Voet D., Voet Ju.G. Biochemistry
Подождите немного. Документ загружается.

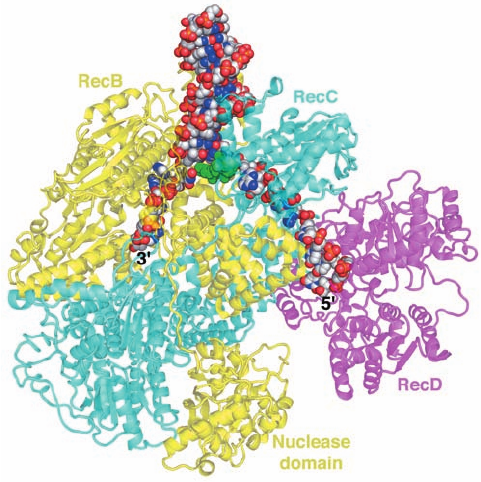
Dale Wigley determined the X-ray structure of E. coli
RecBCD in complex with a 51-nt DNA that could form a
hairpin loop containing an up to 21-bp dsDNA stem (Fig. 30-
75). The structure shows that RecB (1180 residues) and
RecC (1122 residues) are intimately intertwined with
RecB’s C-terminal nuclease domain connected to the rest
of the subunit by an extended 21-residue polypeptide
tether. A 15-bp segment of dsDNA enters the protein
through a tunnel between RecB and RecC. There it en-
counters a loop from RecC that appears to wedge the two
strands apart, with the 6-nt 3¿-ending single strand of the
DNA binding to RecB and the 10-nt 5¿-ending single
strand binding to RecD (608 residues; the 5-nt loop con-
necting the two strands of the dsDNA at the top of Fig. 30-
75 is disordered).The structure explains the different rates
of cleavage of the two DNA strands. The 3¿-ending strand
emerges from a tunnel through RecC in the vicinity of the
RecB nuclease domain, which is positioned to proces-
sively cleave it.The 5¿-ending strand competes with the 3¿-
ending strand for the nuclease site, but since the 5¿-ending
strand is less favorably located, it is cleaved less fre-
quently. However, after RecD has bound a Chi sequence,
the 3¿-ending strand is no longer available for cleavage,
which permits the nuclease to cleave the 5¿-ending strand
more frequently.
RecBCD can only commence unwinding DNA at a free
duplex end. Such ends are not normally present in E. coli,
which has a circular genome, but become available during
such recombinational processes as bacterial transforma-
tion, conjugation, and viral transduction, as well as at col-
lapsed replication forks.
e. RuvABC Mediates the Branch Migration
and the Resolution of the Holliday Junction
The branch migration of the RecA-generated Holliday
junction (Fig. 30-67e,f) requires the breaking and reform-
ing of base pairs as the bases exchange partners in passing
from one double helical stem to the other.Since ⌬G ⫽ 0 for
this process, it was initially assumed that it occurs sponta-
neously. However, such a process would move forward and
backward at random and, moreover, would be blocked by
as little as a single mismatched base pair. In E. coli, and
most other bacteria, branch migration is an ATP-driven
unidirectional process that is mediated by two proteins
whose synthesis is induced by the SOS response (Section
30-5D): RuvB (336 residues; Ruv for repair of UV dam-
age), an ATP-powered pump that drives branch migration
but binds only weakly to DNA; and RuvA (203 residues),
which binds to both a Holliday junction and to RuvB,
thereby targeting RuvB to the DNA.
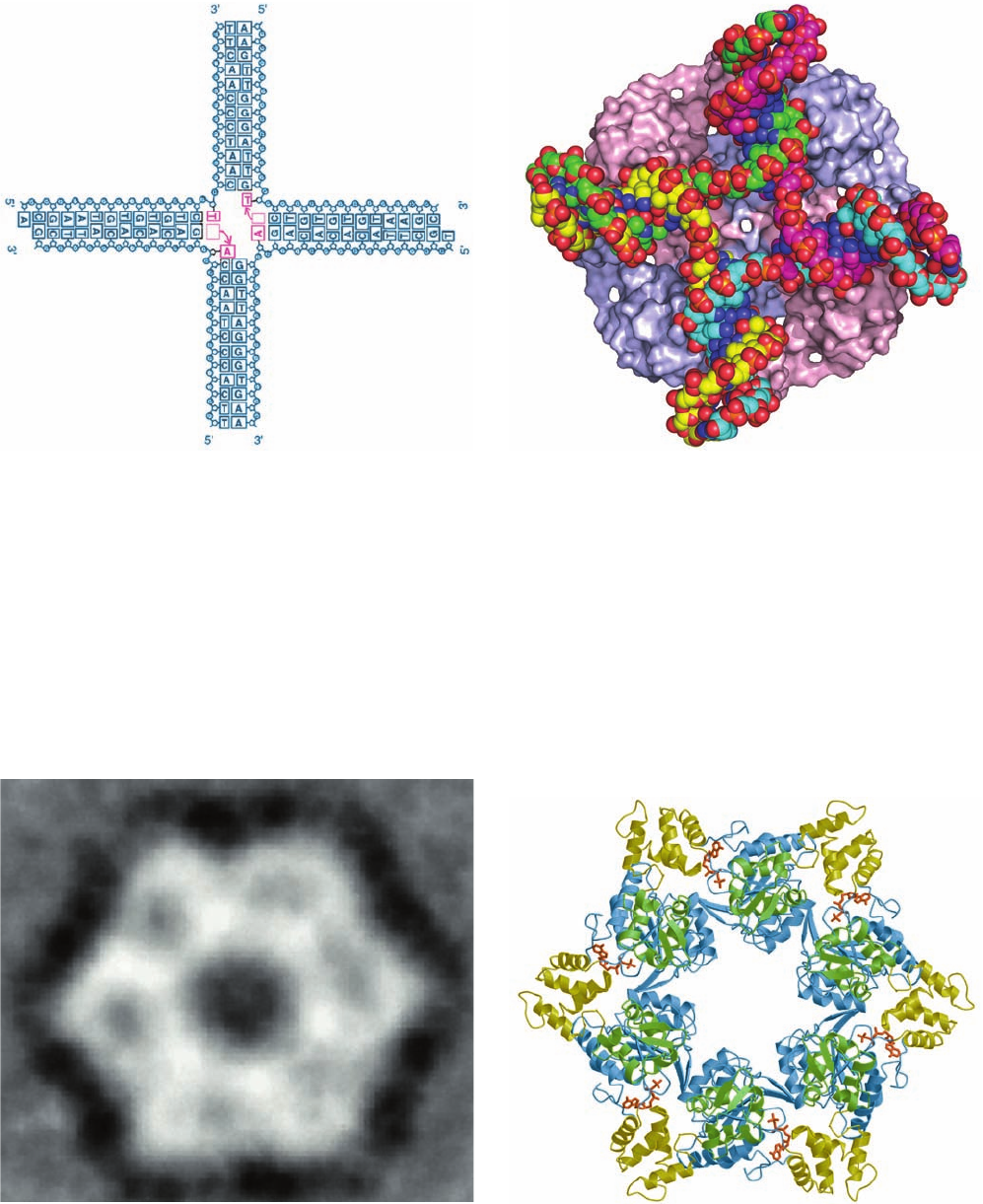
The X-ray structure of Mycobacterium leprae (the cause
of leprosy) RuvA in complex with a synthetic and immo-
bile Holliday junction (Fig. 30-76a), determined by
Morikawa, reveals that RuvA forms a homotetramer to
which the Holliday junction binds in its open-X conforma-
tion (Fig. 30-76b). The RuvA tetramer, which has the ap-
pearance of a four-petaled flower (it has C
4
symmetry
rather than the D
2
symmetry of the vast majority of ho-
motetramers), is relatively flat (80 ⫻ 80 ⫻ 45 Å) with one
square face concave and the other convex. The concave
face (that facing the viewer in Fig. 30-76b), which is highly
positively charged and is studded with numerous con-
served residues, has four symmetry-related grooves that
bind the Holliday junction’s four arms.This face’s centrally
located projection or “pin” is formed by the side chains of
Glu 55 and Asp 56 from each subunit, and hence the repul-
sive forces between them and the Holliday junction’s an-
ionic phosphate groups probably facilitate the separation
of the single-stranded DNA segments and guide them from
one double helix to another.
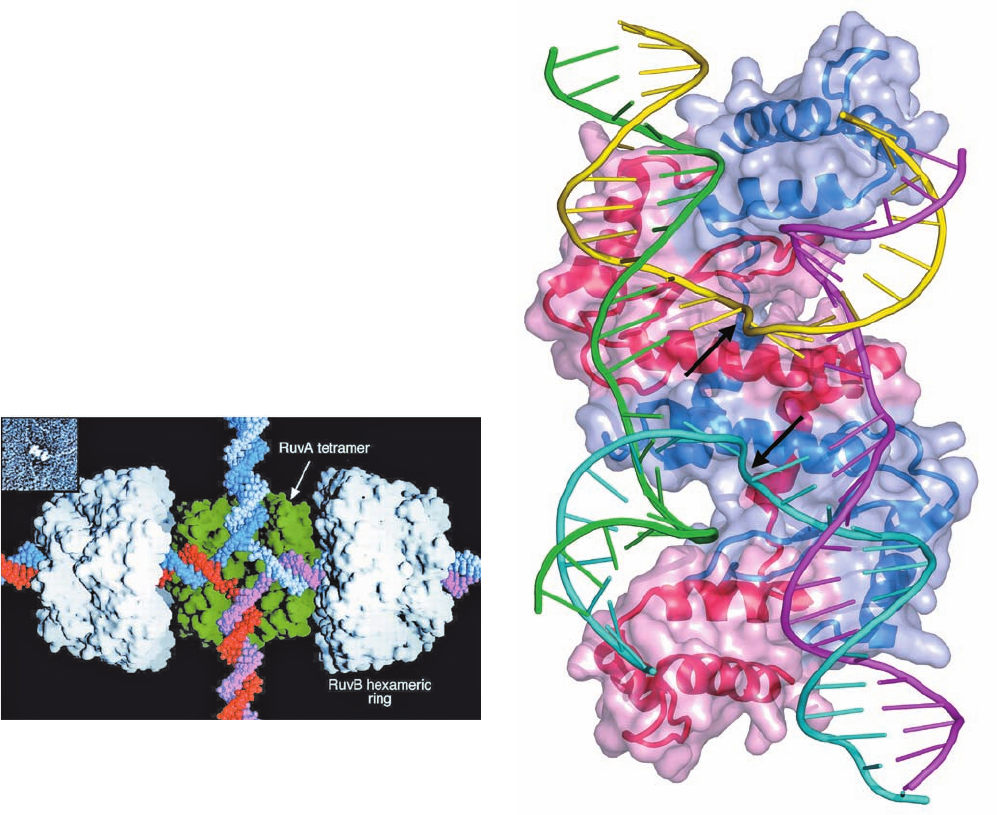
RuvB is a member of the AAA⫹ family of ATPases
(Section 30-2Ca). The X-ray structure of Thermus ther-
mophilus RuvB crystallized in the presence of both ADP
and AMPPNP, determined by Morikawa, reveals two mole-
cules of RuvB with somewhat different conformations: one
binding ADP and the other binding AMPPNP. Each RuvB
molecule consists of three consecutive domains arranged in
a crescentlike configuration with the adenine nucleotides
binding at the interface between its N-terminal and middle
domains. EM studies indicate that, in the presence of ds-
DNA, RuvB oligomerizes to form a hexamer (Fig. 30-77a),
Section 30-6. Recombination and Mobile Genetic Elements 1231
Figure 30-75 X-ray structure of E. coli RecBCD in complex
with a 51-nt DNA capable of forming a 21-bp hairpin loop. The
protein is drawn in semitransparent ribbon form with RecB
yellow, RecC cyan, and RecD magenta. Note how the RecB
nuclease domain is linked to the rest of the subunit by an
extended polypeptide tether.The DNA is shown in space-filling
form with C gray, N blue, O red, and P orange.A loop from
RecC, which is drawn in space-filling form in green, is situated so
as to wedge apart the two strands of the incoming dsDNA with
the 3¿-ending strand binding to the 3¿S5¿ helicase of RecB and
the 5¿-ending strand passing through RecC to bind to the 5’ S 3¿
helicase of RecD. [Based on an X-ray structure by Dale Wigley,
The London Research Institute, Herts, U.K. PDBid 3K70.]
JWCL281_c30_1173-1259.qxd 8/10/10 9:12 PM Page 1231
as do most other AAA⫹ family members, including the D2
domain of NSF (Fig. 12-78). A hexameric model of RuvB
(Fig. 30-77b), constructed by superimposing the N-terminal
domain of the RuvB monomer on the ATPase domains of
the NSF D2 hexamer,agrees well with the EM-based image
and contains no serious steric clashes. This 130-Å-diameter
1232 Chapter 30. DNA Replication, Repair, and Recombination
Figure 30-76 X-ray structure of a RuvA tetramer in complex
with a Holliday junction. (a) A schematic drawing of the
synthetic and immobile Holliday junction in this structure
showing its base sequence. The two A ⴢ T base pairs that are
disrupted at the crossover (and which, if the Holliday junction
consisted of two homologous dsDNAs, as it normally does, would
exchange base pairing partners) are magenta. (b) The
RuvA–Holliday junction complex as viewed along the protein
tetramer’s 4-fold axis. The protein is represented as its molecular
surface with its subunits alternately colored pink and light blue.
The DNA is drawn in space-filling form colored according to
atom type with C atoms in different chains in different colors,
N blue, O red, and P orange. [Part a courtesy of and Part b based
on an X-ray structure by Kosuke Morikawa, Biomolecular
Engineering Research Institute, Osaka, Japan. PDBid 1C7Y.]
Figure 30-77 Proposed structure of the T. thermophilus RuvB
hexamer. (a) An EM-based image reconstruction of RuvB
complexed with a 30-bp DNA (not visible) as viewed along its
6-fold axis. The image resolution is 30 Å. (b) A model of the
RuvB hexamer that was constructed from the X-ray structure of
RuvB monomers by superimposing their N-terminal domains on
the homologous ATPase domains of the NSF D2 homohexamer
(Fig. 12-78). The N-terminal, middle, and C-terminal domains are
blue, yellow, and green, respectively, and its bound AMPPNP is
drawn in stick form in red. [Courtesy of Kosuke Morikawa,
Biomolecular Engineering Research Institute, Osaka, Japan.
PDBid 1HQC.]
(a)
(b)
(b)
(a)
JWCL281_c30_1173-1259.qxd 8/10/10 9:12 PM Page 1232
hexameric model contains a 30-Å-diameter hole through
which a single dsDNA can readily be threaded (see below).
Moreover the six  hairpins, one per monomer, that have
been implicated in binding to RuvA are located on the top
face of the hexamer (as pictured in Fig. 30-77b).
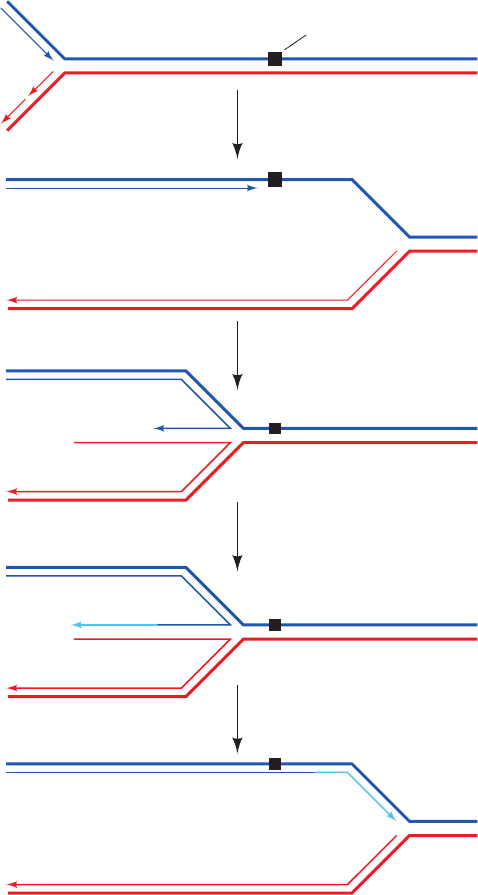
The EM images of the RuvAB–Holliday junction com-
plex indicate that RuvA binds two oppositely located
RuvB hexamers. This has led to the model of their interac-
tion depicted in Fig. 30-78 in which RuvA binds the Holli-
day junction and helps load the RuvB hexameric rings onto
two opposing arms of the Holliday junction.The two hexa-
meric rings are postulated to counter-rotate,each in the an-
ticlockwise direction looking toward the center of the junc-
tion, so as to screw the horizontal DNA strands through
the center of the junction and into the top and bottom dou-
ble helices, thereby effecting branch migration (although
rather than actually rotating relative to RuvA, a RuvB
hexamer might pull the dsDNA through its central hole by
“walking” up its grooves in a manner resembling that pos-
tulated for hexagonal helicases; Section 30-2Ca).The direc-
tion of branch migration depends on which pair of arms the
RuvB hexamers are loaded.
The final stage of homologous recombination is the res-
olution of the Holliday junction into its two homologous
dsDNAs. This process is carried out by RuvC, a homo-
dimeric endonuclease of 173-residue subunits whose X-ray
structure indicates that its active sites are located ⬃30 Å
apart on the same face of the protein. This suggests that
RuvC sits down on the open face of the RuvAB–Holliday
junction complex, that facing the viewer in Fig. 30-78, to
cleave oppositely located strands at the Holliday junction.
The resulting single-strand nicks in the now resolved
dsDNAs are sealed by DNA ligase.
The X-ray structure of RuvC in complex with DNA has
not been determined, although model building studies sug-
gest that it binds Holliday junction DNA in its stacked-X
conformation. However, Dietrich Suck determined the X-
ray structure of bacteriophage T4 endonuclease VII in
complex with a Holliday junction in the stacked-X confor-
mation (Fig. 30-79). RuvC and the 157-residue T4 endonu-
clease VII exhibit no structural similarity but both are
homodimers of relatively small subunits that have similar
Section 30-6. Recombination and Mobile Genetic Elements 1233
Figure 30-78 Model of the RuvAB–Holliday junction
complex. The model is based on electron micrographs such as
that in the inset.The proteins are represented by their surface
diagrams with the RuvA tetramer, as seen in its X-ray structure,
green and the two oppositely oriented RuvB hexamers white. The
DNA of the Holliday junction is drawn in space-filling form with
its homologous blue and pink strands complementary to its light
blue and red strands.The complex is postulated to drive branch
migration via the ATP-driven counter-rotation of the RuvB
hexamers relative to the RuvA tetramer.This pumps (screws)
the horizontal dsDNAs through the RuvB hexamers to the
center of the Holliday junction, where their strands separate and
then base-pair with their homologs to form new dsDNAs, which
are pumped out vertically. [Courtesy of Peter Artymiuk,
University of Sheffield, U.K.]
Figure 30-79 X-ray structure of bacteriophage T4
endonuclease VII resolving a Holliday junction as viewed along
its pseudo-2-fold axis. The Holliday junction DNA is drawn in
ladder form with each of its four different 24-nt strands
differently colored.The protein, a homodimer of 157-residue
subunits, is shown in ribbon form embedded in its semitransparent
molecular surface with one subunit red and the other blue. The
arrows indicate the symmetrically located DNA cleavage sites.
Compare the DNA in this structure to that in Fig. 30-68b. [Based
on an X-ray structure by Dietrich Suck, European Molecular
Biology Laboratory, Heidelberg, Germany. PDBid 2QNC.]
JWCL281_c30_1173-1259.qxd 8/26/10 8:22 PM Page 1233
functions: the resolution of Holliday junctions into two du-
plex DNAs by introducing symmetrically placed nicks in
equivalent strands (Fig. 30-67j).
The forgoing model of the RuvABC resolvosome pro-
vides a satisfying mechanism for branch migration and Holl-
iday junction resolution. However,there is a fly in this partic-
ular ointment. The X-ray structure of an M. leprae
RuvA–Holliday junction complex crystallized under condi-
tions different from that in Fig. 30-76, determined by Lau-
rence Pearl, resembles the complex in Fig. 30-76b but with a
second RuvA tetramer in face-to-face contact with the con-
cave (DNA-binding) side of the first. Hence, the Holliday
junction is contained in two intersecting tunnels running
through the resulting RuvA octamer.Are both RuvA–Holli-
day junction structures biologically relevant, or is one an arti-
fact of crystallization? Pearl argues that the extensive comple-
mentary contacts between the two RuvA tetramers, which are
strongly conserved, are unlikely to be artifactual and that a
single RuvA tetramer is unlikely to withstand the torque ex-
erted by the two (in effect) counter-rotating RuvB hexamers.
However,if the RuvA octamer is biologically relevant, one of
its tetramers would at some point have to dissociate in order
to allow RuvC access to the Holliday junction.Yet, modeling
studies indicate that the RuvC dimer cannot properly contact
the RuvB tetramer-bound Holliday junction without it chang-
ing from its open-X to its stacked-X conformation.Further in-
vestigations are necessary to resolve these inconsistencies.
f. Recombination Repair Reconstitutes Damaged
Replication Forks
Transformation, transduction, and conjugation are such
rare events that the vast majority of bacterial cells never par-
ticipate in these processes. Similarly, the only place in the
metazoan life cycle at which gene shuffling through homolo-
gous recombination occurs is in meiosis (Section 1-4A).Why
then do nearly all cells have elaborate systems for mediating
homologous recombination? It is because damaged replica-
tion forks occur at a frequency of at least once per bacterial
cell generation and perhaps 10 times per eukaryotic cell cy-
cle. The DNA lesions that damage the replication forks can
be circumvented via homologous recombination in a process
named recombination repair [translesion synthesis, which is
highly mutagenic, is a process of last resort (Section 30-
5Db)]. Indeed, the rates of synthesis of RuvA and RuvB are
greatly enhanced by the SOS response.Thus, as Michael Cox
pointed out, the primary function of homologous recombina-
tion is to repair damaged replication forks. In what follows, we
describe recombination repair as it occurs in E. coli.
Recombination repair is called into play when a replica-
tion fork encounters an unrepaired single-strand lesion
(Fig. 30-80):
1. DNA replication is arrested at the lesion but contin-
ues on the opposing undamaged strand for some distance
before the replisome fully collapses (Section 30-5Db).
2. The replication fork regresses to form a type of Hol-
liday junction dubbed a “chicken foot.” This process may
occur spontaneously as driven by the positive supercoiling
that has built up ahead of the replication fork, it may be
mediated by RecA, or it may be promoted by RecG, an
ATP-driven helicase that catalyzes branch migration at
DNA junctions with three or four branches.
3. The single-strand gap at the collapsed replication
fork, now an overhang, is filled in by Pol I.
4. Reverse branch migration mediated by RuvAB or
RecG yields a reconstituted replication fork, which sup-
ports replication restart (see below).
Note that this process does not actually repair the single-
strand lesion that has caused the problem but instead re-
constructs the replication fork in a way that permits the
previously discussed DNA repair systems (Section 30-5) to
eventually eliminate the lesion.
1234 Chapter 30. DNA Replication, Repair, and Recombination
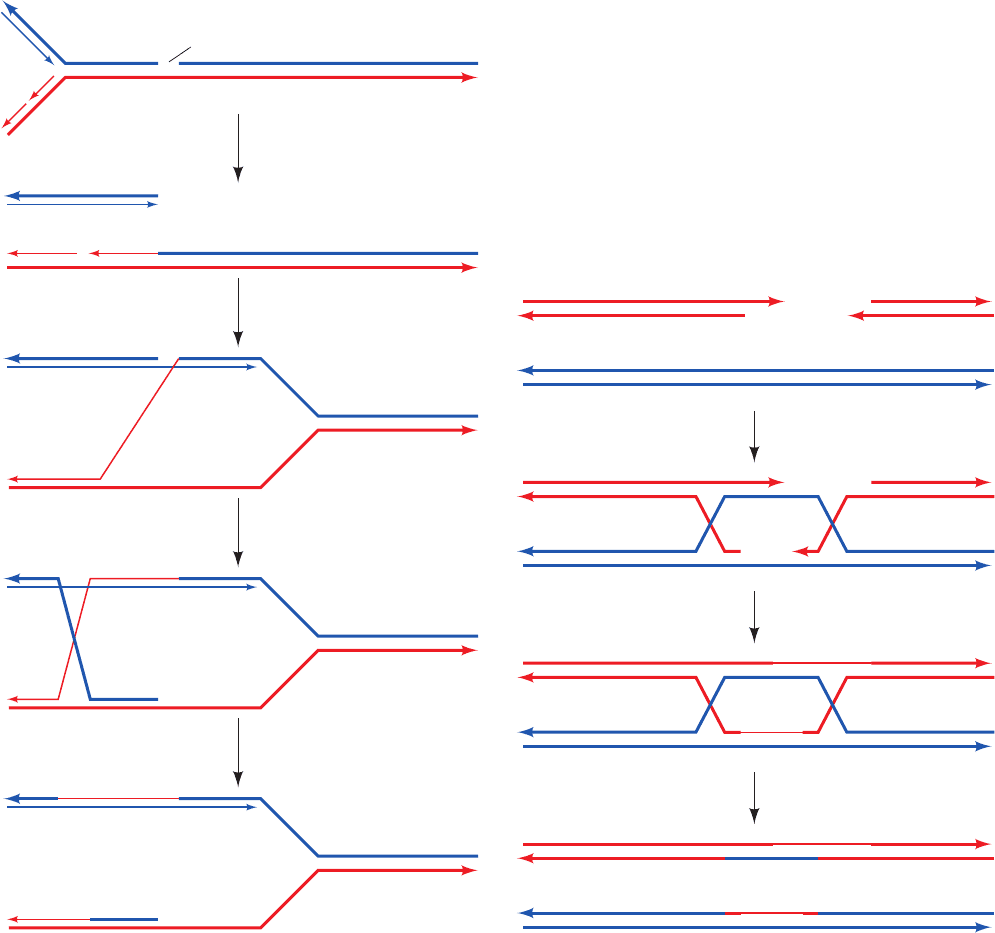
Figure 30-80 The recombination repair of a replication fork
that has encountered a single-strand lesion. Thick lines indicate
parental DNA, thin lines indicate newly synthesized DNA, the
cyan lines indicate DNA that was synthesized by Pol I, and
the arrows point in the 5¿S 3¿ direction. [After Cox, M.M.,
Annu. Rev. Genet. 35, 53 (2001).]
1 Replication fork
collapse
DNA lesion
Chicken foot
2 Fork regressionRecA
3 ReplicationPol I
4 Reverse branch
migration
RuvAB
JWCL281_c30_1173-1259.qxd 8/10/10 9:12 PM Page 1234
1 Rliti fk
DNA nick
2 DNA synthesis and
ligation
1 Formation of two
Holliday junctions
RAD51
3 Holliday junction
resolution
A second situation that requires recombination repair is
the encounter of a replication fork with an unrepaired
single-strand nick (Fig. 30-81):
1. When a single-strand nick is encountered, the repli-
cation fork collapses.
2. The repair process begins via the RecBCD plus
RecA–mediated invasion of the newly synthesized and un-
damaged 3¿-ending strand into the homologous dsDNA
starting at its broken end.
3. Branch migration, as mediated by RuvAB, then
yields a Holliday junction, which exchanges the replication
fork’s 3¿-ending strands.
4. RuvC then resolves the Holliday junction yielding a
reconstituted replication fork ready for replication restart.
Thus, the 5¿-ending strand of the nick has, in effect, become
the 5¿ end of an Okazaki fragment.
The final step in the recombination repair process is the
restart of DNA replication. This process is, of necessity, dis-
tinct from the replication initiation that occurs at oriC (Sec-
tion 30-3Ca).Origin-independent replication restart is medi-
ated by the same seven-protein primosome that initiates the
minus strand replication of bacteriophage X174 (Table 30-
4), which has therefore been named the restart primosome.
g. Recombination Repair Reconstitutes
Double-Strand Breaks
We have seen that double-strand breaks (DSBs) in
DNA can be rejoined, often mutagenically, by nonhomolo-
gous end-joining (NHEJ; Section 30-5E). DSBs may also
be nonmutagenically repaired through a recombination re-
pair process known as homologous end-joining, which oc-
curs via two Holliday junctions (Fig. 30-82):
1. The DSB’s double-stranded ends are resected to pro-
duce single-stranded ends. One of the 3¿-ending strands
invades the corresponding sequence of a homologous
Section 30-6. Recombination and Mobile Genetic Elements 1235
Figure 30-81 The recombination repair of a replication fork
that has encountered a single-strand nick. Thick lines indicate
parental DNA, thin lines indicate newly synthesized DNA, and
the arrows point in the 5¿S3¿ direction. [After Cox, M.M.,
Annu. Rev. Genet. 35, 53 (2001).]
Figure 30-82 The repair of a double-strand break in DNA by
homologous end-joining. Thick lines indicate parental DNA, thin
lines indicate newly synthesized DNA, and the arrows point in
the 5¿S3¿ direction. [After Haber, J.E., Trends Genet. 16, 259
(2000).]
1 Replication fork
collapse
2 Strand invasionRecBCD + RecA
3 Branch migrationRuvAB
4 Holliday junction
resolution
RuvC
JWCL281_c30_1173-1259.qxd 8/10/10 9:12 PM Page 1235

chromosome to form a Holliday junction, a process that, in
eukaryotes, is mediated by the RecA homolog RAD51.
The other 3¿-ending strand pairs with the displaced strand
segment on the homologous chromosome to form a second
Holliday junction.
2. DNA synthesis and ligation fills in the gaps and seals
the joints.
3. Both Holliday junctions are resolved to yield two in-
tact double strands.
Thus, the sequences that may have been expunged in the
formation of the DSB are copied from the homologous
chromosome. Of course, a limitation of homologous end-
joining, particularly in haploid cells, is that a homologous
chromosomal segment may not be available.
The importance of recombination repair in humans is
demonstrated by the observation that defects in the pro-
teins BRCA1 (1863 residues) and BRCA2 (3418
residues), both of which interact with RAD51, are associ-
ated with a greatly increased incidence of breast, ovarian,
prostate, and pancreatic cancers. Indeed, individuals with
mutant BRCA1 or BRCA2 genes have up to an 80% life-
time risk of developing cancer. Recombination can also
function to elongate shortened telomeres without the
need for telomerase.
B. Transposition and Site-Specific Recombination
In the early 1950s, on the basis of genetic analysis, Bar-
bara McClintock reported that the variegated pigmenta-
tion pattern of maize (Indian corn) kernels results from
the action of genetic elements that can move about the
maize genome. This proposal was resoundingly ignored
because it was contrary to the then held genetic ortho-
doxy that chromosomes consist of genes linked in fixed
order. Another 20 years were to pass before evidence of
mobile genetic elements was found in another organism,
E. coli.
It is now known that transposable elements or trans-
posons are common in both prokaryotes and eukaryotes,
where they influence the variation of phenotypic expres-
sion over the short term and evolutionary development
over the long term. Each transposon codes for the enzymes
that specifically insert it into the recipient DNA. This
process has been described as illegitimate recombination
because it requires no homology between donor and
recipient DNAs. Since the insertion site is chosen largely
at random, transposition is a potentially dangerous
process; the insertion of a transposon into an essential
gene will kill a cell together with its resident transposons.
Hence transposition is tightly regulated; it occurs at a rate
of only 10
⫺5
to 10
⫺7
events per element per generation.
The conditions that trigger transposition are, for the most
part, unknown.
a. Prokaryotic Transposons
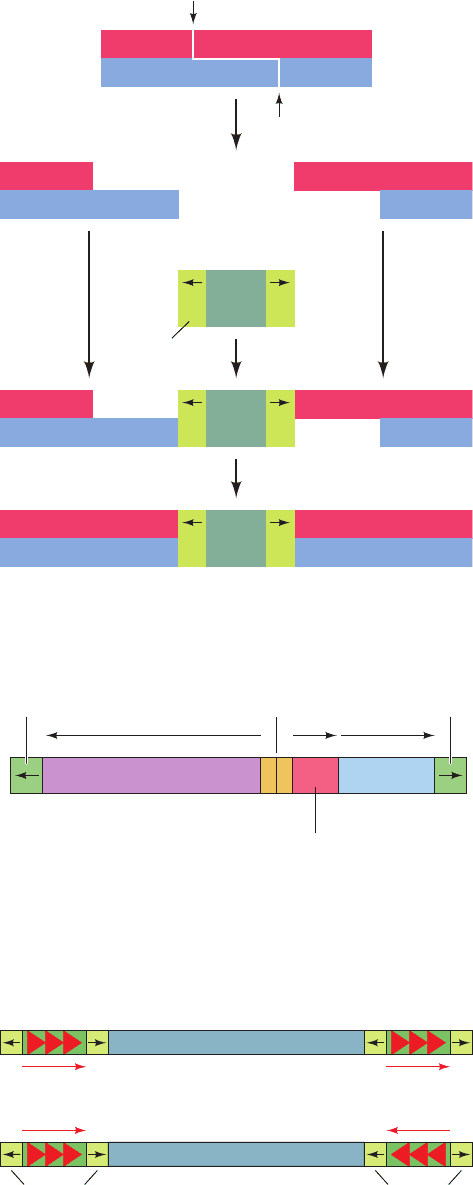
Prokaryotic transposons with three levels of complexity
have been characterized:
1. The simplest transposons, and the first to be char-
acterized, are named insertion sequences or IS elements.
They are designated by “IS” followed by an identifying
number. IS elements are normal constituents of bacterial
chromosomes and plasmids. For example, a common E.
coli strain has eight copies of IS1 and five copies of IS2.
IS elements generally consist of ⬍2000 bp. These com-
prise a so-called transposase gene, and in some cases a
regulatory gene, flanked by short inverted (having oppo-
site orientation) terminal repeats (Fig. 30-83 and Table
30-6). The inverted repeats are essential for transposi-
tion; their genetic alteration invariably prevents this
process. An inserted IS element is flanked by a directly
(having the same orientation) repeated segment of host
DNA (Fig. 30-83). This suggests that an IS element is in-
serted in the host DNA at a staggered cut that is later
filled in (Fig. 30-84). The length of this target sequence
(most commonly 5 to 9 bp), but not its sequence, is char-
acteristic of the IS element.
2. More complex transposons carry genes not involved
in the transposition process, for example, antibiotic resist-
ance genes. Such transposons are designated “Tn” followed
1236 Chapter 30. DNA Replication, Repair, and Recombination
Figure 30-83 Structure of IS elements. These and other
transposons have inverted terminal repeats (numerals) and are
flanked by direct repeats of host DNA target sequences (letters).
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
A
A′
B
B′
C
C′
D
D′
A
A′
B
B′
C
C′
D
D′
1
1′
2
2′
3
3′
4
4′
5
5′
5′
5
4′
4
3′
3
2′
2
1′
1
IS element
Target
sequence
Target
sequence
Inverted Direct Number of
Insertion Length Terminal Repeat at Copies in E. coli
Element (bp) Repeat (bp) Target (bp) Chromosome
IS1 768 23 9 5–8
IS2 1327 41 5 5
IS4 1428 18 11–13 5
IS5 1195 16 4 1–2
Table 30-6 Properties of Some Insertion Elements
Source: Mainly Lewin, B., Genes IX, p. 524, Oxford University Press (2008).
JWCL281_c30_1173-1259.qxd 8/10/10 9:12 PM Page 1236
by an identifying number. For example, Tn3 (Fig. 30-85)
consists of 4957 bp and has inverted terminal repeats of
38 bp each. The central region of Tn3 codes for three pro-
teins: (1) a 1015-residue transposase named TnpA; (2) a
185-residue protein known as TnpR, which mediates the
site-specific recombination reaction necessary to complete
the transposition process (see below) and also functions as
a repressor for the expression of both tnpA and tnpR; and
(3) a -lactamase that inactivates ampicillin (Section 11-3Bb).
The site-specific recombination occurs in an AT-rich region
known as the internal resolution site that is located be-
tween tnpA and tnpR.
3. The so-called composite transposons (Fig. 30-86)
consist of a gene-containing central region flanked by two
identical or nearly identical IS-like modules that have either
the same or an inverted relative orientation. It therefore
seems that composite transposons arose by the association
of two originally independent IS elements. Since the IS-like
modules are themselves flanked by inverted repeats, the
ends of either type of composite transposon must also
be inverted repeats. Experiments demonstrate that com-
posite transposons can transpose any sequence of DNA in
their central region.
There are two modes of transposition: (1) direct or simple
transposition, in which the transposon, as the name im-
plies, physically moves from one DNA site to another;
and (2) replicative transposition, in which the transposon
remains at its original site and a copy of it is inserted at a
target site. The two modes, as we shall see, have similar
mechanistic features and, indeed, some transposons can
move by either mode.
b. Direct Transposition of Tn5 Occurs by a
Cut-and-Paste Mechanism
Tn5 is a 5.8-kb composite transposon that contains the
gene encoding the 476-residue Tn5 transposase together
with three antibiotic resistance genes. It is flanked by in-
verted IS-like modules ending in 19-bp sequences called
outside end (OE) sequences. Tn5 undergoes direct trans-
position via a “cut-and-paste” mechanism that was eluci-
dated in large part by William Reznikoff (Fig. 30-87):
1. Each of Tn5’s two OE sequences on the donor DNA
is bound by a monomer of Tn5 transposase.
2. The transposase dimerizes to form a catalytically ac-
tive synaptic complex in which the transposon is held be-
tween the two transposase subunits.
3. Each transposase subunit activates a water molecule
to nucleophilically attack the outermost nucleotide of its
bound OE sequence, yielding a free 3¿-OH group. This 3¿-
OH group is then activated to attack the opposite strand
on the DNA to form a hairpin structure, thereby excising
the transposon from the DNA. The hairpin is then hy-
drolyzed to yield a blunt-ended dsDNA at each end of the
transposon, thus completing the “cut” portion of the trans-
position mechanism.
4. The synaptic complex binds to the target DNA.
Section 30-6. Recombination and Mobile Genetic Elements 1237
Figure 30-84 A model for the generation of direct repeats of
the target sequence by transposon insertion.
Figure 30-86 A composite transposon. This element consists
of two identical or nearly identical IS-like modules (green)
flanking a central region carrying various genes.The IS-like
modules may have either (a) direct or (b) inverted relative
orientations.
TATTA
ATAAT
T
A
A
T
T
A
T
A
A
T
T
A
A
T
T
A
T
A
A
T
T
A
A
T
T
A
T
A
A
T
TATTA
ATAA
IR
Transposon
Inverted
repeat
Filling in and ligation
Staggered cut
within host DNA
IR
T
IR IR
IR IR
Inverted
repeat
Internal
resolution site
Transposase β-Lactamase
tnpA amptnpR
Inverted
repeat
Repressor and
resolvase
Central region
IS-like
module
(a)
IS-like
module
Central region
Inverted
repeats
Inverted
repeats
(b)
Figure 30-85 A map of transposon Tn3.
JWCL281_c30_1173-1259.qxd 8/10/10 9:12 PM Page 1237
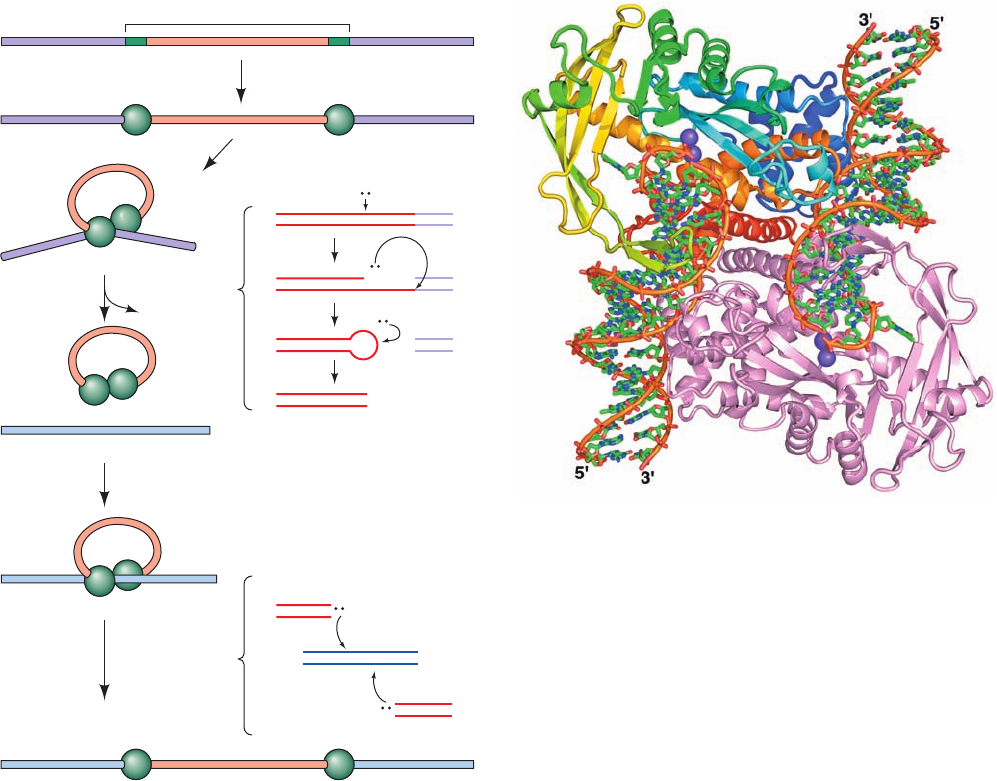
5. The transposon’s 3¿-OH groups nucleophilically at-
tack the target DNA on opposite strands spaced 9 bp apart,
thereby installing the transposon at the target site. Re-
markably, this reaction and the three preceding lytic reac-
tions are all mediated by the same catalytic site. The repair
of the oppositely located single-strand gaps (Fig. 30-84)
completes the “paste” portion of the mechanism.
Although, strictly speaking, not part of the transposition
process, the double-strand break in the donor DNA left by
the excision of the transposon must be repaired if the donor
DNA is to be propagated (in bacteria, the donor DNA is of-
ten a plasmid so that its loss has little effect on the cell since
plasmids are generally present in multiple copies).
The X-ray structure of a Tn5 synaptic complex (Fig. 30-88),
determined by Reznikoff and Ivan Rayment, provides a
model of the synaptic complex at the stage following its
cleavage from the donor DNA (the product of Step 3 in
Fig. 30-87). This 2-fold symmetric complex consists of a
dimer of Tn5 transposase subunits binding two 20-bp DNA
segments containing the Tn5 transposon’s 19-bp OE se-
quence with the outer end of each OE sequence bound to
the protein (and whose opposite ends would, in vivo, be
connected by the looped around transposon; Fig. 30-87).
Both transposase subunits extensively participate in bind-
ing each DNA segment, thereby explaining why the indi-
vidual subunits cannot cleave their bound DNA segments
before forming the synaptic complex.The protein holds the
DNA in a distorted B-DNA conformation with its two end
pairs of nucleotides no longer base paired. Indeed, the
penultimate base on the nontransferred strand is flipped
out of the double helix and binds in a hydrophobic pocket.
The transferred strand’s free 3¿-OH group, which occupies
the active site, is bound in the vicinity of a cluster of three
catalytically essential acidic residues, the so-called DDE
motif, which is shared with other transposases. In the X-ray
structure the DDE motif binds two Mn
2⫹
ions, although
physiologically it probably binds two Mg
2⫹
ions. This sug-
gests that transposases employ a metal-activated catalytic
1238 Chapter 30. DNA Replication, Repair, and Recombination
Figure 30-87 The cut-and-paste transposition mechanism
catalyzed by Tn5 transposase. The reactions comprising Steps 3
and 5 are indicated beside the braces to the right of these steps.
[After Davies, D.R., Goryshin, I.Y., Reznikoff, W.S., and
Rayment, I., Science 289, 77 (2000).]
Tn5 transposon DNA
Cleavage
donor DNA
Target capture
Strand transfer
Transposase binding
Target DNA
Synaptic complex
Donor DNADonor DNA
Dimerization
1
2
3
+
4
5
3′
5′
3′
5′
3′
+
5′
3′
5′
Tn5 Donor
Tn5
Tn5
H–O–H
H–O–H
O–H
Tn5
Tn5
3′
5′
3′
5′
Target
3′
5′
5′
3′
Tn5
O–H
H–O
Figure 30-88 X-ray structure of Tn5 transposase in complex
with a 20-bp DNA containing the OE sequence. The complex,
which represents the product of Step 3 in Fig. 30-87, is viewed
along its 2-fold axis with one of its two identical subunits colored
in rainbow order from N-terminus (blue) to C-terminus (red)
and the other subunit pink.The DNA is drawn in stick form with
C green, N blue, O red, and P orange and with successive P
atoms on the same polynucleotide connected by orange rods.The
bound Mn
2⫹
ions, which mark the enzyme’s active site, are
represented by purple spheres.The DNAs’ reactive 3¿-OH
groups are located at these active sites. [Based on an X-ray
structure by William Reznikoff and Ivan Rayment, University of
Wisconsin. PDBid 1MUS.]
JWCL281_c30_1173-1259.qxd 8/10/10 9:12 PM Page 1238
mechanism similar to that of the DNA polymerases (Sec-
tion 30-2Af).The facing surface of the protein in Fig. 30-88
is positively charged with a prominent groove running
from upper left to lower right that forms the apparent
binding site for the target DNA.
Wild-type Tn5 transposase has such low catalytic activ-
ity that it is undetectable in vitro. However, that in the
X-ray structure is a hyperactive mutant form that contains
the mutations E54K and L372P (an unusual circumstance
in that it is far more common to mutationally inhibit an en-
zyme under crystallographic study so as to trap it at some
specific stage along its reaction pathway). Lys 54 is hydro-
gen bonded to O4 of a thymine base on the transferred
strand. In the wild-type transposase, Glu 54 would proba-
bly have an unfavorable charge–charge repulsion with a
nearby phosphate group, thus providing a structural basis
for the increased activity of the E54K mutant. The L372P
mutation disorders the peptide segment between residues
373 and 391 (it is ordered in the X-ray structure of wild-
type Tn5 transposase lacking its N-terminal 55 residues),
thereby suggesting that this mutation facilitates a confor-
mational change required for substrate binding.
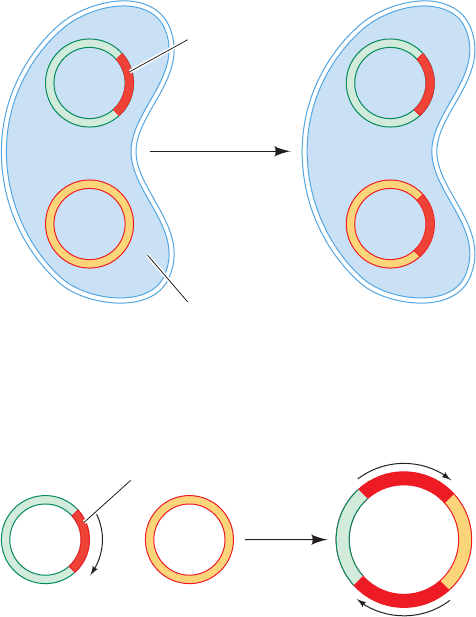
c. Replicative Transposition Occurs via Cointegrates
If a plasmid carrying a transposon resembling Tn3 is in-
troduced into a bacterial cell carrying a plasmid that lacks
the transposon, in some of the progeny cells both types of
plasmid will contain the transposon (Fig. 30-89). Evidently,
such transposition involves the replication of the transposon
into the recipient plasmid rather than its transfer from donor
to recipient.
Two plasmids, one containing a replicative transposon,
will occasionally fuse to form a so-called cointegrate con-
taining like-oriented copies of the transposon at both junc-
tions of the original plasmids (Fig. 30-90). Yet, some of the
progeny of a cointegrate-containing cell lack the cointegrate
and instead contain both original plasmids, each with one
copy of the transposon (Fig. 30-89). The cointegrate must
therefore be an intermediate in the transposition process.
Although the mechanism of replicative transposition has
not been fully elucidated, a plausible model for this process
(and there are several) that accounts for the foregoing ob-
servations consists of the following steps (Fig. 30-91):
1. A pair of staggered single-strand cuts, such as is dia-
grammed in Fig. 30-84, is made by the transposon-encoded
transposase at the target sequence of the recipient plasmid
so as to liberate 3¿-OH ends. Similarly, single-strand cuts
are made on opposite strands to either side of the transpo-
son. Note that these reactions resemble those catalyzed by
Tn5 transposase (Fig. 30-87).
2. Each of the transposon’s free ends is ligated to a pro-
truding single strand at the insertion site.This forms a repli-
cation fork at each end of the transposon.
3. The transposon is replicated, thereby yielding a
cointegrate.
4. Through a site-specific recombination between the
internal resolution sites of the two transposons, the cointe-
grate is resolved into the two original plasmids, each of
which contains a transposon. This crossover process is cat-
alyzed by transposon-encoded recombinases (TnpR in
Tn3; also known as resolvases) rather than RecA; transpo-
sition proceeds normally in recA
⫺
cells (although RecA
will resolve a cointegrate containing a transposon with a
mutant resolvase and/or an altered internal resolution site,
albeit at a much reduced rate).
Site-specific recombinases fall into only two protein
families, serine recombinases and tyrosine recombinases,
which are named after the amino acid residue that forms a
transient covalent linkage to the DNA during the recombi-
nase reaction.As we shall, these two types of recombinases
function via different mechanisms.
d. ␥␦ Resolvase Catalyzes Site-Specific
Recombination
The ␥␦ resolvase, a serine recombinase that forms a ho-
modimer in solution, is a TnpR homolog that is encoded
by the ␥␦ transposon (a member of the Tn3 family of
replicative transposons; Fig. 30-85). It catalyzes a site-
specific recombination event in which a cointegrate con-
taining two copies of the ␥␦ transposon is resolved, via
double-strand DNA cleavage, strand exchange, and
Section 30-6. Recombination and Mobile Genetic Elements 1239
Figure 30-89 Replicative transposition. This type of
transposition inserts a copy of the transposon at the target site
while another copy remains at the donor site.
Figure 30-90 A cointegrate. This structure forms by the fusion
of two plasmids, one carrying a transposon, such that both
junctions of the original plasmid are spanned by transposons
with the same orientation (arrows).
many
generations
Transposon
Bacterium
A
B
A
B'
Transposon
A
+
B
Cointegrate
JWCL281_c30_1173-1259.qxd 8/10/10 9:12 PM Page 1239
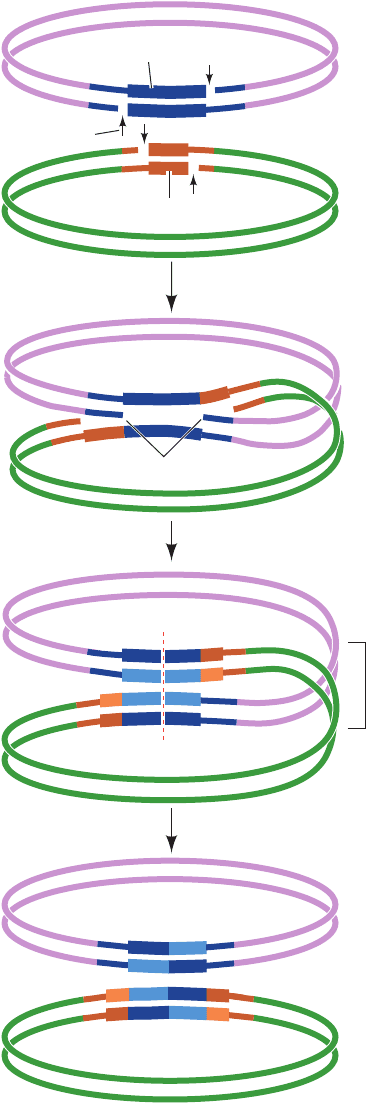
religation (the last step in Fig. 30-91), into two catenated
(linked) dsDNA circles that each contain one copy of the
␥␦ transposon (it also serves as its own transcriptional re-
pressor as does TnpR).The ␥␦ transposon contains a 114-bp
res site that includes three binding sites for ␥␦ resolvase
dimers, each of which contains an inverted repeat of the ␥␦
resolvase’s 12-bp recognition sequence. The resolution of
the cointegrate involves the binding of a ␥␦ resolvase ho-
modimer to all six of these binding sites in the cointegrate
(three from each of its two transposons) as is diagrammed
in Fig. 30-92. The reaction proceeds via the formation of a
transient phosphoSer bond between Ser 10 and the 5¿-
phosphate at each cleavage site.
The X-ray structure of the ␥␦ resolvase homotetramer
in complex with two 34-bp palindromic dsDNA segments
containing an inverted repeat of the 12-bp recognition se-
quence separated by an 4-bp spacer (Fig. 30-93), deter-
mined by Nigel Grindley and Steitz, reveals that this synap-
tic tetramer has D
2
symmetry. Each 183-residue resolvase
monomer consists of an N-terminal catalytic domain
(residues 1–120) and a C-terminal DNA-binding domain
(residues 148–183) connected by an extended arm
(residues 121–147). Both dsDNAs, which are located at the
periphery of the protein core, have been cleaved with each
of the resulting four 5¿ ends in phosphoSer linkage to the
resolvase. This preserves the free energy of the cleaved
phosphodiester bond so that it can later be reformed with
a different partner, much as occurs with topoisomerases
(Section 29-3C).
Each centrally located catalytic domain approaches its
bound DNA from its minor groove side with its C-terminal
helix (helix E) binding over the minor groove (the segment
of the E helix that contacts the DNA is disordered in the
absence of the DNA). Each C-terminal domain binds over
the major groove of its recognition sequence on the oppo-
site side of the DNA from its attached catalytic domain
with the extended arm that connects them running more or
less along the DNA’s minor groove.The two C-terminal do-
mains of the resolvase subunits labeled L and R (and the
symmetry-related L¿ and R¿ subunits) in Fig. 30-93 are
thereby separated by two helical turns along the cleaved
DNA, the segments of which closely assume the B-DNA
conformation. Each C-terminal helix binds in the DNA’s
major groove and, together with its preceding helix, forms
a helix–turn–helix (HTH) motif, a common sequence-spe-
cific DNA-binding motif that occurs mainly in prokaryotic
transcriptional repressors and activators (Section 31-3Da).
The structure of the L–R dimer closely resembles that in
the X-ray structure of the dimer bound to uncleaved site I
DNA (the presynaptic dimer). This, and the short (17 Å)
distance between the free 3¿-OH group in the L subunit
and the phosphoSer bond in the R subunit compared to
other such distances in the complex (L–L¿ and L–R¿), indi-
cates that the L–R and L¿–R¿ dimers correspond to the ini-
tial site I–bound dimers soon after cleavage or just before
religation (Fig. 30-92). Consequently, the interface between
the L–R and L¿–R¿ dimers must be the newly formed
synaptic interface.
How are the DNA strands in the synaptic complex
exchanged, that is, how is the DNA bound to the L sub-
unit ligated to the DNA bound to either the L¿ subunit or
the R¿ subunit (and R to either R¿ or L¿)? In either case,
the free 3¿-OH group on each subunit is ⬃50 Å from the
1240 Chapter 30. DNA Replication, Repair, and Recombination
Transposon
Cleavage
sites
Donor plasmid
A
B
CD
Target sequence
Recipient plasmid
Site-specific cleavage
and ligation
A
B
C
D
5′
5′
3′
3′
Replication forks
Replication
D
A
C
B
Recombination sites
Resolution
Donor plasmid
restored
Recipient plasmid
containing
transposon
and duplicated
target sequence
1
2
3
4
A
B
C
D
Cointegrate
Figure 30-91 A model for transposition involving the
intermediacy of a cointegrate. Here more lightly shaded bars
represent newly synthesized DNA. [After Shapiro, J.A., Proc.
Natl.Acad. Sci. 76, 1934 (1979).]
JWCL281_c30_1173-1259.qxd 8/10/10 9:12 PM Page 1240
