinson, K.D., and Cook,W.J., Three-
dimensional structure of ubiquitin at 2.8 Å resolution, Proc.
Natl.Acad. Sci. 82, 3582–3585 (1985).
Skaug, B., Jiang, X., and Chen, Z.J.,The role of ubiquitin in NF-B
regulatory pathways, Annu. Rev. Biochem. 78, 769–796 (2009).
Sousa, M.C.,Trame, C.B., Tsuruta, H.,Wilbanks, S.M., Reddy, V.J.,
and McKay, D.B., Crystal and solution structures of an HslUV
protease–chaperone complex, Cell 103, 633–643 (2000).
Unno, M., Mizushima, T., Morimoto, Y., Tomisugi, Y., Tanaka, K.,
Yasuoka, N., and Tsukihara, T., The structure of the mam-
malian proteasome at 2.75 Å resolution, Structure 10, 609–618
(2002).
VanDemark, A.P. and Hill, C.P., Structural basis of ubiquitylation,
Curr. Opin. Struct. Biol. 12, 822–830 (2002).
Varshavsky, A., Regulated protein degradation, Trends Biochem.
Sci. 30, 283–286 (2005).
Varshavsky,A.,Turner,G., Du, F., and Xie,Y.,The ubiquitin system
and the N-end rule, Biol. Chem. 381, 779–789 (2000).
Voges, D., Zwickl, P., and Baumeister, W., The 26S proteasome: a
molecular machine designed for controlled proteolysis, Annu.
Rev. Biochem. 68, 1015–1068 (1999).
Wang, J., Hartling, J.A., and Flanagan, J.M., The structure of ClpP
at 2.3 Å resolution suggests a model for ATP-dependent prote-
olysis, Cell 91, 447–456 (1997).
Whitby, F.G., Masters, E.I. Kramer, L., Knowlton, J.R., Yao, Y.,
Wang, C.C., and Hill, C.P., Structural basis for the activation of
20S proteasomes by 11S regulators, Nature 408, 115–120
(2000); and Förster, A.,Whitby, F.G., and Hill, C.P., The pore of
activated 20S proteasomes has an ordered 7-fold symmetric
conformation, EMBO J. 22, 4356–4354 (2003).
Zheng, N., et al., Structure of the Cul1–Rbx1–Skp1–F-box
Skp2
SCF
ubiquitin ligase complex,Nature 41, 703–709 (2002);Schulman,
B.A., et al., Insights into SCF ubiquitin ligases from the struc-
ture of the Skp1–Skp2 complex, Nature 408, 381–386 (2000);
and Zheng, N., Wang, P., Jeffrey, P.D., and Pavletich, N.P., Struc-
ture of a c-Cbl–UbcH7 complex: RING domain function in
ubiquitin-protein ligases, Cell 102, 533–539 (2000).
Zwickl, P., Seemüller, E., Kapelari, B., and Baumeister, W., The
proteasome: A supramolecular assembly designed for con-
trolled proteolysis, Adv. Prot. Chem. 59, 187–222 (2002).
JWCL281_c32_1338-1428.qxd 9/7/10 2:24 PM Page 1427
21. All cells contain an enzyme called peptidyl–tRNA
hydrolase, and cells that are deficient in this enzyme grow very
slowly. What is the probable function of this enzyme and why is
it necessary?
22. An antibiotic named fixmycin, which you have isolated
from a fungus growing on ripe passion fruit, is effective in curing
several types of sexually transmitted diseases. In characterizing
fixmycin’s mode of action, you have found that it is a bacterial
translational inhibitor that binds exclusively to the large subunit
of E. coli ribosomes.The initiation of protein synthesis in the pres-
ence of fixmycin results in the generation of dipeptides that re-
main associated with the ribosome. Suggest a mechanism of
fixmycin action.
23. Heme inhibits protein degradation in reticulocytes by al-
losterically regulating ubiquitin-activating enzyme (E1). What
physiological function might this serve?
24. Genbux Inc., a biotechnology firm, has cloned the gene
encoding an industrially valuable enzyme into E. coli such that the
enzyme is produced in large quantities. However, since the firm
wishes to produce the enzyme in ton quantities, the expense of iso-
lating it would be greatly reduced if the bacterium could be made
to secrete it. As a high-priced consultant, what general advice
would you offer to solve this problem?
1428 Chapter 32. Translation
Aminoacyl–tRNA Dissociation Constant (nM)
Ala–tRNA
Ala
6.2
Gln–tRNA
Ala
0.05
Gln–tRNA
Gln
4.4
Ala–tRNA
Gln
260
Source: LaRiviere, F.J.,Wolfson, A.D., and Uhlenbeck, O.C.,
Science 294, 167 (2001).
Indicate how the mutant could have arisen and give the base se-
quences, as far as possible, of the mRNAs specifying the two pep-
tides. Comment on the function of the peptide in the protein.
4. Explain why the various classes of mutations can reverse a
mutation of the same class but not a different class.
5. Which amino acids are specified by codons that can be
changed to an amber codon by a single point mutation?
6. The mRNA specifying the chain of human hemoglobin
contains the base sequence
The C-terminal tetrapeptide of the normal chain, which is spec-
ified by part of this sequence, is
In hemoglobin Constant Spring,the corresponding region of the
chain has the sequence
Specify the mutation that causes hemoglobin Constant Spring.
7. Explain why a minimum of 32 tRNAs are required to trans-
late the “standard” genetic code.
8. Draw the wobble pairings not in Fig. 32-25a.
9. A colleague of yours claims that by exposing E. coli to
HNO
2
she has mutated a tRNA
Gly
to an amber suppressor. Do you
believe this claim? Explain.
*10. Deduce the anticodon sequences of all suppressors listed
in Table 32-6 except UGA-1 and indicate the mutations that
caused them.
11. How many different types of macromolecules must be
minimally contained in a cell-free protein synthesizing system
from E. coli? Count each type of ribosomal component as a differ-
ent macromolecule.
12. Why do oligonucleotides containing Shine–Dalgarno se-
quences inhibit translation in prokaryotes? Why don’t they do so
in eukaryotes?
13. Why does m
7
GTP inhibit translation in eukaryotes? Why
doesn’t it do so in prokaryotes?
14. What would be the distribution of radioactivity in the com-
pleted hemoglobin chains on exposing reticulocytes to
3
H-labeled
leucine for a short time followed by a chase with unlabeled leucine?
15. Design an mRNA with the necessary prokaryotic control
sites that codes for the octapeptide Lys-Pro-Ala-Gly-Thr-Glu-
Asn-Ser.
*16. Indicate the translational control sites in and the amino
acid sequence specified by the following prokaryotic mRNA.
5¿-CUGAUAAGGAUUUAAAUUAUGUGUCAAUCACGA-
AUGCUAAUCGAGGCUCCAUAAUAACACUU CGAC-3¿
-Ser-Lys-Tyr-Arg-Gln-Ala-Gly-
p
-Ser-Lys-Tyr-Arg
p
UCCAAAUACCGUUAAGCUGGA
p
17. What is the energetic cost, in ATP equivalents, for the E.
coli synthesis of a polypeptide chain of 100 residues starting from
amino acids and mRNA? Assume that no losses are incurred as a
result of proofreading.
18. It has been suggested that Gly–tRNA synthetase does not
require an editing mechanism.Why?
19. Explain why prokaryotic ribosomes can translate a circu-
lar mRNA molecule,whereas eukaryotic ribosomes normally can-
not, even in the presence of the required cofactors.
20. EF-Tu binds all aminoacyl–tRNAs with approximately
equal affinity so that it can deliver them to the ribosome with the
same efficiency. Based on the experimentally determined binding
constants for EF-Tu and correctly charged and mischarged
aminoacyl–tRNAs (see table), explain how the tRNA–EF-Tu
recognition system could prevent the incorporation of the wrong
amino acid during translation.
JWCL281_c32_1338-1428.qxd 8/19/10 10:06 PM Page 1428

A
A77, 1116
AAA
domain, 1184–1185
AAA
family of ATPases, 1197, 1206
AAA
proteins, 1197–1198
AADP
(3-aminopyridine adenine
dinucleotide phosphate), 1125
A antigen, 415
aaRSs, see Aminoacyl-tRNA synthetase
Abasic sites, 1218
Abbé refractometer, 156
AB blood type, 22
ABCA1 (ATP-cassette binding
protein A1), 458
ABC transporters, 766–768, 767F
Abe (abequose), 379F
Abelson, J., 1312, 1351
Abequose (Abe), 379F
A (amyloid- protein), 311–312
A precursor protein (PP), 311–312
Ab initio methods, 305
Abl, 708
A blood type, 22
ABO blood group system, 22, 414–415
Abortive initiation, 1267–1268
Abrin, 1395T
Absolute configuration, 74–75
Absolute rate theory, 484–487
Absorbance:
in Beer–Lambert law, 90
of DNA and nucleic acids, 90, 92, 92F
Absorbance spectrum, 92
AC, see Adenylate cyclase
ac
4
C (N
4
-acetylcytidine), 1347F
ACAT, see Acyl-CoA:cholesterol
acyltransferase
ACC (acetyl-CoA carboxylase), 962–964
Accepted names, enzymes, 479
Acceptor, tRNA, 1346
Acceptor control, 862
Accessory pigments, 908–909
Accessory proteins:
G proteins, 692
GroEL/ES system, 294–302
peptidyl prolyl cis-trans isomerases, 292
protein disulfide isomerases, 290–292,
290F, 291F
Accommodation, 1388
ACE (angiotensin converting
enzyme), 547F
ACE inhibitors, 547F
Acetal, 520F
Acetaldehyde, re and si faces, 77F
Acetaminophen, 544–545, 544F, 1000
Acetate, as cholesterol precursor, 975F,
976–977
Acetate ion, 45, 45T, 48F
Acetic acid:
creation in Miller-Urey experiments, 32T
distribution curve, 48, 48F
titration curve, 47, 47F
Acetimidoquinine, 544–545, 544F
Acetoacetate, 959, 960F
decarboxylation, 510–511
and degradation of leucine/lysine,
1040–1041
and degradation of
phenylalanine/tyrosine, 1043–1047
degradation of tryptophan, 1041–1042
Acetoacetyl-CoA, 960
-Aceto--hyroxybutyrate, 1075
Acetolactate, 1075
Acetone, 959
and cell wall lysis, 130
dielectric constant and dipole
moment, 43T
protein solubility in, 134
Acetonel, 679F
Acetonyl-CoA, 806–807
Acetylcholine (ACh), 527, 778–780
Acetylcholine receptor (AChR),
780–781, 780F
Acetylcholinesterase, 781–782, 782F
diffusion-limited, 490, 491
inactivation by DIPF, 527
Michaelis–Menten kinetic constants, 489T
Acetyl-CoA, 561, 561F
in citric acid cycle, 789, 792–796, 792F,
795F, 816–819
and degradation of leucine/lysine,
1040–1041
and integration of pathways, 1090
in ketogenesis, 960–961, 960F
pyruvate carboxylase regulation, 873–874
Acetyl-CoA acetyltransferase, 960–961
Acetyl-CoA carboxylase (ACC), 962–964
Acetyl-coenzyme A, see Acetyl-CoA
N
4
-Acetylcytidine (ac
4
C), 1347F
N-Acetyl-D-galactosamine, 392F
Acetyl-dihydrolipamide-E2, 795
N-Acetylglucosamine, see NAG
N-Acetylglutamate, 1025, 1029, 1070F, 1071
N-Acetylglutamate-5-semialdehyde,
1070F, 1071
N-Acetylglutamate synthase, 1029,
1070F, 1071
N-Acetylimidazolium, 513
ε-N-Acetyllysine, 79F
N-Acetylmuramic acid, see NAM
N-Acetylneuraminic acid, see NANA
N-Acetylneuraminidate, 392F
Acetyl phosphate, 580, 607
Acetylsalicylic acid, 998
N-Acetylserine, 79F
O-Acetylserine, 1071
Acetyl transferase (AT), 967, 968
N-Acetylxylosamine (XylNAc) residue, 523
ACh, see Acetylcholine
AChR, see Acetylcholine receptor
Acids, 45
dissociation constants, 45–47, 46T
polyprotic, 48–50, 49F
Acid-base buffers, 48
Acid-base catalysis, 506–510
Acid-base indicators, 51
Acid-base reactions, 45–47
and amino acids, 72–73
for protein assays, 131
and standard state conventions, 60
Acid chain hypothesis, 446
Acidic solutions, 47
Acidosis, 955
Ackee fruit, 948
Ackers, G., 353
Aconitase, 789, 790F, 808–809, 808F
cis-Aconitate, 789
ACP (acyl-carrier protein), 961
ACP synthase, 961
Acquired immune deficiency syndrome,
see AIDS
Acridine orange, 157
Acromegaly, 685
Acrosomal protease, 525T
Acrylamide, 147, 147F
ACTH, see Adrenocorticotropic hormone
Actin, 10, 10F, 301, 857
in erythrocyte membranes, 413F, 414
helical symmetry, 269
Actinomycin D, 79, 1272, 1272F
Action potentials, 775–777, 775F, 776F
Activated complex, 485
Activation barrier, 486
Activators, 96, 351
Active, rolling mechanism (DNA
unwinding), 1185–1186, 1186F
Active transport, 420, 747
ATP-driven, 758–768
ion-driven, 768–771
Activity, 59–61
Activity coefficient, 61
Acute intermittent porphyria, 1056
Acute lymphoblastic leukemia, 1034
Acute pancreatitis, 537
Acyl-carnitine, 946
Acyl-carrier protein (ACP), 961
Acyl-CoA:cholesterol acyltransferase
(ACAT), 455, 984
Acyl-CoA dehydrogenase, 948F
I-1
INDEX
Page references in bold face refer to a major discussion of the entry. Positional and configurational designations in chemical
names (e.g., 3-, , N-, p-, trans, D-, sn-) are ignored in alphabetizing. Numbers and Greek letters are otherwise alphabetical as if
they were spelled out.
Note: Chapters 33–35, which are only available on our book companion website, www.wiley.com/college/voet, are separately
indexed at this website.
JWCL281_ndx_I1-I53.qxd 10/28/10 12:02 PM Page I-1
I-2 Index
Acyl-CoA dehydrogenase (AD), 947, 948
Acyl-CoA oxidase, 958
Acyl-CoA synthases, 945–946
Acyl-dihydroxyacetone phosphate, 971
1-Acyldihydroxyacetone phosphate, 1007
Acyl-enzyme intermediate, 533
2-Acylglycerol, 941
1-Acylglycerol-3-phosphate acyltransferase,
971
Acyl group transfer, 565
Acyl phosphates, 580
N-Acylsphingosine phosphocholine, 1008
Acyltransferase (AT), 968
AD, see Acyl-CoA dehydrogenase;
Alzheimer’s disease
ADA, see Adenosine deaminase
ada gene, 1216
Adair, G., 348
Adair constants, 349
Adair equation, 348–349
Ada protein, 1216
Adapter proteins, see APs
Adaptive enzymes, 1261
Adaptor, 705–711, 709
Adaptor hypothesis, 1345
ADAR1, 1150–1151, 1150F
and RNA editing, 1322–1323, 1323F
ADAR2:
and RNA editing, 1322, 1323F
Addison’s disease, 681
Adducin, 412F–413F
Ade, see Adenine
Adenine, 18, 83, 83T. See also Watson-Crick
base pairs
and genetic code, 100T, 1343T
IR spectra of derivatives, 1155F
modified nucleosides of, 1347F
and point mutations, 1339–1340
Adenine phosphoribosyltransferase
(APRT), 1114
Adenohypophysis, 682
Adenosine, 83T, 578, 1130F
Adenosine-3¿,5¿-cyclic monophosphate,
see cAMP
Adenosine-5¿-(-amino) diphosphate
(AMPPN), 763, 763F
Adenosine-5¿-(,-imido) triphosphate,
see AMPPNP
Adenosine-5¿-(,-methylene) triphosphate
(AMPPCP), 763, 763F, 764
Adenosine-5¿-phosphosulfate, 183
Adenosine-5¿-phosphosulfate (APS)
kinase, 1072
Adenosine deaminase (ADA), 1130F, 1131
Adenosine diphosphate, see ADP
Adenosine monophosphate, see AMP
Adenosine triphosphate, see ATP
S-Adenosylhomocysteine, 1034
S-Adenosylmethionine, see SAM
Adenyl-3¿,5¿-uridyl-3,¿5¿-cytidyl-3,¿5¿-
guanylyl-3¿-phosphate, 84F
Adenylate cyclase (AC), 653, 697–698, 1286
Adenylate kinase (AK), 582
and glycolysis control in muscle,
627–628, 628F
X-ray structure of porcine, 254, 255F
Adenylic acid, see AMP
Adenylosuccinate, 1112
Adenylosuccinate lyase (PurB), 1109F,
1111, 1112
Adenylosuccinate synthetase, 1112F
Adenyltransferase, 1069, 1071
ADH, see Alcohol dehydrogenase;
Antidiuretic hormone
Adhesins, 382
Adipocytes, 388–389, 388F, 1096, 1097F
Adiponectin, 1096, 1097F
Adiponectin receptors, 1096
Adipose tissue, 389
brown, 860
glucose uptake regulation in, 751F
and other organs, 1091F, 1093
A-DNA, 1146F, 1147F, 1148–1149, 1148T, 1151
AdoCbl, see 5¿-Deoxyadenosylcobalamin
AdoMet, see SAM
ADP (adenosine diphosphate), 17, 82
in Calvin cycle, 931
in catabolism, 561, 561F
in citric acid cycle, 816–817
in glycolysis, 594F, 595
in GroEL/ES system, 295–302
RuvB binding of, 1231
and thermodynamics of life, 578–583
ADP/ATP carrier, 771
ADP-glucose, 645
ADP-glucose pyrophosphorylase, 931
ADPNP, see AMPPNP
ADP-ribosylated diphthalamide residue,
eEF2, 1398
ADP-ribosylation, 1398
ADP-ribosylation factor, see ARF1
Adrenal glands, 672F
Adrenaline, see Epinephrine
-Adrenergic receptors, 664, 680
Adrenocortical steroids, 680
Adrenocorticotropic hormone (ACTH),
673T, 682–683
Adrenoleukodystrophy (ALD), 767, 958
-Adrenoreceptors, 660, 680
Adriamycin, 1169
Adsorption chromatography, 132T, 135,
143–144
Adult-onset diabetes, 1102–1103
Aequorea victoria, 120
Aerobes, 6. See also specific organisms
Aerobic fermentation, 594F
Aerobic metabolism, 864–865
Aerobic oxidation, 594F
Affinity chromatography, 132T
ion exchange, 136
metal chelation, 145
mRNA, 158
for proteins, 118, 141–143, 141F
Affinity labeling, 527
Aflatoxin B
1
, 1225
AFM (atomic force microscopy), 376
Africa, sickle-cell anemia in, 188, 343
African clawed toad, 1202
African sleeping sickness, 799
Agard, D., 456
Agarose gel electrophoretogram, 106F
Agarose gels:
in affinity chromatography, 142, 142F
in gel electrophoresis, 147–149
in gel filtration chromatography,
139, 140T, 141
in ion exchange chromatography, 137
AGC family of protein kinases, 730
A
gene, 1261–1262, 1264
a gene (fruit fly), 25F
Agglutinin, 366
Aggrecan, 373–375, 373T, 374F
Aging, telomere length and, 1210–1211
Aglycone, 366
AGO, see Argonaute
Agonist, 539, 680
Agouti related peptide (AgRP), 1099
Agrin, 373T
Agrobacterium radiobacter, 106F
AgRP (agouti related peptide), 1099
AICAR, see 5-Aminoimidazole-4-
carboxamide ribotide
AICAR transformylase (PurH), 1109F, 1111
AIDS (acquired immune deficiency
syndrome), 123, 545–546. See also HIV
AIR, see 5-Aminoimidazole ribotide
AIR carboxylase, 1109F, 1111
AIR synthetase (PurM), 1109F, 1110
AK, see Adenylate kinase
AKAPs (A-kinase anchoring proteins), 714
Akey, C., 424
A-kinase anchoring proteins (AKAPs), 714
Akt, see Protein kinase B
Akt1/PKB, 734
Akt2/PKB, 734
Akt3/PKB, 734
Ala, see Alanine
ALA (-aminoleuvulinic acid), 1048
ALAD (-aminoleuvulinic acid
dehydratase), 1048
Alanine:
biosynthesis, 1067–1073
degradation of, 1030–1034
and degradation of tryptophan,
1041–1042
genetic codes for, 100T, 1343T
half-life, 1413T
helix/ sheet propensity, 302T, 304
hydropathy scale for, 264T
in Miller-Urey experiments, 32T
in native unfolded proteins, 283
nonessential amino acid, 1065T
nonpolar side chain, 70
Ramachandran diagram, 224F, 225
structure and general properties, 68T, 71F
(S)-Alanine, 76, 77F
-Alanine:
creation in Miller-Urey experiments, 32T
in pyrimidine catabolism, 1136
L-Alanine, 76, 77F
Alanine tRNA, 176, 1345
AlaRS, 1351
ALAS-1, 1056
ALAS-2, 1056
ALA synthase, 1056
Ala-tRNA
Cys
, 1359
Albumin, 154F, 542, 677. See also
Serum albumin
Alcaptonuria, 569
Alcohols:
effect on protein denaturation, 265
reactions to form hemiacetals and
hemiketals, 361F
Alcohol dehydrogenase (ADH), 479
in fermentation, 618–619
and methanol oxidation, 505
molecular mass of, 140F
stereospecificity of, 470–473
Alcoholic fermentation, 470, 593–594,
616–619, 616F
Alcohol:NAD
oxidoreductase, 479
ALD, see Adrenoleukodystrophy
Aldehydes, hemiacetal formation of, 361F
Aldehyde dehydrogenase, 447F
Alditols, 364
Aldohexoses, 360F
Aldolase, 568
binding to erythrocyte membrane, 412
in glycolysis, 596F, 600–603, 600F, 602F
half-life, 1408T
molecular mass determination by gel
filtration chromatography, 140F
sedimentation coefficient, 154F
Aldol condensation, 568
Aldol histidine, 239F
JWCL281_ndx_I1-I53.qxd 10/28/10 12:02 PM Page I-2
