Voet D., Voet Ju.G. Biochemistry
Подождите немного. Документ загружается.

Phillips first determined the X-ray structure of met re-
pressor in the absence of DNA. Model building studies
aimed at elucidating how met repressor binds to its palin-
dromic target DNA assumed that the 2-fold rotation axes
of both molecules would be coincident, as they are in all
prokaryotic protein–DNA complexes of known structure.
There were, consequently, two reasonable choices: (1) The
protein could dock to the DNA with the above pairs of 
ribbons entering successive major grooves; or (2) a symme-
try-related pair of protruding ␣ helices on the opposite face
of the protein could do so in a manner resembling the way
in which the recognition helices of HTH motifs interact
with DNA. A variety of structural criteria suggested that
the ␣ helices make significantly better contacts with the
DNA than do the  ribbons.Thus, the observation that it is,
in fact, the  ribbons that bind to the DNA provides an im-
portant lesson: The results of model building studies must
be treated with utmost caution. This is because our impre-
cise understanding of the energetics of intermolecular in-
teractions (Sections 8-4 and 29-2) prevents us from reliably
predicting how associating macromolecules conform to
one another. In the case of the met repressor, unpredicted
mutual structural accommodations of the protein and
DNA yielded a significantly more extensive interface than
had been predicted by simply docking the uncomplexed
Met repressor to canonical B-DNA.
The numerous prokaryotic transcriptional regulators
of known structure either contain an HTH motif or pairs
of  ribbons like the met repressor (although numerous
prokaryotic DNA-binding proteins, including CAP, con-
tain an elaboration of the HTH motif known as the
winged helix motif in which two protein loops, one of
which contacts the DNA’s minor groove, flank the HTH
recognition helix like the wings of a butterfly). Moreover,
most of these proteins are homodimers that bind to palin-
dromic or pseudopalindromic DNA target sequences.
However, eukaryotic transcription factors, as we shall see
in Section 34-3B, employ a much wider variety of struc-
tural motifs to bind their target DNAs, many of which
lack symmetry.
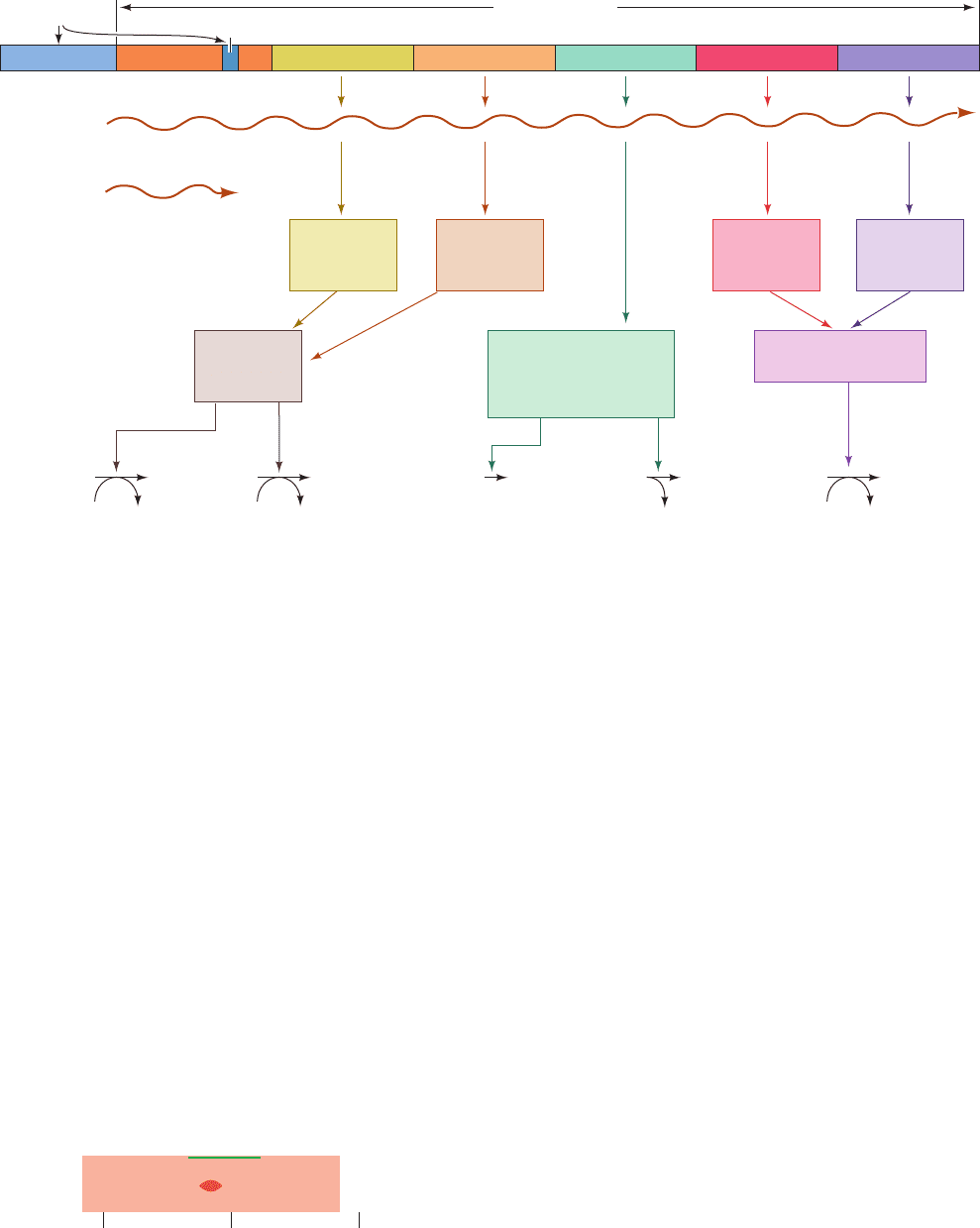
E. araBAD Operon: Positive and Negative Control
by the Same Protein
Humans neither metabolize nor intestinally absorb the
plant sugar
L-arabinose. Hence, the E. coli that normally in-
habit the human gut are periodically presented with a ban-
quet of this pentose. Three of the five E. coli enzymes that
Section 31-3. Control of Transcription in Prokaryotes 1291
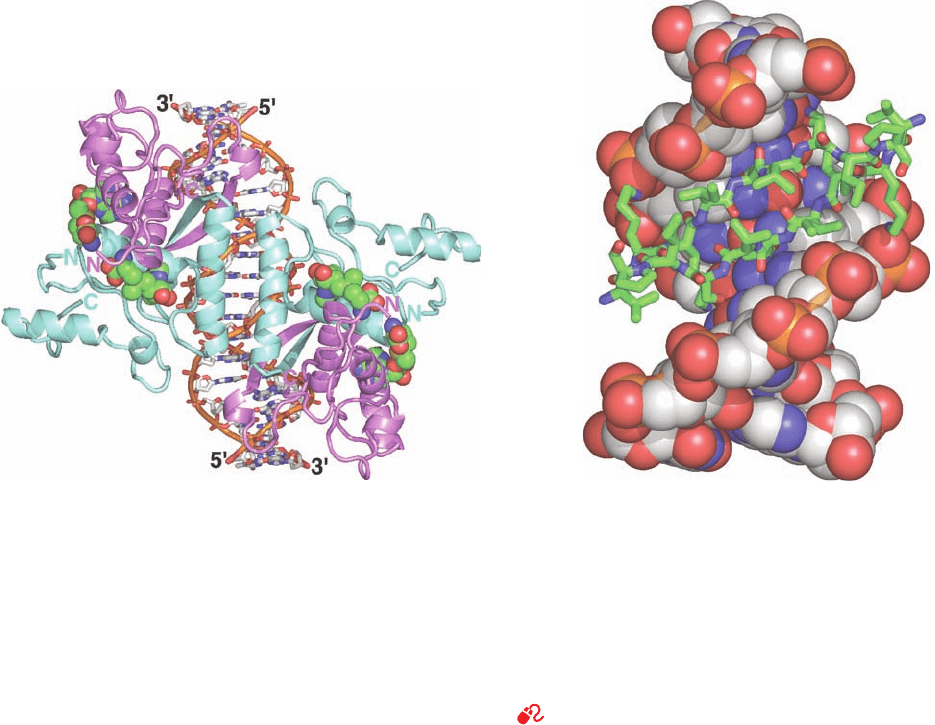
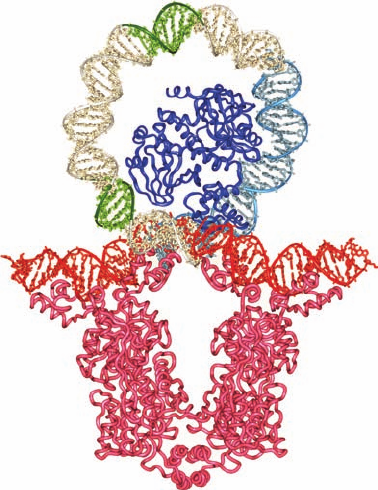
Figure 31-35 X-ray structure of the E. coli met
repressor–SAM–operator complex. (a) The overall structure of
the complex as viewed along its 2-fold axis of symmetry. The
self-complementary 18-bp DNA is drawn in stick form, and
SAM, which must be bound to the repressor for it to also bind
DNA, is shown in space-filling form with the DNA C white,
SAM C green, N blue, O red, P orange, and S yellow. The DNA
binds four identical 104-residue repressor subunits. Pairs of
subunits (light cyan and lavender) form symmetric dimers in
which each subunit donates one strand of the 2-stranded
antiparallel  ribbon that is inserted in the DNA’s major groove
(upper left and lower right).Two such dimers pair across the
complex’s 2-fold axis via their antiparallel N-terminal helices,
which contact one another over the DNA’s minor groove. (b)
Detailed view of the lower half of Part a showing the 2-stranded
antiparallel  ribbon (residues 21–29) inserted into the DNA’s
major groove, as viewed along its local 2-fold axis (rotated
relative to Part a by 50° about the vertical axis).The DNA is
shown in space-filling form and the polypeptide chains are
drawn in stick form with C green. [Based on an X-ray structure
by Simon Phillips, University of Leeds, U.K. PDBid 1CMA.]
See Interactive Exercise 41
(b)
(a)
JWCL281_c31_1260-1337.qxd 10/19/10 11:26 AM Page 1291

metabolize arabinose are products of the catabolite re-
pressible araBAD operon (Fig. 31-36).
The araBAD operon, as Robert Schleif has shown, con-
tains, moving upstream from its transcriptional start site,
the araI, araO
1
, and araO
2
control sites (Fig. 31-37a). The
araI site (I for inducer) consists of two closely similar 17-bp
half-sites, araI
1
and araI
2
, that are direct repeats separated
by 4 bp and are oriented such that araI
2
, which overlaps the
⫺35 region of the araBAD promoter, is downstream of
araI
1
. Likewise, araO
1
consists of two directly repeating
half-sites, O
1L
and O
1R
. Intriguingly, however, araO
2
con-
sists of a single half-site that is located in a noncoding up-
stream region of the araC gene (see below), at position
⫺270 relative to the araBAD start site.
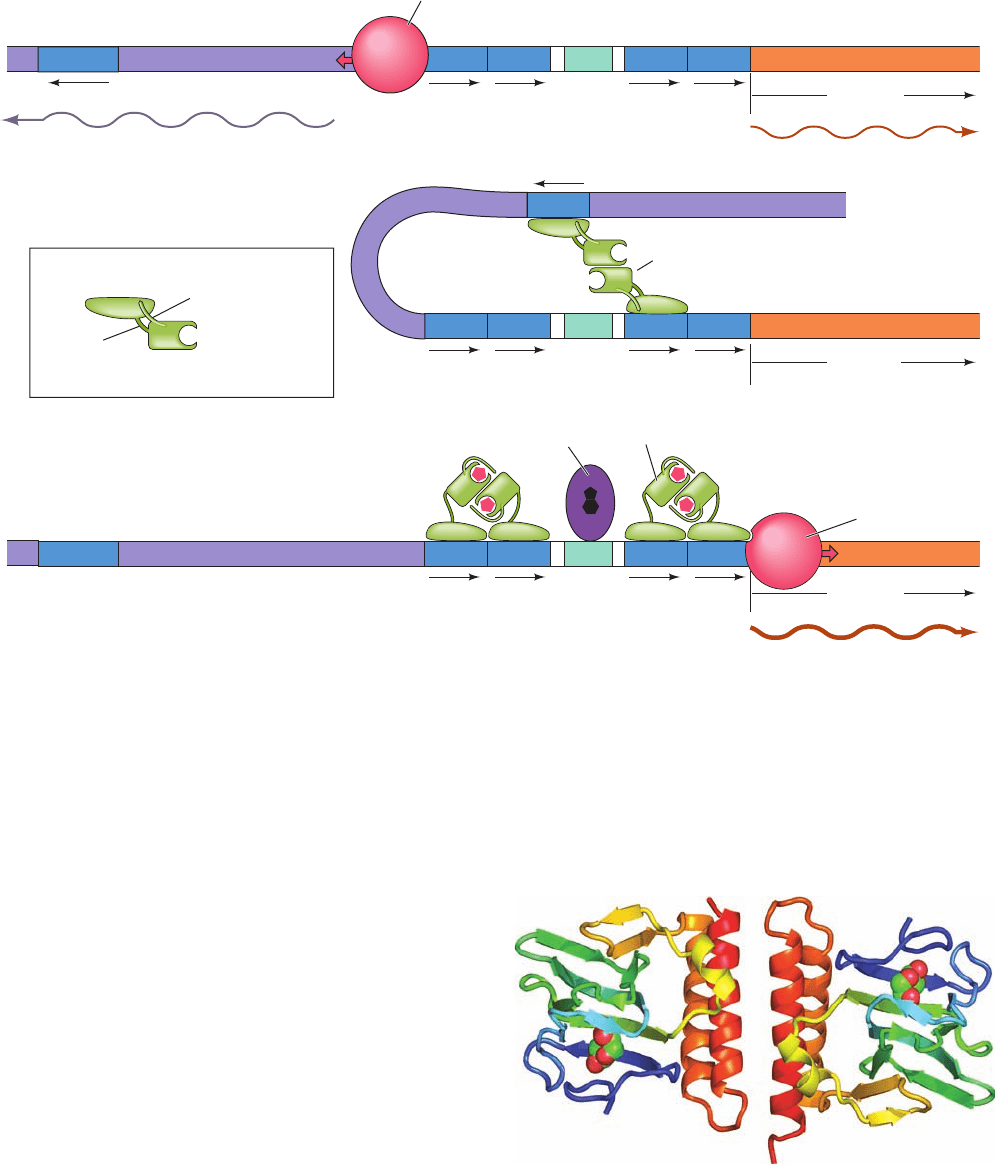
The transcription of the araBAD operon is regulated by
both CAP–cAMP and the arabinose-binding protein
AraC. Each 292-subunit of the homodimeric AraC con-
sists of an N-terminal, arabinose-binding, dimerization do-
main (residues 1–170) connected via a flexible linker to a
C-terminal DNA-binding domain (residues 178–292).
Regulation of the araBAD operon occurs as follows
(Fig. 31-37):
1. In the absence of AraC, RNA polymerase initiates
transcription of the araC gene in the direction away from
its upstream neighbor, araBAD. The araBAD operon is ex-
pressed at a low basal level.
2. When AraC is present, but neither arabinose nor
CAP–cAMP (high glucose),AraC binds to araO
2
and araI
1
via two HTH motifs in each of its subunits. The binding of
AraC to araI
1
prevents RNAP from initiating transcription
of the araBAD operon (negative control).A series of dele-
tion mutations indicate that the presence of araO
2
is also
required for the repression of araBAD. The remarkably
large 210-bp separation between araO
2
and araI
1
therefore
strongly suggests that the DNA between them is looped
such that a dimeric molecule of AraC protein simultane-
ously binds to both araO
2
and araI
1
.This is corroborated by
the observation that the level of repression is greatly
diminished by the insertion of 5 bp (half a turn) of DNA
between these two sites, thereby transferring araO
2
to the
opposite face of the DNA helix relative to araI
1
in the
putative loop.Yet, the insertion of 11 bp (one turn) of DNA
has no such effect. Moreover, looping does not readily oc-
cur unless the DNA is supercoiled, which presumably
drives the looping process. The AraC dimer also binds to
araO
1
, the operator of the araC gene, so as to block the
transcription of araC but only at high concentrations.Thus,
it is likely that DNA looping itself represses the transcrip-
tion of araC. In either case, the expression of araC is
autoregulatory.
3. When arabinose is present, it allosterically induces
the AraC subunit bound to araO
2
to instead bind to araI
2
.
This activates RNAP to transcribe the araBAD genes (pos-
itive control). When the cAMP level is high (low glucose),
CAP–cAMP, whose presence is required to achieve the
maximum level of transcriptional activation, binds to a site
between araO
1
and araI
1
, where it functions to help break
the loop between araO
2
and araI
1
and hence to increase the
affinity of AraC for araI
2
.The orientation of araO
1
with re-
spect to araC is opposite to that of araI with respect to
araBAD, and hence the binding of AraC–arabinose at
araO
1
blocks RNAP binding at the araC promoter, that is,
it represses the expression of AraC.
If the araI
2
subsite is mutated so as to increase AraC’s
affinity for it, arabinose is no longer required for transcrip-
tional activation.This suggests that arabinose does not con-
formationally transform AraC to an activator but, rather,
weakens its binding affinity for araO
2
. If the araI site is
1292 Chapter 31. Transcription
Activator
L
Repressor
-Arabinose
araC mRNA
araC araO
2
araO
1
araI araB araA araD
Structural genes
Control sites
Regulatory gene
araBAD mRNA
L-ribulokinase
L-ribulose-5-P
epimerase
L-arabinose
isomerase
permease
system
L-Arabinose
(external)
L-Arabinose
(internal)
L-Ribulose L-Ribulose-5-P D -Xylulose-5-P
ADPATP
CAP
Figure 31-36 A genetic map of the E. coli araC and araBAD operons. The map indicates the
proteins these operons encode and the reactions in which these proteins participate. The permease
system, which transports arabinose into the cell, is the product of the araE and araF genes,
which occur in two independent operons.The pathway product, xylulose-5-phosphate, is
converted, via the pentose phosphate pathway, to the glycolytic intermediates fructose-6-
phosphate and glyceraldehyde-3-phosphate (Section 23-4). [After Lee, N., in Miller, J.H. and
Rezinkoff, W.S. (Eds.), The Operon, p. 390, Cold Spring Harbor Laboratory Press (1979).]
JWCL281_c31_1260-1337.qxd 8/11/10 9:48 PM Page 1292
turned around or if it is moved upstream so that araI
2
does
not overlap the araBAD promoter, AraC cannot stimulate
transcription. Evidently, AraC activates RNAP through spe-
cific and relatively inflexible protein–protein interactions.
The X-ray structures of the N-terminal domain of AraC
(residues 2–178), in both the presence and the absence of
arabinose, were determined by Schleif and Cynthia Wol-
berger. In the presence of arabinose, this domain consists
of an 8-stranded  barrel followed by two antiparallel ␣
helices (Fig. 31-38). Two such domains associate via an an-
tiparallel coiled coil between each of their C-terminal
helices to form the protein’s dimerization interface. An
arabinose molecule binds in a pocket of each  barrel via a
network of direct and water-mediated hydrogen bonds
with side chains that line the pocket. Residues 7 to 18 of the
N-terminal arm lie across the mouth of the sugar-binding
pocket (residues 2–6 are disordered), thereby fully enclos-
ing the arabinose. The structure of the N-terminal domain
in the absence of arabinose is largely superimposable on
that in the complex with arabinose, with the exception that
Section 31-3. Control of Transcription in Prokaryotes 1293
araO
1R
araO
1L
araO
1R
araO
1L
araO
1R
araO
1L
(a) When AraC is absent,
araC is transcribed and
araBAD is transcribed
at a basal level
araO
araI
2
araBAD
araC
mRNA
araO
2
AraC
C-terminal
DNA-binding domain
N-terminal arm
Linker
Arabinose-binding
pocket
N-terminal
dimerization domain
araI
1
araI
1
araBAD
(b)
When cAMP and L-arabinose
are low, AraC represses
araBAD transcription
(c) When cAMP and L-arabinose
are
abundant, araBAD
transcription is activated
araO
2
araC
araI
2
araI
1
araI
2
araBAD
RNA polymerase
araBAD mRNA
araBAD mRNA (basal level)
2
RNA polymerase
CAP
araC
AraC–arabinose
CAP
CAP
CAP–cAMP
araC
Figure 31-37 The mechanism of araBAD regulation. (a) In
the absence of AraC, RNAP initiates the transcription of araC.
araBAD is also expressed but at a low basal level. (b) When
AraC is present, but not
L-arabinose or cAMP, AraC links
together araO
2
and araI
1
to form a DNA loop, thereby repressing
both araC and araBAD.(c) When AraC and
L-arabinose are
Figure 31-38 X-ray structure of E. coli AraC in complex with
L-arabinose. The homodimeric protein is viewed along its 2-fold
axis with each of its subunits colored in rainbow order from
N-terminus (blue) to C-terminus (red).The arabinose is drawn in
space-filling form with C green and O red. [Based on an X-ray
structure by Robert Schleif and Cynthia Wolberger, Johns
Hopkins University. PDBid 2ARC.]
both present and cAMP is abundant, the resulting AraC–
arabinose complex releases araO
2
and instead binds araI
2
,
thereby activating araBAD transcription. This process is
facilitated by the binding of CAP–cAMP. araC is repressed by
the binding of AraC–arabinose to araO
1
.
JWCL281_c31_1260-1337.qxd 8/11/10 9:48 PM Page 1293
Hinge helix
N-subdomain
C-subdomain
Tetramerization helix
C
N
Headpiece
Inducer-binding
pocket
the N-terminal arm is disordered, a not unexpected obser-
vation considering that it interacts with bound arabinose
via a series of hydrogen bonds.
How does arabinose binding induce the AraC subunit
bound at araO
2
to instead bind to araI
2
? Several lines of
evidence indicate that, in the absence of arabinose, AraC’s
N-terminal arm binds to its DNA-binding domain in a
way that favors loop formation: (1) the deletion of the
N-terminal arm beyond its sixth residue makes AraC act as
if arabinose is present; (2) mutations to surface residues on
the DNA-binding domain that presumably eliminate its
binding of the N-terminal arm also constitutively activate
AraC; and (3) mutations in the DNA-binding domain that
weaken the binding of arabinose to the protein, presum-
ably by strengthening the binding of the N-terminal arm,
can be suppressed by a second mutation in the N-terminal
arm or by the deletion of its five N-terminal residues. Evi-
dently, the binding of the N-terminal arms to the DNA-binding
domains in the absence of arabinose rigidifies the AraC
dimer such that it cannot simultaneously bind to the directly
repeated araI
1
and araI
2
and hence induce the transcription
of araBAD. This is corroborated by the observations that
(1) joining two AraC DNA-binding domains by flexible
polypeptide linkers yields proteins that behave like AraC
in the presence of arabinose, and (2) a construct consisting
of two double-stranded araI
1
half-sites flexibly connected
by a 24-nt segment of ssDNA binds wild-type AraC with an
affinity that is unaffected by arabinose.
F. lac Repressor II: Structure
Here we continue our discussions of the lac repressor, but
now in terms of the concepts learned in Sections 31-3C–E.
a. Loop Formation Is Important in the Expression
of the lac Operon
DNA loop formation, which is now known to occur in
numerous bacterial and eukaryotic systems, apparently
permits several regulatory proteins and/or regulatory sites
on one protein to simultaneously influence transcription
initiation by RNAP. In fact, the lac repressor has three bind-
ing sites on the lac operon: the primary operator (Fig. 31-
28), now known as O
1
, and two so-called pseudo-operators
(previously thought to be nonfunctional evolutionary fos-
sils), O
2
and O
3
, which are located 401 bp downstream and
92 bp upstream of O
1
(within the lacZ gene and overlap-
ping the CAP binding site, respectively). Müller-Hill deter-
mined the relative contributions of these various operators
to the repression of the lac operon through the construc-
tion of a set of eight plasmids: Each contained the lacZ
gene under the control of the natural lac promoter as well
as the three lac operators (O
1
, O
2
, and O
3
), which were ei-
ther active or mutagenically inactive in all possible combi-
nations. When all three operators are active, lacZ expres-
sion is repressed 1300-fold relative to when all three
operators are inactive.The inactivation of only O
1
results in
almost complete loss of repression whereas the inactiva-
tion of only O
2
or O
3
causes only a ⬃2-fold loss in repres-
sion. However, when O
2
and O
3
are both inactive, repres-
sion is decreased ⬃70-fold. These results suggest that effi-
cient repression requires the formation of a DNA loop be-
tween O
1
and either O
2
or O
3
. Indeed, such loop formation,
and/or the cooperativity of repressor binding arising from
it, appears to be a greater contributor to repression than
repressor binding to O
1
alone, which provides only 19-fold
repression.
b. The lac Repressor Is a Dimer of Dimers
Ponzy Lu and Mitchell Lewis determined the X-ray
structures of the lac repressor alone, in its complex with
IPTG, and in its complex with a 21-bp duplex DNA seg-
ment whose sequence is a palindrome of the left half of O
1
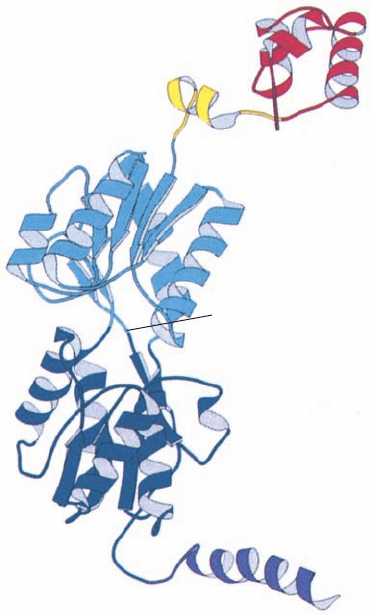
(Fig. 31-28). Each repressor subunit consists of five func-
tional units (Fig. 31-39): (1) an N-terminal DNA-binding
domain (residues 1–49) which is known as the “headpiece”
because it is readily proteolytically cleaved away from the
remaining still tetrameric “core” protein; (2) a hinge helix
(residues 50–58) that also binds to the DNA; (3 and 4) a
sugar-binding domain (residues 62–333) that is divided into
an N-subdomain and a C-subdomain; and (5) a C-terminal
tetramerization helix (residues 340–360).
1294 Chapter 31. Transcription
Figure 31-39 X-ray structure of the lac repressor subunit. The
DNA-binding domain (the headpiece), which contains an HTH
motif, is red, the DNA-binding hinge helix is yellow, the
N-subdomain of the sugar-binding domain is light blue, its
C-subdomain is dark blue, and the tetramerization helix is
purple. [Courtesy of Ponzy Lu and Mitchell Lewis, University
of Pennsylvania. PDBid 1LBI.]
JWCL281_c31_1260-1337.qxd 8/11/10 9:48 PM Page 1294
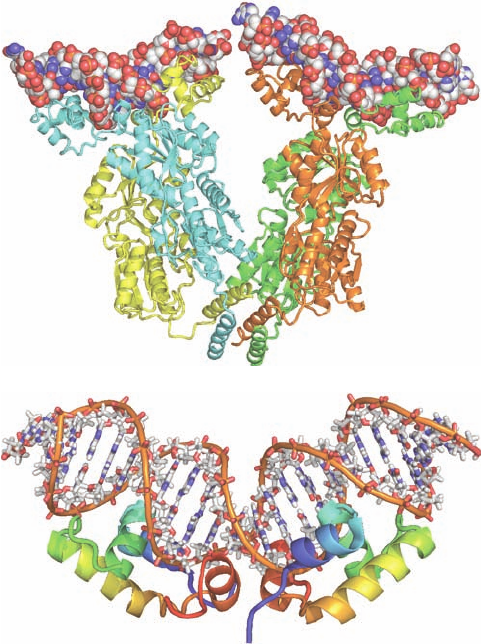
The lac repressor has an unusual quaternary structure
(Fig. 31-40a).Whereas nearly all homotetrameric nonmem-
brane proteins of known structure have D
2
symmetry
(three mutually perpendicular 2-fold axes; Fig. 8-65b), lac
repressor is a V-shaped protein that has only 2-fold symme-
try. Each leg of the V consists of a locally symmetric dimer
of closely associated repressor subunits. Two such dimers
associate rather tenuously, but with 2-fold symmetry, at the
base (point) of the V to form a dimer of dimers.
In the structures of lac repressor alone and that of its
IPTG complex, the DNA-binding domain is not visible,
apparently because the hinge region that loosely tethers it
to the rest of the protein is disordered. However, in the
DNA complex, in which one DNA duplex binds to each of
the two dimers forming the repressor tetramer, the DNA
domain forms a compact globule containing three helices,
the first two of which form a helix–turn–helix (HTH) mo-
tif. The two DNA-binding domains extending from each
repressor dimer (at the top of each leg of the V) bind in
successive major grooves of a DNA molecule via their
HTH motifs, much as is seen, for example, in the com-
plexes of 434 phage repressor and trp repressor with their
target DNAs (Figs. 31-32 and 31-34). The binding of the
lac repressor distorts the operator DNA such that it
bends away from the DNA-binding domain with an ⬃60
Å radius of curvature due to an ⬃45° kink at the center of
the operator that widens the DNA’s minor groove to over
11 Å and reduces its depth to less than 1 Å. These distor-
tions permit the now ordered hinge helix to bind in the
minor groove so as to contact the identically bound hinge
helix from the other subunit of the same dimer. NMR
structures by Robert Kaptein and Rolf Boelens reveal
that the DNA-binding domain, when cleaved from the re-
pressor, binds to the lac operator without distorting the
DNA, but that the DNA-binding domain together with
the hinge helix forms a complex with the lac operator in
which the hinge helix binds in the DNA’s distorted minor
groove (Fig. 31-40b) as in the X-ray structure. Thus, the
binding of the two hinge helices to the lac operator ap-
pears necessary for DNA distortion. The two DNA du-
plexes that are bound to each repressor tetramer are ⬃25
Å apart and do not interact.
The sugar-binding domain consists of two topologi-
cally similar subdomains that are bridged by three
polypeptide segments (Fig. 31-39).The two sugar-binding
domains of a dimer make extensive contacts (Fig. 31-
40a). IPTG binds to each sugar-binding domain between
its subdomains. This does not significantly change the
conformations of these subdomains, but it changes the
angle between them.Although the hinge helix is not visi-
ble in the IPTG complex, model building indicates that,
since the dimer’s two hinge helices extend from its sugar-
binding domains, this conformation change levers apart
these hinge helices by 3.5 Å such that they and their at-
tached HTH motifs can no longer simultaneously bind to
their operator half-sites. Thus, inducer binding, which is
allosteric within the dimer (has a positive homotropic ef-
fect; Section 10-4), greatly loosens the repressor’s grip on
the operator.
The C-terminal helices from each subunit, which are lo-
cated on the opposite end of each subunit from the DNA-
binding portion (at the point of the V), associate to form a
bundle of four parallel helices that holds together the two
repressor dimers, thereby forming the tetramer (Fig. 31-
40a). The allosteric effects of inducer binding within each
dimer are apparently not transmitted between dimers.
Section 31-3. Control of Transcription in Prokaryotes 1295
Figure 31-40 The structure of the lac repressor in complex
with DNA. (a) The X-ray structure of the lac repressor tetramer
bound to two 21-bp segments of symmetric lac operator DNA.
The protein subunits are shown in ribbon form in yellow, cyan,
green, and orange and the dsDNA segments are drawn in space-
filling form with C white, N blue, O red, and P orange. [Courtesy
of Ponzy Lu and Mitchell Lewis with coordinates generated by
Benjamin Weider, University of Pennsylvania. PDBid 1LBG.]
(b) The NMR structure of the 23-bp O
1
lac operator DNA in
complex with two identical segments of the lac repressor
consisting of its DNA-binding domain and its hinge helix. Each
of the protein subunits is drawn in ribbon form colored in
rainbow order from its N-terminus (blue) to its C-terminus (red).
The DNA is represented in stick form with C white, N blue, O
red, and P orange and with successive P atoms in the same chain
connected by orange rods.The complex is viewed with its 2-fold
axis vertical. Note that the protein dimer’s two HTH motifs are
inserted in successive major grooves at the periphery of the
complex and that the insertion of the two centrally located hinge
helices into the DNA’s minor groove greatly widens and flattens
the minor groove at this point and kinks the DNA in an upward
bend. [Based on an NMR structure by Robert Kaptein and Rolf
Boelens, Utrecht University, The Netherlands. PDBid 2KEI.]
(b)
(a)
JWCL281_c31_1260-1337.qxd 8/11/10 9:48 PM Page 1295
Moreover, the E. coli purine repressor (PurR), which is
homologous to the lac repressor but lacks its C-terminal
helix, crystallizes as a dimer whose X-ray structure closely
resembles that of the lac repressor dimer.What then is the
function of lac repressor tetramerization?
Model building suggests that when the lac repressor
tetramer simultaneously binds to both the O
1
and O
3
oper-
ators, the 93-bp DNA segment containing them forms a
loop ⬃80 Å in diameter (Fig. 31-41). Furthermore, the
CAP–cAMP binding site is exposed on the inner surface
of the loop. Adding the CAP–cAMP at its proper position
to this model reveals that the ⬃90° curvature which
CAP–cAMP binding imposes on DNA (Fig. 31-31) has the
correct direction and magnitude to stabilize the DNA
loop, thereby stabilizing this putative CAP–cAMP–lac
repressor–DNA complex.It may seem paradoxical that the
binding of CAP–cAMP, a transcriptional activator, stabi-
lizes the repressor–DNA complex. However, when both
glucose and lactose are in short supply, it is important that
the bacterium lower its basal rate of lac operon expression
in order to conserve energy. The binding site (promoter)
for RNAP is also located on the inner surface of the loop.
Thus, the large size of the RNAP molecule would prevent
it from fully engaging the promoter in this looped complex,
thereby maximizing repression.
c. Combining Genetic and Structural Studies of
the lac Repressor Reveals Its Allosterically
Important Residues
The phenotypes of 4042 point mutations of the lac re-
pressor, which encompass nearly all of its 360 residues
(making the lac repressor the most exhaustively mutation-
ally characterized protein known) have been mapped onto
its X-ray structure. Mutations with an I
⫺
phenotype (lac re-
pressors that fail to bind to the lac operator, so that -
galactosidase is constitutively synthesized) are located at
the lac repressor’s DNA-binding interface, at its dimer in-
terface, or at internal residues of its inducer-binding core
domain. Residues whose mutations result in the I
S
pheno-
type (S for super-repressed; lac repressors that, in the pres-
ence of inducer, continue to repress the synthesis of -
galactosidase) appear to be of two types: (1) residues that
are in direct contact with the inducer, whose alteration
therefore interferes with inducer binding; and (2) residues
at the dimer interface that are ⬎8 Å from (not in direct
contact with) the inducer-binding site. These latter muta-
tions reveal which residues mediate the lac repressor’s
allosteric mechanism rather than directly binding the inducer
or the DNA. Most of the allosterically important residues
are located at the dimer interface and are members of the
N-subdomain of the core domain, which links the inducer-
binding sites to the operator DNA-binding sites.This is con-
sistent with the observation that inducer binding causes a
relative twist and translation of the N-subdomain, a move-
ment which is propagated to the hinge helix and DNA-
binding domain. This study demonstrates the power of
combining genetic analysis with structural studies to eluci-
date structure–function relationships.
G. trp Operon: Attenuation
We now discuss a sophisticated transcriptional control
mechanism named attenuation through which bacteria reg-
ulate the expression of certain operons involved in amino
acid biosynthesis. This mechanism was discovered through
the study of the E. coli trp operon (Fig. 31-42), which en-
codes five polypeptides comprising three enzymes that me-
diate the synthesis of tryptophan from chorismate (Section
26-5Bc). Charles Yanofsky established that the trp operon
genes are coordinately expressed under the control of the
trp repressor, a dimeric protein of identical 107-residue sub-
units that is the product of the trpR gene (which forms an
independent operon). The trp repressor binds
L-tryptophan,
the pathway’s end product, to form a complex that specifi-
cally binds to trp operator (trpO, Fig. 31-43) so as to reduce
the rate of trp operon transcription 70-fold. The X-ray struc-
ture of the trp repressor–operator complex (Section 31-3Da)
indicates that tryptophan binding allosterically orients trp
repressor’s two symmetry related helix–turn– helix “DNA
reading heads” so that they can simultaneously bind to
trpO (Fig. 31-34). Moreover, the bound tryptophan forms a
hydrogen bond to a DNA phosphate group, thereby
1296 Chapter 31. Transcription
Figure 31-41 Model of the 93-bp DNA loop formed when lac
repressor binds to O
1
and O
3
. The proteins are represented by
their C
␣
backbones and the DNA is drawn in stick form with its
sugar–phosphate backbones traced by helical ribbons.The model
was constructed from the X-ray structure of the lac repressor
(magenta) in complex with two 21-bp operator DNA segments
(red) and the X-ray structure of CAP–cAMP (blue) in complex
with its 30-bp target DNA (cyan; Fig. 31-28).The remainder of
the DNA loop was generated by applying a smooth curvature to
canonical B-DNA (white) with the ⫺10 and ⫺35 regions of the
lac promoter highlighted in green. [Courtesy of Ponzy Lu and
Mitchell Lewis, University of Pennsylvania.]
JWCL281_c31_1260-1337.qxd 8/11/10 9:48 PM Page 1296
strengthening the repressor–operator association. Trypto-
phan therefore acts as a corepressor; its presence prevents
what is then superfluous tryptophan biosynthesis (SAM
similarly functions as a corepressor with the met repressor;
Fig. 31-35a). The trp repressor also controls the synthesis
of at least two other operons: the trpR operon and the
aroH operon (which encodes one of three isozymes that
catalyze the initial reaction of chorismate biosynthesis;
Section 26-5Bc).
a. Tryptophan Biosynthesis Is Also Regulated
by Attenuation
The trp repressor–operator system was at first thought
to fully account for the regulation of tryptophan biosynthe-
sis in E. coli. However, the discovery of trp deletion mu-
tants located downstream from trpO that increase trp
operon expression 6-fold indicated the existence of an ad-
ditional transcriptional control element. Sequence analysis
established that trpE, the trp operon’s leading structural
gene, is preceded by a 162-nucleotide leader sequence
(trpL). Genetic analysis indicated that the new control ele-
ment is located in trpL, ⬃30 to 60 nucleotides upstream of
trpE (Fig. 31-42).
When tryptophan is scarce, the entire 6720-nucleotide
polycistronic trp mRNA, including the trpL sequence, is
synthesized. As the tryptophan concentration increases,
the rate of trp transcription decreases as a result of the
trp repressor–corepressor complex’s consequent greater
abundance. Of the trp mRNA that is transcribed, how-
ever, an increasing proportion consists of only a 140-nu-
cleotide segment corresponding to the 5¿ end of trpL. The
availability of tryptophan therefore results in the prema-
ture termination of trp operon transcription. The control
element responsible for this effect is consequently termed
an attenuator.
b. The trp Attenuator’s Transcription Terminator Is
Masked when Tryptophan Is Scarce
What is the mechanism of attenuation? The attenuator
transcript contains four complementary segments that can
form one of two sets of mutually exclusive base-paired
Section 31-3. Control of Transcription in Prokaryotes 1297
anthranilate
synthase
component I
anthranilate
synthase
component II
tryptophan
synthase
β subunit
tryptophan
synthase
α subunit
anthranilate
synthase
N-(5′-phosphoribosyl)-
anthranilate isomerase,
Indole-3-glycerol
phosphate synthase
tryptophan synthase
(α
2
β
2
)
(CoI
2
CoII
2
)
Chorismate Anthranilate N
-(5′-Phosphoribosyl)-
anthranilate
Enol-1-o-carboxy-
phenylamino-
1-deoxyribulose
phosphate
Indole-3-glycerol-P
Glutamine Glutamate
Pyruvate
+
PRPP PP
i
CO
2
L-Tryptophan
L-Serine Glyceraldehyde-3-P
Control
sites
trpP,O trpL trpE trpD trpC trpB trpA
Attenuator
Structural genes
mRNA
Leader
mRNA
or
CG A T GT A GAATGACTCAGAAC T CA
GC T A CA T CTTACTGACTTGTG A GT
⫺20 ⫺10 ⫹1
Figure 31-42 A genetic map of the E. coli trp operon
indicating the enzymes it specifies and the reactions they
catalyze. The gene product of trpC catalyzes two sequential
Figure 31-43 The base sequence of the trp operator. The
nearly palindromic sequence is boxed and its ⫺10 region is
overscored.
reactions in the synthesis of tryptophan. [After Yanofsky, C., J.
Am. Med. Assoc. 218, 1027 (1971).]
JWCL281_c31_1260-1337.qxd 8/11/10 9:48 PM Page 1297
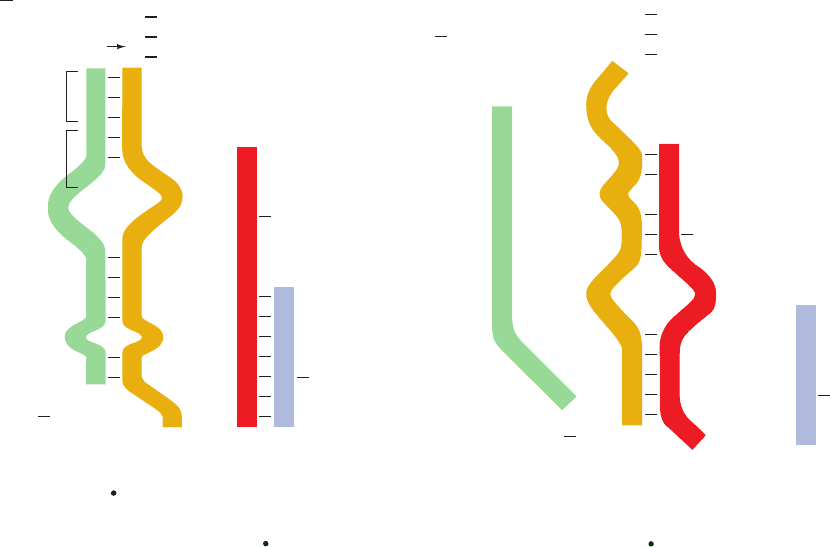
hairpins (Fig. 31-44). Segments 3 and 4 together with the suc-
ceeding residues comprise a normal intrinsic transcription
terminator (Section 31-2Da): a G ⫹ C–rich sequence that
can form a self-complementary hairpin structure followed
by several sequential U’s (compare with Fig. 31-18). Tran-
scription rarely proceeds beyond this termination site unless
tryptophan is in short supply.
A section of the leader sequence, which includes seg-
ment 1 of the attenuator, is translated to form a 14-residue
polypeptide that contains two consecutive Trp residues
(Fig. 31-44, left). The position of this particularly rare
dipeptide segment (1.1% of the residues in E. coli proteins
are Trp;Table 4-1) provided an important clue to the mech-
anism of attenuation.An additional essential aspect of this
mechanism is that ribosomes commence the translation of
a prokaryotic mRNA shortly after its 5¿ end has been syn-
thesized.
The above considerations led Yanofsky to propose the
following model of attenuation (Fig. 31-45).An RNA poly-
merase that has escaped repression initiates trp operon
transcription. Soon after the ribosomal initiation site of the
trpL gene has been transcribed, a ribosome attaches to it
and begins translation of the leader peptide. When trypto-
phan is abundant, so that there is a plentiful supply of tryp-
tophanyl–tRNA
Tr p
(the transfer RNA specific for Trp with
an attached Trp residue; Section 32-2C), the ribosome fol-
lows closely behind the transcribing RNA polymerase so as
to sterically block the formation of the 2 ⴢ 3 hairpin. In-
deed, RNA polymerase pauses past position 92 of the tran-
script and only continues transcription on the approach of
a ribosome, thereby ensuring the proximity of these two
entities at this critical position.The prevention of 2 ⴢ 3 hair-
pin formation permits the formation of the 3 ⴢ 4 hairpin,
the transcription terminator pause site, which results in the
termination of transcription (Fig. 31-45a). When trypto-
phan is scarce, however, the ribosome stalls at the tandem
UGG codons (which specify Trp; Table 5-3) because of the
lack of tryptophanyl–tRNA
Tr p
. As transcription continues,
the newly synthesized segments 2 and 3 form a hairpin be-
cause the stalled ribosome prevents the otherwise compet-
itive formation of the 1 ⴢ 2 hairpin (Fig. 31-45b).The forma-
tion of the transcriptional terminator’s 3 ⴢ 4 hairpin is
thereby pre-empted for sufficient time for RNA poly-
merase to transcribe through it and consequently through
the remainder of the trp operon. The cell is thus provided
with a regulatory mechanism that is responsive to the tryp-
tophanyl–tRNA
Tr p
level, which, in turn, depends on the
protein synthesis rate as well as on the tryptophan supply.
There is considerable evidence supporting this model of
attenuation. The trpL transcript is resistant to limited
1298 Chapter 31. Transcription
Figure 31-44 The alternative secondary structures of trpL
mRNA. The formation of the base paired 2 ⴢ 3 (antiterminator)
hairpin (right) precludes the formation of the 1 ⴢ 2 and 3 ⴢ 4
(terminator) hairpins (left) and vice versa.Attenuation results in
the premature termination of transcription immediately after
U
G
G
U
G
C
G
U
A
C
C
A
C
C
A
C
U
70
U
G
U
G
A
C
C
G
G
G
C
G
U
U
C
U
U
A
Trp
Trp
G
C
C
U
A
A
U
A
G
U
G
U
U
UUUUUU
A
A
A
U
U
U
A
A
A
A
A
A
A
A
G
AA
A
50
G
C
A
U
G
A
GA
AU
G
A
A
C
A
G
A
U
A
C
C
C
A
G
C
C
C
G
C
U
U
C
G
G
G
C
G
U U U U U U
130
130
110
110
12
34
23
“terminator”
“antiterminator”
G
G
G
U
U
G
G
U
G
G
C
G
C
A
C
U
A
U
U
G
AA
A
C
C
70
50
C
G
G
G
C
G
C
C
C
G
A
U
G
U
A
C
C
U
C
G
U
G
C
A
G
A
C
C
C
C
C
C
C
U
C
G
G
G
C
G
A
G
nucleotide 140 when the 3 ⴢ 4 hairpin is present.The arrow
indicates the mRNA site past which RNA polymerase pauses
until approached by an active ribosome. [After Fisher, R.F. and
Yanofsky, C., J. Biol. Chem. 258, 8147 (1983).]
JWCL281_c31_1260-1337.qxd 8/11/10 9:48 PM Page 1298
RNase T1 digestion, indicating that it has extensive second-
ary structure. The significance of the tandem Trp codons in
the trpL transcript is corroborated by their presence in trp
leader regions of several other bacterial species. Moreover,
the leader peptides of the five other amino acid–biosynthe-
sizing operons known to be regulated by attenuation (most
exclusively so) are all rich in their corresponding amino
acid residues (Table 31-3). For example, the E. coli his
operon, which specifies enzymes synthesizing histidine
(Fig. 26-65), has seven tandem His residues in its leader
peptide whereas the ilv operon, which specifies enzymes
participating in isoleucine, leucine, and valine biosynthesis
(Fig. 26-61), has five Ile’s, three Leu’s, and six Val’s in its
leader peptide. Finally, the leader transcripts of these oper-
ons resemble that of the trp operon in their capacity to
form two alternative secondary structures, one of which
contains a trailing termination structure.
H. Riboswitches Are Metabolite-Sensing RNAs
We have just seen how the formation of secondary struc-
ture in a growing RNA transcript can regulate gene expres-
sion though attenuation. The conformational flexibility of
mRNA also allows it to regulate genes by directly interacting
Section 31-3. Control of Transcription in Prokaryotes 1299
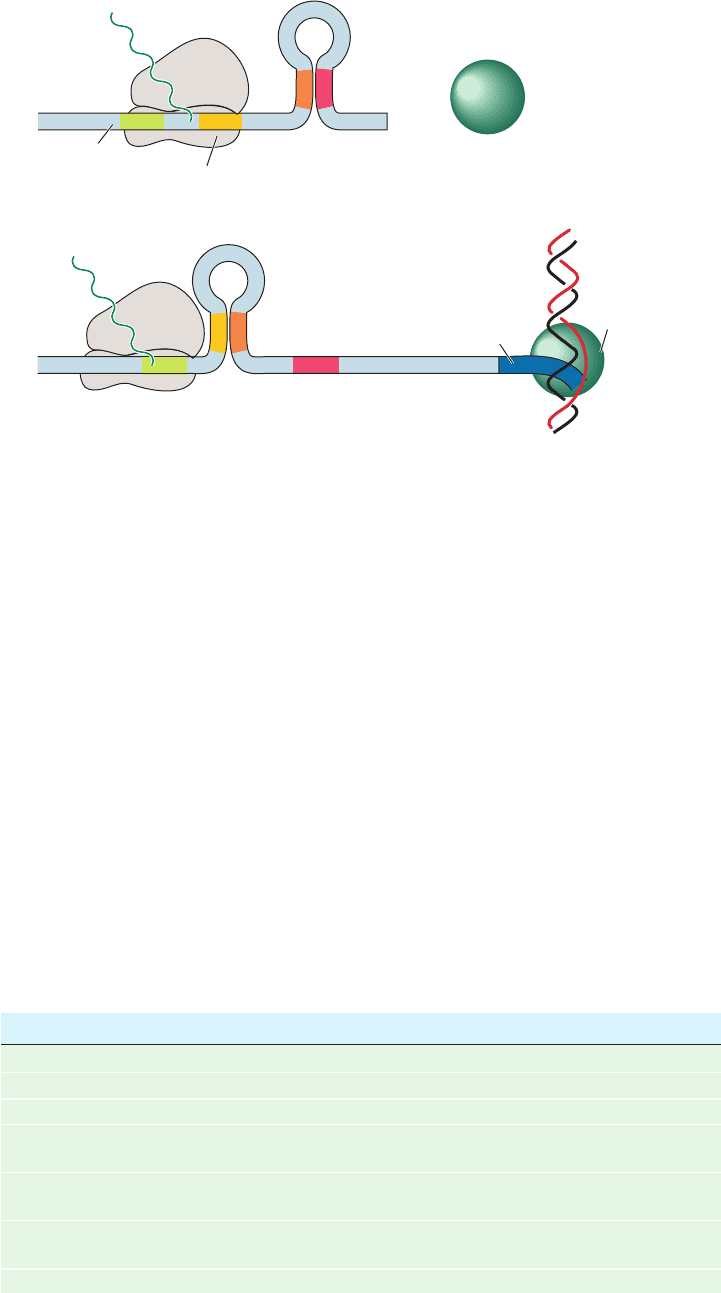
Figure 31-45 Attenuation in the trp operon. (a) When
tryptophanyl–tRNA
Tr p
is abundant, the ribosome translates trpL
mRNA. The presence of the ribosome on segment 2 prevents the
formation of the base-paired 2 ⴢ 3 hairpin.The 3 ⴢ 4 hairpin, an
essential component of the transcriptional terminator, can
thereby form, thus aborting transcription. (b) When tryptophanyl–
High tryptophan(a)
(b)
Leader
peptide
34
Ribosome transcribing
the leader peptide mRNA
Transcription
terminator
+
“Terminated”
RNA polymerase
trpL mRNA
Low tryptophan
Antiterminator
Ribosome stalled
at tandem
Trp codons
1
3
4
2
trp operon mRNA
Transcribing
RNA
polymerase
DNA encoding
operontrp
12
Table 31-3 Amino Acid Sequences of Some Leader Peptides in Operons
Subject to Attenuation
Operon Amino Acid Sequence
a
trp Met-Lys-Ala-Ile-Phe-Val-Leu-Lys-Gly-TRP-TRP-Arg-Thr-Ser
pheA Met-Lys-His-Ile-Pro-PHE-PHE-PHE-Ala-PHE-PHE-PHE-Thr-PHE-Pro
his Met-Thr-Arg-Val-Gln-Phe-Lys-HIS-HIS-HIS-HIS-HIS-HIS-HIS-Pro-Asp
leu Met-Ser-His-Ile-Val-Arg-Phe-Thr-Gly-LEU-LEU-LEU-LEU-Asn-Ala-
Phe-Ile-Val-Arg-Gly-Arg-Pro-Val-Gly-Gly-Ile-Gln-His
thr Met-Lys-Arg-ILE-Ser-THR-THR-ILE-THR-THR-THR-ILE-THR-ILE-
THR-THR-Gln-Asn-Gly-Ala-Gly
ilv Met-Thr-Ala-LEU-LEU-Arg-VAL-ILE-Ser-LEU-VAL-VAL-ILE-Ser-
VAL-VAL-VAL-ILE-ILE-ILE-Pro-Pro-Cys-Gly-Ala-Ala-Leu-Gly-Arg-
Gly-Lys-Ala
a
Residues in uppercase are synthesized in the pathway catalyzed by the operon’s gene products.
Source: Yanofsky, C., Nature 289, 753 (1981).
tRNA
Tr p
is scarce, the ribosome stalls on the tandem Trp codons
of segment 1.This situation permits the formation of the 2 ⴢ 3
hairpin which, in turn, precludes the formation of the 3 ⴢ 4
hairpin. RNA polymerase therefore transcribes through this
unformed terminator and continues trp operon transcription.
JWCL281_c31_1260-1337.qxd 8/11/10 9:48 PM Page 1299
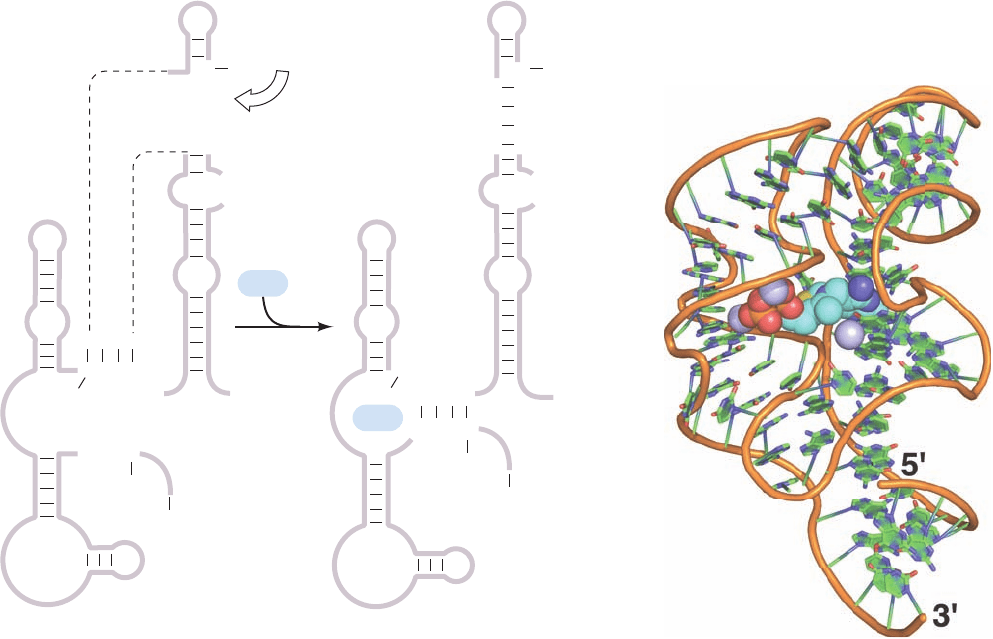
with certain cellular metabolites, thereby eliminating the
need for sensor proteins such as the lac repressor,CAP, and
the trp repressor.
In E. coli, the biosynthesis of thiamine pyrophosphate
(TPP; Section 17-3Ba) requires the action of several pro-
teins whose levels vary according to the cell’s need for TPP.
In at least two of the relevant genes the untranslated re-
gions at the 5¿ end of the mRNA include a highly conserved
sequence called the thi box. The susceptibility of the thi
box to chemical or enzymatic cleavage, as Ronald Breaker
showed, differs in the presence and absence of TPP, sug-
gesting that the RNA changes its secondary structure when
TPP binds to it (the binding of a metabolite by RNA is not
unprecedented; synthetic oligonucleotides known as ap-
tamers bind specific molecules with high specificity and
affinity; Section 7-6C). The TPP-sensing mRNA element
has been dubbed a riboswitch.
The predicted secondary structure of the TPP-sensing
riboswitch and its proposed mechanism are shown in Fig.
31-46a. In the absence of TPP, the mRNA assumes a con-
formation that allows a ribosome to begin translation. In
the presence of TPP, an alternative secondary structure
masks the sequence that identifies its translational initia-
tion site to the ribosome (its so-called Shine–Dalgarno
sequence; Section 32-3Cb) so that the ribosome cannot ini-
tiate the mRNA’s translation. Thus, the concentration of a
metabolite can regulate the expression of genes required for
its synthesis. The X-ray structure of the 80-nt TPP-binding
domain from the E. coli TPP-sensing riboswitch, deter-
mined by Breaker and Dinshaw Patel, reveals an intri-
cately folded RNA that binds TPP in an extended confor-
mation (Fig. 31-46b).
Over 20 classes of riboswitches have as yet been identi-
fied, including those that regulate the expression of en-
zymes involved in the metabolism of coenzyme B
12
(Fig.
25-21), riboflavin (Fig. 16-8), S-adenosylmethionine (SAM;
Fig. 26-18), lysine, and adenine. In general, they consist of
two components, an aptamer that binds an effector and a
so-called expression platform that transduces effector
binding to a change in gene expression. In some cases, the
1300 Chapter 31. Transcription
Figure 31-46 Structure of the TPP-sensing riboswitch from
E. coli. (a) The predicted secondary structure of a 165-residue
segment at the 5¿ end of the thiM gene is shown in the absence
(left) and presence (right) of TPP. The TPP-binding conformation
masks the Shine–Dalgarno sequence (orange) required by the
ribosome to initiate translation at the AUG start codon (red) just
downstream. [After Winkler,W., Nahvi,A., and Breaker, R.R.,
Nature 419, 952 (2002).] (b) The X-ray structure of the
riboswitch’s 90-nt TPP-sensing domain. The RNA is drawn in
A
A
GC
GU
U
AU
G
A
G
G
A
G
G
A
U
G
CUGA
GACU
CCUUUUCC
G
U
C
AAG
GACU
A
G
C
TPP
Ribosome
83
83
125
125
11
11
3´
3´
5´
5´
TPP
(a)
(b)
cartoon form with its sugar–phosphate backbone represented
by an orange rod and its bases represented by paddles with
C green, N blue, and O red. The TPP is drawn in space-filling
form with C cyan, N blue, O red, and S yellow. Mg
2⫹
ions are
represented by lavender spheres. [Based on an X-ray structure
by Ronald Breaker,Yale University, and Dinshaw Patel,
Memorial Sloan-Kettering Cancer Center, New York, New York.
PDBid 2GDI.]
JWCL281_c31_1260-1337.qxd 8/11/10 9:48 PM Page 1300
