Voet D., Voet Ju.G. Biochemistry
Подождите немного. Документ загружается.

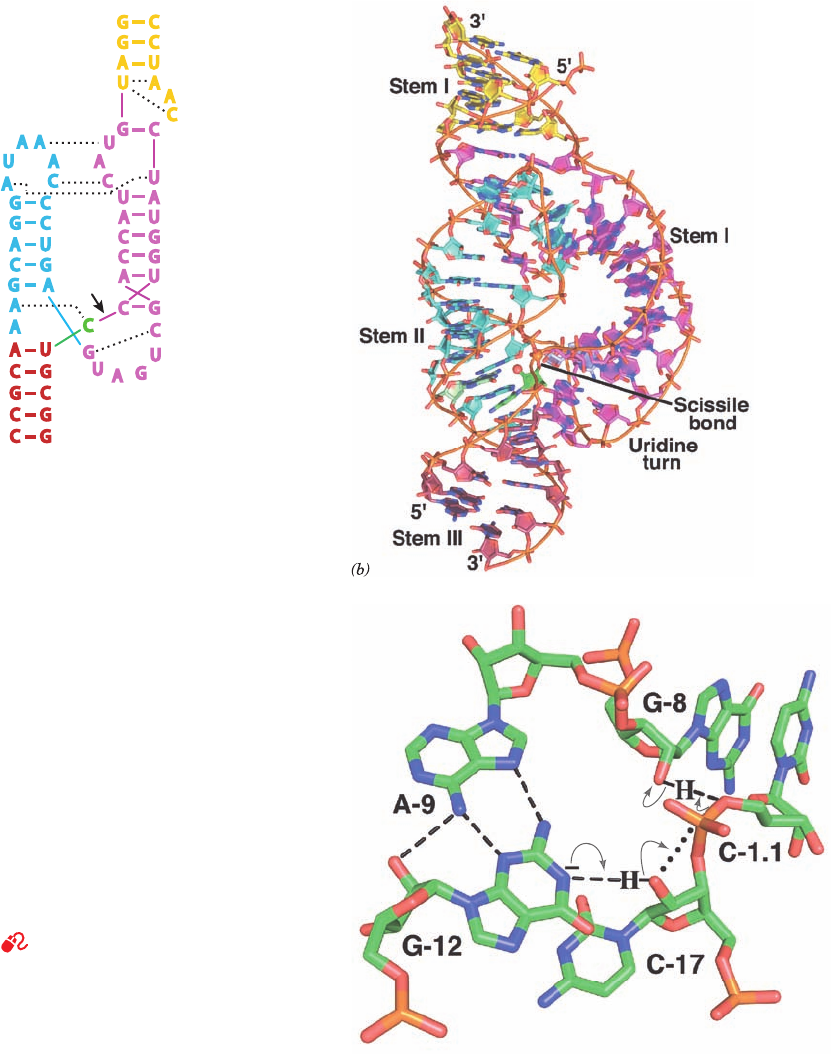
nucleophilic attack on the P atom that links atom O3¿ of
C-17 to atom O5¿ of C-1.1. The N1 of the invariant G-12
when deprotonated and the 2¿-OH of the invariant G-8 ap-
pear to be properly positioned to respectively act as base
and acid catalysts in this reaction (Fig. 31-57c), which
strongly suggests that the reaction occurs via a concerted
acid–base catalyzed mechanism. This reaction mechanism
does not involve the participation of metal ions and none
are observed in the ribozyme’s catalytic core. However, in
solution, the presence of divalent metal ions provides an
⬃50-fold enhancement rate over the presence of only
monovalent metal ions. Perhaps divalent metal ions stabi-
lize the negative charge on the trigonal bipyramidal inter-
mediate (Fig. 16-6b) and/or they may help position and
orient reactive groups.
h. Splicing of Pre-mRNAs Is Mediated by snRNPs
in the Spliceosome
How are the splice junctions of pre-mRNAs recognized
and how are the two exons to be joined brought together in
the splicing process? Part of the answer to this question
was established by Joan Steitz going on the assumption
that one nucleic acid is best recognized by another.The eu-
karyotic nucleus, as has been known since the 1960s,
contains numerous copies of several highly conserved
60- to 300-nucleotide RNAs called small nuclear RNAs
Section 31-4. Post-Transcriptional Processing 1311
Figure 31-57 Structure of the Schistosoma mansoni
hammerhead ribozyme. (a) The sequence and schematic
structural organization of the ribozyme colored to match the
X-ray structure drawn in Part b. Base pairs and tertiary
interactions are represented by dashes and dotted lines,
respectively. Nucleotides are labeled according to the universal
numbering system for hammerhead ribozymes. (b) X-ray
structure of the ribozyme drawn in paddle form with C atoms the
same color as in Part a except that those of G-12 are light green
and those of C-1.1 are light blue, N blue, O red, and P orange.
Adjacent P atoms in the same strand are connected by thin
orange rods. The P atom of the scissile phosphate group
(bridging C-17 and C-1.1) and the nucleophile (O2¿ of C-17) are
represented by small spheres. (c) The ribozyme’s active site
residues, which are drawn in stick form with C green, O red,
N blue, and P orange. Hydrogen bonds are represented by
dashed black lines.The proposed reaction mechanism is
indicated by the curved arrows with the dotted black line
marking the in-line trajectory taken by atom O2¿ of C-17 in
nucleophilically attacking the P atom of the scissile phosphate
group. [Part a courtesy of and Parts b and c based on an X-ray
structure by William Scott, University of California at Santa
Cruz. PDBid 2GOZ.]
See Interactive Exercise 43
3
33
6
66
5
55
8
9
99
7
77
17
10.1
10.110.111.1
12
1212
13
1313
14
1414
15.1
16.1
16.116.1
2.1
2.12.1
1.1
1.11.1
L1
L3
L3L3
L2
L2L2
L6
L6L6
L5
L5L5
L4
L4L4
B8
B5
B5B5
B2
B6
B4
B4B4
B3
B1
B7
11.2
11.3
10.2
10.210.2
10.3
10.310.3
scissile
bond
Uridine
turn
Stem I
Stem III
Stem I
Stem II
5'3'
5'
3'
(a)
(
c
)
JWCL281_c31_1260-1337.qxd 10/19/10 10:41 AM Page 1311
(snRNAs), which form protein complexes termed small
nuclear ribonucleoproteins (snRNPs; pronounced “snurps”).
Steitz recognized that the 5¿ end of one of these snRNAs,
U1-snRNA (so called because it is a member of a U-rich
subfamily of snRNAs), is partially complementary to the
consensus sequence of the 5¿ splice site. The consequent
hypothesis, that U1-snRNA recognizes the 5¿ splice site, was
corroborated by the observations that splicing is inhibited
by the selective destruction of the U1-snRNA sequences
that are complementary to the 5¿ splice site or by the pres-
ence of anti-U1-snRNP antibodies (produced by patients
suffering from systemic lupus erythematosus, an often fatal
autoimmune disease). Three other snRNPs are also impli-
cated in splicing: U2-snRNP, U4–U6-snRNP (in which
the U4- and U6-snRNAs associate via base pairing), and
U5-snRNP.
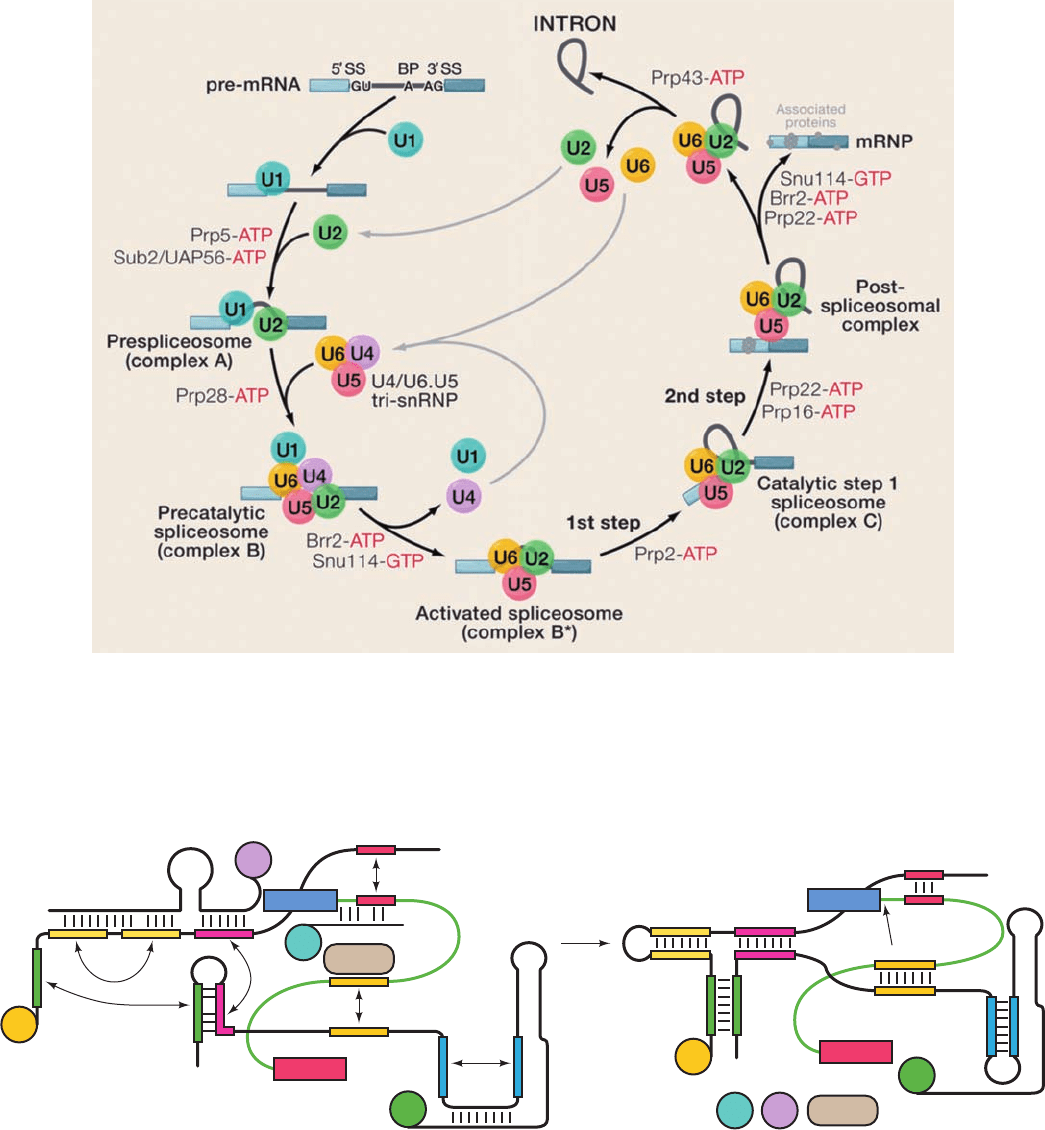
Splicing takes place in an as yet poorly characterized
⬃2700 kD particle dubbed the spliceosome (Fig. 31-58).
The spliceosome brings together a pre-mRNA, the forego-
ing four snRNPs, and a variety of pre-mRNA binding pro-
teins. Note that the spliceosome, which consists of 5 RNAs
and ⬃150 polypeptides, is comparable in size and complex-
ity to the ribosome (which in E. coli consists of 3 RNAs and
52 polypeptides with an aggregate mass of ⬃2500 kD; Sec-
tion 32-3A).
In addition to its size and complexity, the spliceosome is a
highly dynamic entity, with its various components associ-
ating and dissociating during specific stages of the splicing
reaction (Fig. 31-59), while undergoing a variety of ATP-
driven conformational changes. For example, to carry out
the first transesterification reaction yielding the lariat
structure (Fig. 31-53), the spliceosome undergoes a com-
plex series of rearrangements that are schematically dia-
grammed in Fig. 31-60. Similarly extensive rearrangements
are required to carry out the second transesterification
reaction and to recycle the spliceosome for subsequent
splicing reactions.
Although spliceosomal transesterification reactions
were initially assumed to be mediated by protein catalysts,
their chemical resemblance to the reactions carried out by
the self-splicing group II introns suggests, as is noted
above, that it is really the snRNAs that catalyze the splicing
of pre-mRNAs (pre-mRNA introns have such varied se-
quences outside of their splice and branch sites that they
are unlikely to play an active role in splicing). In fact, James
Manley has shown that, in the absence of protein, segments
of human U2- and U6-snRNAs catalyze an Mg
2⫹
-depend-
ent reaction in an intron branch site sequence–containing
RNA that resembles splicing’s first transesterification
reaction.
i. Splicing Also Requires the Participation of
Splicing Factors
Around 170 different proteins known as splicing-associ-
ated factors that are extrinsic to spliceosomes also partici-
pate in splicing, with individual assembly intermediates
(e.g., complexes A, B, and C in Fig. 31-59) each associated
with ⬃125 such proteins. Among them are branch
point–binding protein [BBP; also known as splicing factor
1 (SF1)] and U2-snRNP auxiliary factor (U2AF), which
cooperate to select the intron’s branch point. U2AF binds
to the polypyrimidine tract upstream of the 3¿ splice site
(Fig. 31-52), whereas BBP recognizes the nearby branch
point sequence (Figs. 31-53 and 31-60). The NMR structure
of the 131-residue RNA-binding segment of the 638-residue
BBP in complex with an 11-nt RNA containing a branch
1312 Chapter 31. Transcription
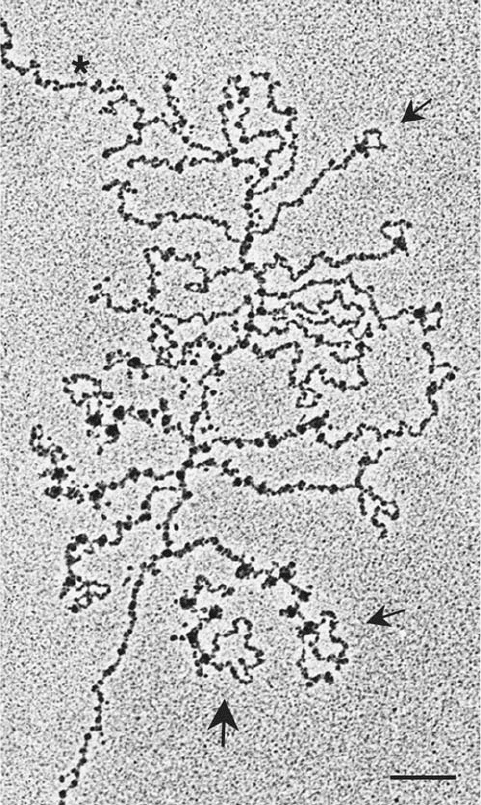
Figure 31-58 An electron micrograph of spliceosomes in
action. A Drosophila gene that is ⬃6 kb long enters from the
upper left of the micrograph and exits at the lower left.
Transcription initiates near the point marked by an asterisk. The
growing RNA chains appear as fibrils of increasing lengths that
emanate from the DNA. The transcripts are undergoing
cotranslational splicing as revealed by the progressive formation
and loss of intron loops near the 5¿ ends of the RNA transcripts
(arrows).The beads at the base of each intron loop as well as
elsewhere on the transcripts are the spliceosomes.The large
arrow points to a transcript near the 3¿ end of the gene that is no
longer attached to the DNA template and hence appears to have
recently been terminated and released.The bar is 200 nm long.
[Courtesy of Ann Beyer and Yvonne Osheim, University of
Virginia.]
JWCL281_c31_1260-1337.qxd 8/11/10 9:48 PM Page 1312
Section 31-4. Post-Transcriptional Processing 1313
Figure 31-59 The spliceosomal assembly/disassembly cycle.
The sequential actions of the spliceosomal snRNPs (colored
circles), but not the non-snRNP proteins, are diagrammed in the
process of excising an intron from a pre-RNA containing two
exons (blue). Here 5¿SS, BP, and 3¿SS stand for the pre-mRNA’s
5¿ splice site, its branch point, and its 3¿ splice site, respectively.
Figure 31-60 Schematic diagram of six rearrangements
that the spliceosome undergoes in mediating the first
transesterification reaction in pre-mRNA splicing. The RNA is
color coded to indicate segments that become base-paired.The
black and green lines represent snRNA and pre-mRNA, and
BBP stands for branch point–binding protein. U5, which
participates in the second transesterification reaction, has been
omitted for clarity. (1) Exchange of U1 for U6 in base pairing to
the intron’s 5¿ splice site. (2) Exchange of BBP for U2 in binding
to the intron’s branch site. (3) Intramolecular rearrangement in
5'
5'
5'
5
5'
6
4
2
3
3
2
1
5
6
4
1
5'
A
5'
U4
U2
U6
U1
U4
U6
U2
3' exon
5' exon
5' exon
3' exon
BBP
BBP
U1
Eight conserved DExD/H box–containing RNA-dependent
RNA ATPases/helicases as well as the GTPase Snu114 act in
specific steps of the splicing cycle to motivate RNA–RNA
rearrangements and RNP remodeling reactions. [Courtesy of
Reinhard Lührmann, Max-Planck-Institut für biophysikalische
Chemie, Göttingen, Germany.]
U2. (4) Disruption of a base-paired stem between U4 and U6 to
form a stem–loop in U6. (5) Disruption of a second stem
between U4 and U6 to form a stem between U2 and U6.
(6) Disruption of a stem–loop in U2 to form a second stem
between U2 and U6.The order of these rearrangements is
unclear.The transesterification reaction is represented by the
arrow from the A in the yellow segment of the pre-mRNA (right
panel) to the 3¿ end of the 5¿ exon. [Adapted from Staley, J.P. and
Guthrie, C., Cell 92, 315 (1998).]
JWCL281_c31_1260-1337.qxd 8/11/10 9:48 PM Page 1313
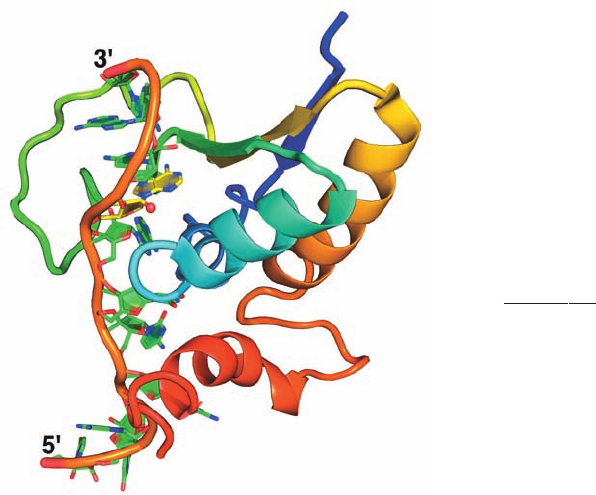
point sequence, determined by Michael Sattler, reveals that
the RNA assumes an extended conformation and is largely
buried in a groove that is lined with both aliphatic and basic
residues (Fig. 31-61). The branch point adenosine, whose
mutation abolishes BBP binding,is deeply buried and binds
to BBP via hydrogen bonds that mimic Watson–Crick base
pairing with uracil.
Other splicing factors include SR proteins and several
members of the heterogeneous nuclear ribonucleoprotein
(hnRNP) family. SR proteins each have one or more
RRMs (RNA recognition motifs) near their N-terminus
and a distinctive C-terminal RS domain that contains nu-
merous Ser-Arg (SR) repeats and which participates in
protein–protein interactions. SR proteins, when appropri-
ately phosphorylated on their RS domains, specifically
bind to their corresponding exonic splicing enhancers
(ESEs) via their RRMs and thereby recruit the splicing
machinery to the flanking 5¿ and 3¿ splice sites. The hnRNP
proteins, which are highly abundant RNA-binding pro-
teins, lack RS domains and hence cannot recruit the splic-
ing machinery. Instead, they bind to their corresponding
ESSs and ISSs (exonic and intronic splicing silencers) so as
to block the binding of the splicing machinery at the flank-
ing splice sites.
A simplistic interpretation of Fig. 31-53 suggests that
any 5¿ splice site could be joined with any following 3¿
splice site, thereby eliminating all the intervening exons
together with the introns joining them. However, such
exon skipping does not normally take place (but see be-
low). Rather, all of a pre-mRNA’s introns are individually
excised in what appears to be a largely fixed order that
more or less proceeds in the 5¿S3¿ direction.This occurs,
at least in part, because splicing takes place cotranscription-
ally (Fig. 31-58).Thus, as a newly synthesized exon emerges
from an RNAP II, it is bound by splicing factors that are
also bound to the RNAP II’s highly phosphorylated C-
terminal domain (CTD; Section 31-2E). This tethers the
exon and its associated spliceosome to the CTD so as to
ensure that splicing occurs when the next exon emerges
from the RNAP II.
j. Spliceosomal Structures
All four snRNPs involved in pre-mRNA splicing con-
tain the same so-called snRNP core protein, which consists
of seven Sm proteins (so called because they react with au-
toantibodies of the Sm serotype from patients with sys-
temic lupus erythematosis), which are named B, D1, D2,
D3, E, F, and G proteins. Each of these Sm proteins con-
tains two conserved segments, Sm1 and Sm2, that are sepa-
rated by a linker of variable length.The seven Sm proteins
collectively bind to a conserved RNA sequence, the Sm
RNA motif, which occurs in U1-,U2-,U4-, and U5-snRNAs
and which has the single-stranded sequence AAUU-
UGUG. However, in the absence of a U-snRNA, the Sm
proteins form three stable complexes D1–D2, D3–B, and
E–F–G. None of these complexes alone bind U-snRNA.
However, the D1–D2 and E–F–G complexes form a stable
subcore snRNP with U-snRNA, to which D3–B binds to
form the complete Sm core domain.
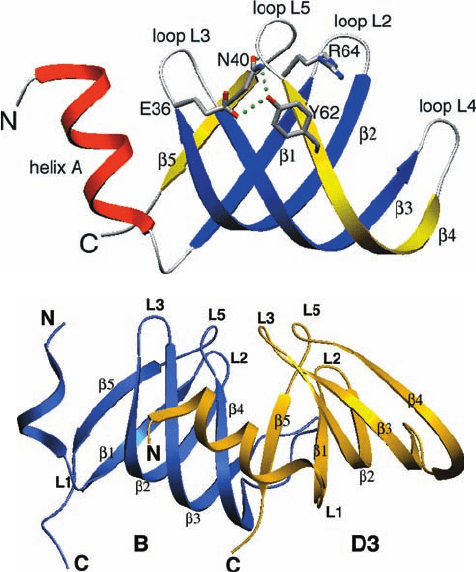
The X-ray structures of the D3–B and D1–D2 het-
erodimers, determined by Reinhard Lührmann and
Kiyoshi Nagai, reveals that these four proteins share a
common fold which consists of an N-terminal helix fol-
lowed by a 5-stranded antiparallel  sheet that is strongly
bent so as to form a hydrophobic core (Fig. 31-62a). The
subunits of both dimers associate in a similar manner
with the 5 strands of D3 and D1 binding to the 4
strands of B and D2, respectively, so as to join their 
1314 Chapter 31. Transcription
Figure 31-61 The NMR structure of the RNA-binding portion
of human branch point–binding protein (BBP) in complex with
its target RNA. The 11-nt RNA contains the sequence
5¿-UAU
ACUAACAA-3¿ in which the branch site sequence for
both yeast and vertebrates is underlined and the branch point A
is in bold.The protein is drawn in cartoon form colored in
rainbow order from its N-terminus (blue) to its C-terminus (red).
The RNA is drawn in paddle form with C green except for the C
atoms of the branch point A, which are yellow, N blue, and O red
and with successive P atoms connected by an orange rod.The
branch point O2¿ is represented by a small red sphere. [Based on
an NMR structure by Michael Sattler, European Molecular
Biology Laboratory, Heidelberg, Germany. PDBid 1K1G.]
JWCL281_c31_1260-1337.qxd 8/11/10 9:48 PM Page 1314
sheets (Fig. 31-62b). This, together with biochemical and
mutagenic experiments, indicates that the Sm proteins
form a closed heteroheptameric ring whose subunits are
arranged in the order –B–D3–G–E–F–D2–D1–. This
model is corroborated by the X-ray structure of an Sm-
like protein from the hyperthermophilic archeon Py-
robaculum aerophilum, determined by David Eisenberg,
that forms a homoheptameric ring that is structurally
similar to the heteroheptameric model. This structure
also supports the hypothesis that the seven eukaryotic
Sm proteins arose through a series of duplications of an
archaeal Sm-like protein gene.
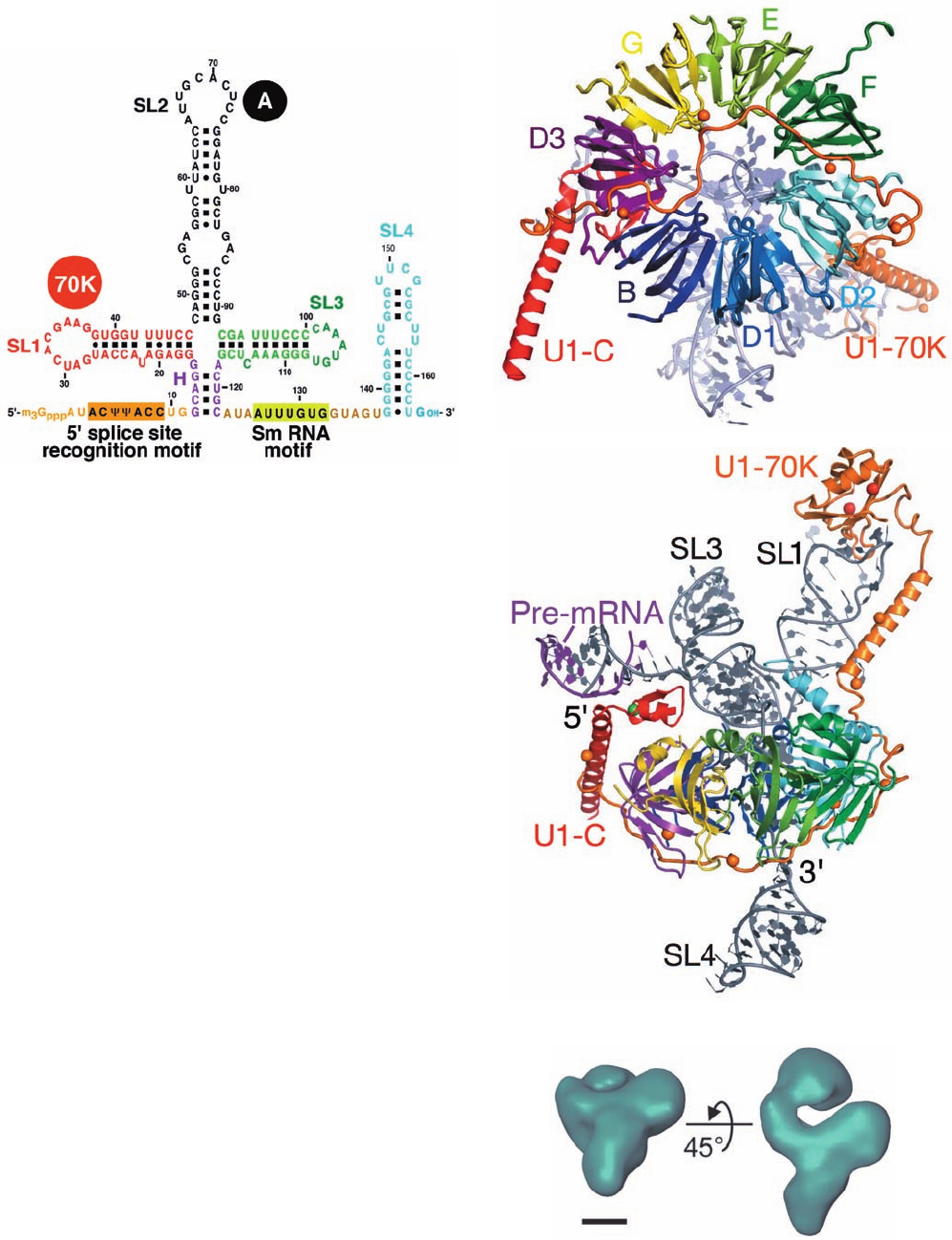
Mammalian U1-snRNP consists of U1-snRNA and ten
proteins, the seven Sm proteins that are common to all
U-snRNPs as well as three that are specific to U1-snRNP:
U1-70K, U1-A, and U1-C (437, 282, and 159 residues, re-
spectively, in humans). The predicted secondary structure
of the 165-nt U1-snRNA contains five double helical stems,
four of which come together at a 4-way junction (Fig.31-63a).
U1-70K and U1-A bind directly to RNA stem–loops 1 and
2 (SL1 and SL2), respectively, whereas U1-C is bound by
other proteins.
Nagai determined the X-ray structure of human U1
snRNP at 5.5 Å resolution.At this low resolution the major
and minor grooves of the double-stranded RNA stems are
visible. Moreover, the helices and sheets of the proteins are
apparent, which allows the placement of protein folds of
known structure. SL2 of the U1-snRNA had been altered
and shortened to promote crystallization. This eliminated
the binding site for U1-A. However, U1 snRNP in which
U1-A is depleted is active in a splicing assay.
SL1 and SL2 are stacked coaxially as are SL3 and helix
H, and these two stacked helices cross at an angle of ⬃90⬚.
The Sm proteins form the predicted heteroheptameric
ring, which is ⬃70 Å in diameter (Fig. 31-63b). The Sm
RNA motif together with SL4 are threaded through the
ring’s funnel-shaped central hole such the Sm RNA motif
interacts with the Sm proteins (Fig. 31-63c). U1-C associ-
ates with the Sm ring via an interaction between its zinc fin-
ger domain and the D3 subunit (Fig. 31-63c). An RRM in
the C-terminal segment of U1-70K interacts with the loop
of SL1 and this protein’s N-terminal segment is draped
across the external face of the Sm ring.
Fortuitously, the 5¿ ends of two neighboring U1-snRNAs
in the crystal form a double helical segment. Since, in the
spliceosome, the 5¿ end of U1-snRNA base-pairs with the 5¿
splice site of the pre-RNA (Fig. 31-60), this interaction pro-
vides a model of how the spliceosome recognizes the 5¿
splice site. The zinc finger domain of U1-C interacts with
this double helix (left side of Fig. 31-63c) and presumably
stabilizes it. This is consistent with the observation that
mutants of U1-C in this region cannot initiate spliceosome
formation (the first reaction in Fig. 31-59).
Difficulties in obtaining homogeneous preparations of
spliceosomal subassemblies and their poor stabilities
have hindered their crystallization and limited the resolu-
tion of their cryo-EM–based images. Nevertheless, the
cryo-EM–based structures of a variety of spliceosomal
components have been reported. For example, the ⬃40-Å
resolution structure of human complex B after it has re-
leased U1-snRNP but before it has released U4-snRNP
(Fig. 31-59) was determined by Lührmann and Holger
Section 31-4. Post-Transcriptional Processing 1315
Figure 31-62 X-ray structures of Sm proteins. (a) The
structure of D3 protein.The N-terminal helix and the  strands
of its Sm1 domain are red and blue and the  strands of its Sm2
domain are yellow. The B, D1, and D2 Sm proteins have similar
structures with their L4 loops and N-terminal segments,
including helix A, comprising their most variable portions.
Several highly conserved residues are shown in stick form (with
C gray, N blue, and O red), and a conserved hydrogen bonding
network is represented by green dotted lines. (b) The D3–B
dimer with D3 gold and B blue. The 5 strand of D3 associates
with the 4 strand of B to form a continuous antiparallel  sheet.
Note that their corresponding loops extend in similar directions.
[Courtesy of Kiyoshi Nagai, MRC Laboratory of Molecular
Biology, Cambridge, U.K. PDBid 1D3B.]
(a)
(b)
JWCL281_c31_1260-1337.qxd 8/11/10 9:48 PM Page 1315
Stark (Fig. 31-64). This 370 ⫻ 270 ⫻ 170 Å particle, which
is named B¢U1, has a roughly triangular body connected
to a head domain. Other splicesome components are
equally irregular.
k. The Significance of Gene Splicing
The analysis of the large body of known DNA se-
quences reveals that introns are rare in prokaryotic struc-
tural genes, uncommon in lower eukaryotes such as yeast
(which has a total of 239 introns in its ⬃6600 genes and,
with two exceptions, only one intron per polypeptide), and
abundant in higher eukaryotes (the only known vertebrate
structural genes lacking introns are those encoding his-
tones and the antiviral proteins known as interferons). Pre-
mRNA introns, as we have seen, can be quite long and
many genes contain large numbers of them. Consequently,
unexpressed sequences constitute ⬃80% of a typical verte-
brate structural gene and ⬃99% of a few of them.
1316 Chapter 31. Transcription
Figure 31-64 Cryo-EM–based structure of the human
spliceosome at its B⌬U1 stage of assembly/disassembly at 40 Å
resolution. The bar represents 100 Å. [Courtesy of Holger Stark,
Max-Planck Institute for Biophysical Chemistry, Göttingen,
Germany.]
Figure 31-63 X-ray structure of human U1-snRNP. (a) The
predicted secondary structure of U1-snRNA with the RNA
segments at which the proteins U1-70K and U1-A bind
indicated. ⌿ is the symbol for pseudouridine residues (Section
30-5Be). (b) View of the X-ray structure along the axis of the Sm
ring.The proteins are drawn in ribbon form in different colors
and the RNA is shown in paddle form in gray. SL4 has been
deleted for clarity. The yellow spheres represent the selenium
atoms in selenoMet residues (Met with its S atom replaced by
Se), which were mutagenically inserted into the proteins to aid in
solving the structure. Their known positions in the polypeptide
chains helped trace the polypeptides’ paths. (c) The structure as
viewed from above in Part a.The Zn
2⫹
ion bound to the zinc
finger motif of U1-C is represented by a green sphere. The 5¿
terminal portion of the U1-snRNA forms a double helical
segment with the 5¿ end of a neighboring U1-snRNA, which is
drawn in paddle form in purple. [Part a modified from a drawing
by, and Parts b and c courtesy of, Kiyoshi Nagai, MRC
Laboratory of Molecular Biology, Cambridge, U.K. PDBid
3CW1.]
(a)
(b)
(c)
JWCL281_c31_1260-1337.qxd 8/11/10 9:48 PM Page 1316
The argument that introns are only molecular parasites
(junk DNA) seems untenable since it would then be difficult
to rationalize why the evolution of complex splicing machin-
ery offered any selective advantage over the elimination of
the split genes. What then is the function of gene splicing?
Although, since its discovery, the significance of gene splic-
ing has been often vehemently debated, two important roles
for it have emerged: (1) It is an agent for rapid protein evo-
lution; and (2) through alternative splicing, it permits a sin-
gle gene to encode several (sometimes many) proteins that
may have significantly different functions. In the following
paragraphs, we discuss these aspects of gene splicing.
l. Many Eukaryotic Proteins Consist of Modules
That Also Occur in Other Proteins
The 839-residue LDL receptor is a plasma membrane
protein that functions to bind low-density lipoprotein
(LDL) to coated pits for transport into the cell via endocyto-
sis (Section 12-5Bc). LDL receptor’s 45-kb gene contains 18
exons, most of which encode specific functional domains of
the protein. Moreover, 13 of these exons specify polypeptide
segments that are homologous to segments in other proteins:
1. Five exons encode a 7-fold repeat of a 40-residue se-
quence that occurs once in complement C9 (an immune
system protein; Section 35-2F).
2. Three exons each encode a 40-residue repeat similar
to that occurring four times in epidermal growth factor
(EGF; Section 19-3A) and once each in three blood clot-
ting system proteins: factor IX, factor X, and protein C
(Section 35-1).
3. Five exons encode a 400-residue sequence that is
33% identical with a polypeptide segment that is shared
only with EGF.
Evidently, the LDL receptor gene is modularly constructed
from exons that also encode portions of other proteins. Nu-
merous other eukaryotic proteins are similarly constituted
including, as we have seen, many of the proteins involved
in signal transduction (e.g., those containing SH2 and SH3
domains; Section 19-3C). Moreover, many exons encode
complete domains that frequently have independent func-
tions. It therefore appears that the genes encoding these
modular proteins arose by the stepwise collection of exons
that were assembled by (aberrant) recombination between
their neighboring introns.
m. Alternative Splicing Greatly Increases the Number
of Proteins Encoded by Eukaryotic Genomes
The expression of numerous cellular genes is modulated
by the selection of alternative splice sites. Thus, certain exons
in one type of cell may be introns in another. For example,
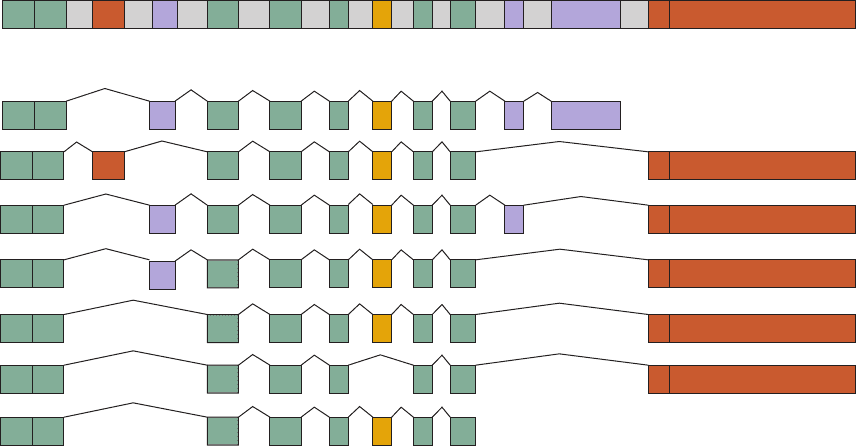
a single rat gene encodes seven tissue-specific isoforms
(splice variants) of the muscle protein ␣-tropomyosin (Sec-
tion 35-3Ca) through the selection of alternative splice
sites (Fig. 31-65).
Section 31-4. Post-Transcriptional Processing 1317
Figure 31-65 The organization of the rat ␣-tropomyosin gene
and the seven alternative splicing pathways that give rise to
cell-specific ␣-tropomyosin isoforms. The thin kinked lines
indicate the positions occupied by the introns before they are
spliced out to form the mature mRNAs. Tissue-specific exons are
indicated together with the amino acid (aa) residues they
encode:“constitutive” exons (those expressed in all tissues) are
1–385′ UT
Striated muscle
mRNA transcripts
Smooth muscle
Striated muscle′
Myoblast
Nonmuscle/
fibroblast
Hepatoma
Brain
3
′ UT
STR
STR
SM STR
3′ UT39–80 39–80 81–125 126–164 188
165–
213
189–
234
214–
257
235–
284
258–
284
258–
aa
green, those expressed only in smooth muscle (SM) are brown,
those expressed only in striated muscle (STR) are purple, and
those variably expressed are yellow. Note that the smooth and
striated muscle exons encoding amino acid residues 39 to 80 are
mutually exclusive; likewise, there are alternative 3¿-untranslated
(UT) exons. [After Breitbart, R.E., Andreadis, A., and
Nadal-Ginard, B., Annu. Rev. Biochem. 56, 481 (1987).]
JWCL281_c31_1260-1337.qxd 8/11/10 9:48 PM Page 1317
Alternative splicing occurs in all metazoa and is espe-
cially prevalent in vertebrates. In fact, microarray-based
comparisons of the cDNAs obtained from various tissues
indicate that ⬃95% of human structural genes are subject
to at least one alternative splicing event. This rationalizes
the discrepancy between the ⬃23,000 genes identified in
the human genome (Section 7-2Bc) and earlier estimates
that it contains 50,000 to 140,000 structural genes.
The variation in mRNA sequence can take several dif-
ferent forms: Exons can be retained in an mRNA or they
can be skipped; introns may be excised or retained; and the
positions of 5¿ and 3¿ splice sites can be shifted to make ex-
ons shorter or longer. Alterations in the transcriptional
start site and/or the polyadenylation site can further con-
tribute to the diversity of the mRNAs that are transcribed
from a single gene. In a particularly striking example, the
Drosophila protein Dscam (for Down syndrome cell-adhe-
sion molecule), which functions in neuronal development,
is encoded by 24 exons of which there are 12 mutually ex-
clusive variants of exon 4, 48 of exon 6, 33 of exon 9, and 2
of exon 17 (which are therefore known as cassette exons)
for total of 38,016 possible isoforms of this protein (com-
pared to ⬃14,000 identified genes in the Drosophila
genome).Although it is unknown if all possible Dscam iso-
forms are produced, experimental evidence suggests that
the Dscam gene expresses many thousands of them.
[Dscam is a membrane-anchored cell-surface protein of
the immunoglobulin superfamily. The specific isoform ex-
pressed in a given neuron binds to itself but rarely to other
isoforms. This permits the neuron to distinguish its own
processes (axons and dendrites) from those of other neu-
rons and thereby plays an essential role in neural pattern-
ing. However, the precise identity of a given isoform ap-
pears to be unimportant.] Clearly, the number of genes in
an organism’s genome does not by itself provide an ade-
quate assessment of its protein diversity. Indeed, it has
been estimated that, on average, each human structural
gene encodes three different proteins.
The types of changes that alternative splicing confers on
expressed proteins spans the entire spectrum of protein
properties and functions. Entire functional domains or
even single amino acid residues may be inserted into or
deleted from a protein, and the insertion of a stop codon
may truncate the expressed polypeptide. Splice variations
may, for example, control whether a protein is soluble or
membrane bound, whether it is phosphorylated by a spe-
cific kinase, the subcellular location to which it is targeted,
whether an enzyme binds a particular allosteric effector,
and the affinity with which a receptor binds a ligand.
Changes in an mRNA, particularly in its noncoding re-
gions, may also influence the rate at which it is transcribed
and its susceptibility to degradation. Since the selection of
alternative splice sites is both tissue- and developmental
stage-specific,splice site choice must be tightly regulated in
both space and time. In fact, it is estimated that from ⬃15%
to 50% of human genetic diseases are caused by point muta-
tions that result in pre-mRNA splicing defects. Some of these
mutations delete functional splice sites, thereby activating
nearby pre-existing cryptic splice sites. Others generate
new splice sites that are used instead of the normal ones
and yet others are in the genes encoding components of
the splicing machinery. In addition, tumor progression is
correlated with changes in levels of proteins implicated in
alternative splice site selection.
How are alternative splice sites selected? Well-understood
examples of such processes occur in the pathway responsible
for sex determination in Drosophila, two of which we dis-
cuss here:
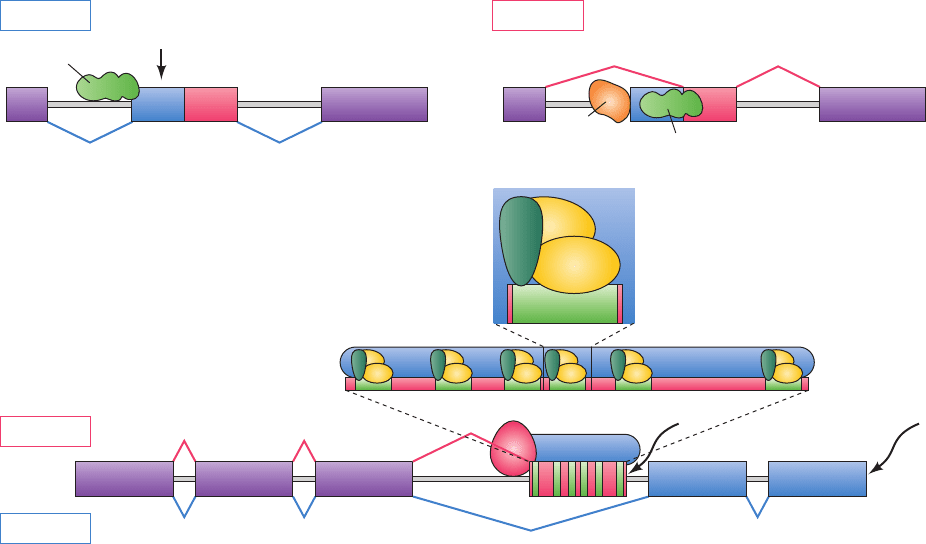
1. Exon 2 of transformer (tra) pre-mRNA contains two
alternative 3¿ splice sites (which succeed the excised in-
tron),with the proximal (close; to exon 1) site used in males
and the distal (far) site used in females (Fig. 31-66a). The
region between these two sites contains a Stop codon
(UAG). In males, the splicing factor U2AF binds to the
proximal 3¿ splice site to yield an mRNA containing this
premature stop codon, which thereby directs the synthesis
of truncated and hence nonfunctional TRA protein. In fe-
males, however, the proximal 3¿ splice site is bound by the
female-specific SXL protein, the product of the sex-lethal
(sxl) gene (which is only expressed in females), so as to
block the binding of U2AF, which then binds to the distal 3¿
splice site, thereby excising the UAG and inducing the ex-
pression of functional TRA protein (U2AF and TRA both
contain RS domains but not RRMs so that neither is an SR
protein).
2. In doublesex (dsx) pre-mRNA, the first three exons
are constitutively spliced in both males and females. How-
ever, the branch site immediately upstream of exon 4 has a
suboptimal pyrimidine tract to which U2AF does not bind
(Fig. 31-66b). Hence in males, exon 4 is not included in dsx
mRNA, leading to the synthesis of male-specific DSX-M
protein that functions as a repressor of female-specific
genes. However, in females, TRA protein promotes the co-
operative binding of the SR protein RBP1 and the SR-like
protein
TRA2 [the product of the transformer 2 (tra-2)
gene] to six copies of an exonic splice enhancer (ESE)
within exon 4. This heterotrimeric complex recruits the
splicing machinery to the upstream 3¿ splice site of exon 4,
leading to its inclusion in dsx mRNA.The resulting female-
specific DSX-F protein is a repressor of male-specific
genes.
Thus, the synthesis of functional TRA protein involves the
repression of a splice site, whereas the synthesis of female-
specific DSX-F protein involves the activation of a splice
site. Similar mechanisms of alternative splice site selection
have been identified in vertebrates.
In general, the decision as to whether an alternative
exon is kept or eliminated is determined by the activities
and concentrations of its various regulators, many of which
are SR proteins and hnRNPs. Hence the tissue-specific ex-
pression of these regulators and the phosphorylation state
of the SR proteins are important contributors to the com-
plex regulation of mRNA splicing. Moreover, extensive
analysis of the sequences of numerous alternative splice
sites has revealed the existence of a “splicing code” that
uses combinations of over 200 RNA features that are
1318 Chapter 31. Transcription
JWCL281_c31_1260-1337.qxd 8/11/10 9:48 PM Page 1318
present in both introns and exons and which are recog-
nized by the foregoing regulators.
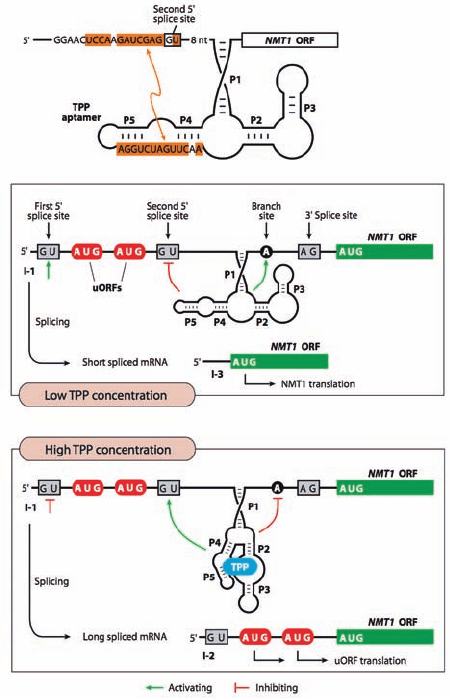
Riboswitches (Section 31-3H) have been implicated in
controlling alternative splicing in eukaryotes. For example,
as Breaker has shown, in the NMT1 gene of the bread mold
Neurospora crassa [which expresses an enzyme that partic-
ipates in the metabolism of TPP (thiamine pyrophos-
phate)], a TPP-sensing riboswitch is contained in an intron
that is located upstream of the mRNA’s normal AUG
translational start codon. This intron contains one 3¿ splice
site and two 5¿ splice sites with two AUG start codons be-
tween them. When the concentration of TPP is low so that
it does not bind to the riboswitch, one strand of the
riboswitch’s P4–P5 segment base-pairs with the second
(downstream) splice site so as to inactivate it (Fig. 31-67a).
The spliceosome then efficiently excises the entire intron
yielding an mRNA that is readily translated (I-3; Fig.31-67b).
However, when the concentration of TPP is high so that it
binds to the riboswitch, the riboswitch assumes a confor-
mation that activates the second splice site but occludes the
branch point A (Fig. 31-67c). Consequently, the spliceo-
some inefficiently excises only the downstream portion of
the intron.The two upstream AUG codons, which are pres-
ent in both the unspliced mRNA (I-1) and in the spliced
mRNA containing only the upstream portion of the intron
(I-2), compete for ribosomes with mRNA’s normal AUG
start codon and thereby repress the mRNA’s translation
(Fig. 31-67c).
n. AU–AC Introns Are Excised by
a Novel Spliceosome
A small fraction of introns (⬃0.3%) have AU rather
than GU at their 5¿ ends and AC rather than AG at their 3¿
ends, but are nevertheless excised via a lariat structure to
an internal intron A. These so-called AU–AC introns (al-
ternatively, AT–AC introns after their DNA sequences),
which occur in organisms as diverse as Drosophila, plants,
and humans, are excised by a novel so-called AU–AC
spliceosome (alternatively, an AT–AC spliceosome) that
has one snRNP, U5, in common with the major (GU–AG)
spliceosome, and three others, U11, U12, and
U4atac–U6atac, which are distinct from but structurally
and functionally analogous to U1, U2, and U4–U6. Curi-
ously, all genes known to contain AU–AC introns also con-
tain multiple major class introns. Moreover, AU–AC in-
trons are not conserved in either length or position in their
host genes. Thus, the functional and evolutionary signifi-
cance of the AU–AC spliceosome and introns is obscure.
o. Trans-Splicing
The types of splicing we have so far considered occur
within single RNA molecules and hence are known as
Section 31-4. Post-Transcriptional Processing 1319
Figure 31-66 Mechanisms of alternative splice site selection in
the Drosophila sex-determination pathway as described in the
text. In all panels, exons are represented by colored rectangles
and introns are shown as pale gray lines. (a) Alternative splicing
in tra pre-mRNA. UAG is a Stop codon. (b) Alternative splicing
in dsx pre-mRNA. The six ESEs (exonic splice enhancers) in
UAG
U2AF
Male
Female
Male
Female
SXL
U2AF
TRA
RBP1
TRA2
S
123
112233
456
pA pA
(a)
(b)
exon 4 are indicated by green rectangles and S represents the
splicing machinery. In females, polyadenylation (pA) of dsx
mRNA occurs downstream of exon 4, whereas in males, it occurs
downstream of exon 6. [After a drawing by Maniatis, T. and Tasic,
B., Nature 418, 236 (2002).]
JWCL281_c31_1260-1337.qxd 8/11/10 9:48 PM Page 1319
cis-splicing. The chemistry of the spliceosomal cis-splicing
reaction, however, is the same as would occur if the two ex-
ons to be joined initially resided on two different RNA
molecules, a process called trans-splicing. This, in fact, oc-
curs in trypanosomes (kinetoplastid protozoa; the cause of
African sleeping sickness).Trypanosomal mRNAs all have
the same 35-nt noncoding leader sequence, although this
leader sequence is not present in the corresponding genes.
Rather, this sequence is part of a so-called spliced leader
(SL) RNA that is transcribed from an independent gene.
The 5¿ splice site that succeeds the SL RNA leader se-
quence, and the branch site and 3¿ splice site that precede
the exon sequence have the same consensus sequences as
occur in the RNAs spliced by the major spliceosome. Con-
sequently, the SL RNA leader and the pre-mRNA are
joined in a trans-splicing reaction that resembles the
spliceosomal cis-splicing reaction (Fig. 31-53) with the ex-
ception that the product of the first transesterification re-
action is necessarily Y-shaped rather than lariat-shaped
(Fig. 31-68). Trypanosomes, whose pre-mRNAs lack in-
trons, nevertheless have U2- and U4–U6-snRNPs but lack
U1- and U5-snRNPs. However, the SL RNA, which is pre-
dicted to fold into three stem–loops and a single-stranded
Sm RNA–like motif as does U1-snRNA (Fig. 31-63a), ap-
parently carries out the functions of U1-snRNA in the
trans-splicing reaction.
Trans-splicing has been shown to occur in nematodes
(roundworms; e.g., C. elegans) and flatworms. These organ-
isms also carry out cis-splicing and, indeed, perform both
types of splicing on the same pre-mRNA. There are also
several reports that trans-splicing occurs in higher eukary-
otes such as Drosophila and vertebrates, but if it does occur,
it does so in only a few pre-mRNAs and at a very low level.
p. mRNA Is Methylated at Certain
Adenylate Residues
During or shortly after the synthesis of vertebrate pre-
mRNAs, ⬃0.1% of their A residues are methylated at their
N6 atoms. These m
6
A’s tend to occur in the sequence
RRm
6
ACX, where X is rarely G. Although the functional
significance of these methylated A’s is unknown, it should
be noted that a large fraction of them are components of
the corresponding mature mRNAs.
q. RNA Can Be Edited by the Insertion or Deletion
of Specific Nucleotides
Certain mRNAs from a variety of eukaryotic organisms
have been found to differ from their corresponding genes
in several unexpected ways, including C S U and U S C
changes, the insertion or deletion of U residues, and the in-
sertion of multiple G or C residues. The most extreme ex-
amples of this phenomenon, which occur in the mitochon-
1320 Chapter 31. Transcription
Figure 31-67 Control of translation by the N. crassa
TPP-sensing riboswitch through alternative splicing. (a) The
predicted secondary structure of the TPP aptamer, which resides
in the 5¿ untranslated region of the NMT1 mRNA. The sequence
of one strand of the P4-P5 stem is complementary (orange
shading) to a segment that overlaps the second 5¿ splice site.
(b) At low TPP concentrations, the aptamer inhibits (red tee)
splicing from the second 5¿ splice site while activating (green
arrow) the branch point A so that the spliceosome excises the
RNA between the first 5¿ splice site and the 3¿ splice site, yielding
an ORF (open reading frame; I-3) that is normally translated
from its AUG start codon. (c) At high TPP concentrations, the
binding of TPP to the aptamer activates the second 5¿ splice site
but occludes the branch point A.The spliceosome therefore
inefficiently excises the intron’s downstream portion yielding an
mRNA (I-2) that contains two uORFs (upstream ORFs) that
compete with the translation of the primary ORF. The unspliced
mRNA (I-1) also contains the two uORFs and hence is likewise
inefficiently translated. [Courtesy of Ronald Breaker,Yale
University.]
JWCL281_c31_1260-1337.qxd 8/11/10 9:48 PM Page 1320
