Voet D., Voet Ju.G. Biochemistry
Подождите немного. Документ загружается.

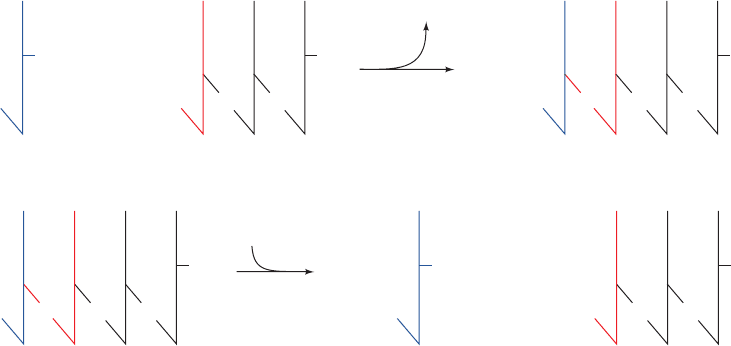
RNA is eventually replaced by DNA under conditions that
permit accurate base pairing to be achieved.
One might wonder why cells have evolved the complex
system of discontinuous lagging strand synthesis rather
than a DNA polymerase that could simply extend DNA
chains in their 3¿S5¿ direction. Consideration of the chem-
istry of DNA chain extension also leads to the conclusion
that this system promotes high-fidelity replication.The link-
ing of 5¿-deoxynucleotide triphosphates in the 3¿S5¿ direc-
tion would require the retention of the growing chain’s 5¿-
terminal triphosphate group to drive the next coupling step
(Fig. 30-39a). On editing a mispaired 5¿-terminal nucleotide
(Fig. 30-39b), this putative polymerase would—in analogy
with Pol I, for example—excise the offending nucleotide,
leaving either a 5¿-OH or a 5¿-phosphate group. Neither of
these terminal groups is capable of energizing further chain
extension. A proofreading 3¿S5¿ DNA polymerase would
therefore have to be capable of reactivating its edited prod-
uct. The inherent complexity of such a system has presum-
ably selected against its evolution.
4 EUKARYOTIC REPLICATION
There is a remarkable degree of similarity between eukary-
otic and prokaryotic DNA replication mechanisms. Never-
theless, there are important differences between these two
replication systems as a consequence of the vastly greater
complexity of eukaryotes in comparison to prokaryotes.
For example, eukaryotic chromosomes are structurally
complicated and dynamic complexes of DNA and protein
(Section 34-1) with which the replication machinery must
interact in carrying out its function. Consequently, as is true
of most aspects of biochemistry, our knowledge of how
DNA is replicated in eukaryotes has lagged well behind
that for prokaryotes, although in recent years there has
Section 30-4. Eukaryotic Replication 1201
at aberrantly high levels is more likely to be misincorpo-
rated and, conversely, one present at low levels is more
likely to be replaced by the dNTPs present at higher levels.
2. The polymerase reaction itself has extraordinary fi-
delity. This is because, as we have seen (Section 30-2Ae),
the polymerase reaction occurs in two stages: (1) a binding
step in which the incoming dNTP base-pairs with the tem-
plate while the enzyme is in an open conformation that
cannot catalyze the polymerase reaction; and (2) a catalysis
step in which the polymerase forms a closed conformation
about the newly formed base pair,which properly positions
its catalytic residues (induced fit). Since the formation of
the closed conformation requires that the incoming dNTP
form a Watson–Crick-shaped base pair with the template,
the conformation change constitutes a double check for
correct base pairing.
3. The 3¿S5¿ exonuclease functions of Pol I and Pol III
detect and eliminate the occasional errors made by their
polymerase functions. In fact, mutations that increase a
DNA polymerase’s proofreading exonuclease activity de-
crease the rates of mutation of other genes.
4. A remarkable battery of enzyme systems, contained
in all cells, function to repair residual errors in the newly
synthesized DNA as well as any damage that it may incur
after its synthesis through chemical and/or physical insults.
We discuss these DNA repair systems in Section 30-5.
In addition, the inability of a DNA polymerase to initiate
chain elongation without a primer is a feature that increases
DNA replication fidelity. The first few nucleotides of a
chain to be coupled together are those most likely to be
mispaired because of the cooperative nature of base pairing
interactions (Section 29-2). The editing of a short duplex
oligonucleotide is similarly an error-prone process.The use
of RNA primers eliminates this source of error since the
OH
ppp ppp
(a)
+
…
ppp
H
2
O
pp
PP
i
…
pp p
OH
ppp
(b)
+
…
pp pppp
…
pp p
3⬘5⬘
3⬘5⬘
Figure 30-39 Chemical consequences if a DNA polymerase
could synthesize DNA in its 3ⴕ S 5ⴕ direction. (a) The coupling
of each nucleoside triphosphate to the growing chain would be
driven by the hydrolysis of the previously appended nucleoside
triphosphate. (b) The editorial removal of an incorrect 5¿-
terminal nucleoside triphosphate would render the DNA chain
incapable of further extension.
JWCL281_c30_1173-1259.qxd 8/10/10 9:11 PM Page 1201
1202 Chapter 30. DNA Replication, Repair, and Recombination
cer; Sections 19-3B and 34-4C); by the surgical removal of
a tissue, which results in its rapid regeneration; or by pro-
teins known as mitogens, which bind to cell-surface recep-
tors and induce cell division (Section 34-4D).
a. The Cell Cycle Is Controlled by Cyclins and
Cyclin-Dependent Protein Kinases
The progression of a cell through the cell cycle is regu-
lated by proteins known as cyclins and cyclin-dependent
protein kinases (Cdks). Cyclins are so named because they
are synthesized during one phase of the cell cycle and com-
pletely degraded during a succeeding phase (protein degra-
dation is discussed in Section 32-6). A particular cyclin
specifically binds to and thereby activates its corresponding
Cdk(s) to phosphorylate its target proteins, thus activating
these proteins to carry out the processes comprising that
phase of the cell cycle. In order to enter a new phase in the
cell cycle, a cell must satisfy a corresponding checkpoint,
which monitors whether the cell has satisfactorily completed
the preceding phase [e.g., the attachment of all chromo-
somes to the mitotic spindle must precede mitosis (Section
1-4Aa); if this were not the case for even one chromosome,
one daughter cell would lack this chromosome and the other
would have two, both deleterious if not lethal conditions]. If
the cell has not met the criteria of the checkpoint, the cell cy-
cle is slowed or even arrested until it does so.We further dis-
cuss cell cycle control in Section 34-4C.
B. Eukaryotic Replication Mechanisms
Much of what we know about eukaryotic DNA replication
has been learned from studies on budding yeast (Saccha-
romyces cerevisiae) and fission yeast (Schizosaccharomyces
pombe), the simplest eukaryotes, and on simian virus 40
(SV40), which has a 5243-bp circular DNA chromosome
that has only one replication origin. However, studies of
DNA replication in the cells of metazoa (multicellular ani-
mals), particularly Drosophila, Xenopus laevis (an African
clawed toad, whose eggs are easily studied), and humans,
have also led to important advances in our knowledge.
a. Eukaryotic Cells Contain Numerous
DNA Polymerases
The many known DNA polymerases can be classified
into six families based on phylogenetic relationships. Mem-
bers of families A (e.g., E. coli Pol I), B (e.g., E. coli Pol II),
and C (e.g., E. coli Pol III) encompass all replicative poly-
merases as well as some repair polymerases, family D occurs
only in archaea where its functions are poorly understood,
and families X and Y participate in DNA repair. The fingers
and thumb domains have structures that are unique to each
family, whereas the catalytic residue–containing palm do-
mains are similar in families A, B, and Y. Animal cells ex-
press at least four distinct types of DNA polymerases that
are implicated in DNA replication (Table 30-5). They are
designated, in the order of their discovery, DNA poly-
merases (pols) ␣, ␥, ␦, and ε (alternatively, POLA, POLG,
POLD1, and POLE), of which pol ␥ is a member of family A
and the others are members of family B.
been significant progress in our understanding of this es-
sential process. In this section, we outline what is known
about DNA replication in eukaryotes. We also discuss two
DNA polymerases that are peculiar to eukaryotic systems:
reverse transcriptase and telomerase.
A. The Cell Cycle
The cell cycle, the general sequence of events that occur
during the lifetime of a eukaryotic cell, is divided into four
distinct phases (Fig. 30-40):
1. Mitosis and cell division occur during the relatively
brief M phase (for mitosis).
2. This is followed by the G
1
phase (for gap),which cov-
ers the longest part of the cell cycle.This is the main period
of cell growth.
3. G
1
gives way to the S phase (for synthesis), which in
contrast to events in prokaryotes, is the only period in the
cell cycle when DNA is synthesized.
4. During the relatively short G
2
phase, the now
tetraploid cell prepares for mitosis. It then enters M phase
once again and thereby commences a new round of the cell
cycle.
The cell cycle for cells in culture typically occupies a 16-
to 24-h period. In contrast, cell cycle times for the different
types of cells of a multicellular organism may vary from as
little as 8 h to ⬎100 days. Most of this variation occurs in
the G
1
phase. Moreover, many terminally differentiated
cells, such as neurons or muscle cells, never divide; they as-
sume a quiescent state known as the G
0
phase.
A cell’s irreversible “decision” to proliferate is made
during G
1
. Quiescence is maintained if, for example, nutri-
ents are in short supply or the cell is in contact with other
cells (contact inhibition). Conversely, DNA synthesis may
be induced by various agents such as carcinogens or tumor
viruses, which trigger uncontrolled cell proliferation (can-
Figure 30-40 The eukaryotic cell cycle. Cells in G
1
may enter
a quiescent phase (G
0
) rather than continuing about the cycle.
Preparation
for mitosis
(2–6 h)
Mitosis
and cell
division
(1 h)
DNA
replication
(6–8 h)
Commitment
to DNA
replication
(~10 h)
Quiescence
(variable)
G
2
M
G
1
G
0
S
JWCL281_c30_1173-1259.qxd 8/10/10 9:11 PM Page 1202
3. There is a large N-terminal domain that A-family
polymerases lack.
4. The template strand enters the active site from a cleft
between the N-terminal and exonuclease domains,
whereas in A-family polymerases, it does so from the fin-
gers domain.
5. The newly formed dsDNA nearest the active site has
a B-DNA-like conformation instead of the A-DNA-like
conformation observed in A-family polymerases (Section
30-2Ae).
Section 30-4. Eukaryotic Replication 1203
Pol ␣ occurs only in the cell nucleus where it partici-
pates in the replication of chromosomal DNA. This func-
tion was largely established through the use of its specific
inhibitor aphidicolin
and by the observation that pol ␣ activity varies with the rate
of cellular proliferation. Pol ␣, as do all DNA polymerases,
replicates DNA by extending a primer 5¿S3¿ under the di-
rection of a single-stranded DNA template. This hetero-
tetramer, which lacks exonuclease activity, consists of a 167-
kD polymerase subunit, a 48-kD primase subunit, a 62-kD
subunit that is required for full primase activity, and a 79-kD
subunit that is implicated in the regulation of initiation,all of
which are collectively known as pol ␣/primase.
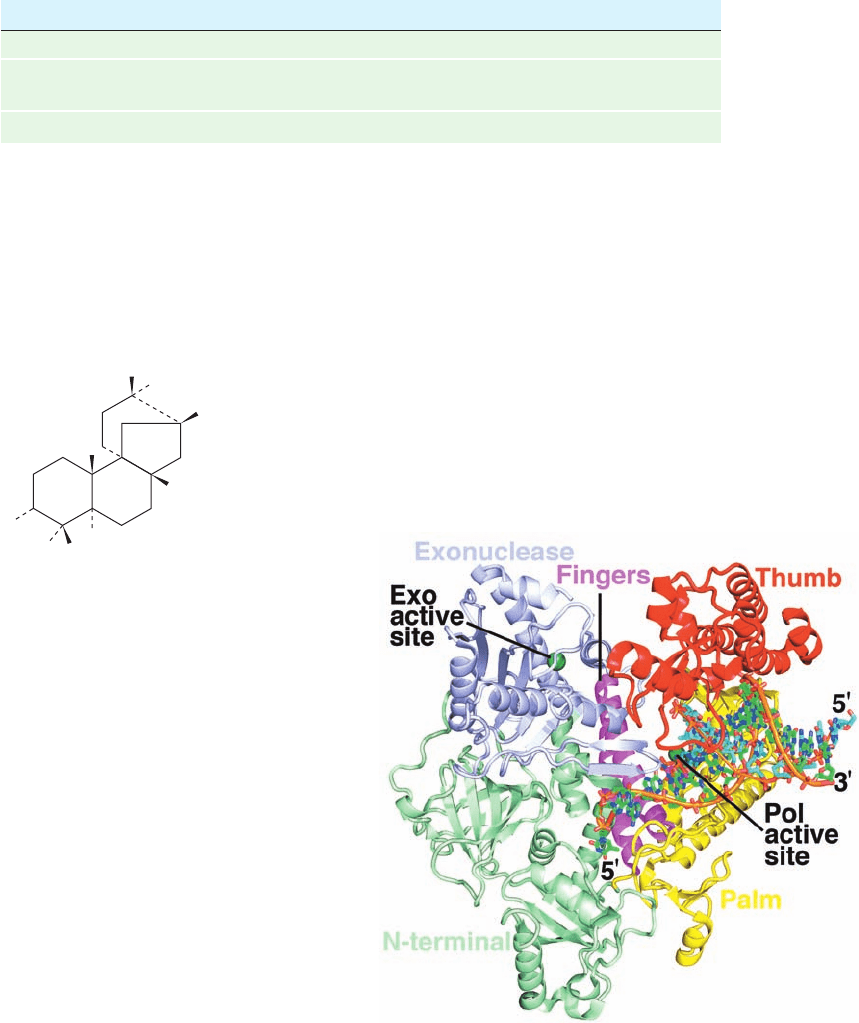
Pol ␦ is a heterotrimer whose 125-kD catalytic subunit
lacks an associated primase but contains a proofreading
3¿S5¿ exonuclease domain. The X-ray structure of the
yeast pol ␦ catalytic subunit (also called pol ␦), determined
by Aneel Aggarwal, reveals that this enzyme consists of
five domains arranged around a central hole that is near its
polymerase active site (Fig. 30-41). It has the right-hand-
like architecture first seen in A-family DNA polymerases
(Figs. 30-8 and 30-9), and its palm domain has a structurally
similar core that contains the two invariant Asp residues
implicated in the nucleotidyl transfer mechanism (Fig.30-10).
However, there are major differences between A-family
polymerases (e.g., Fig. 30-8) and pol ␦, which is representa-
tive of B-family polymerases. Most notably, in pol ␦:
1. The fingers domain, which consists of only a pair of
antiparallel helices, is rotated by ⬃60° relative to that in A-
family polymerases.
2. The exonuclease domain projects from the top of the
fingers domain rather than from the bottom of the palm
domain, as it does in A-family polymerases.
Aphidicolin
H
H
OH
H
HO
HOH
2
C CH
3
CH
2
OH
H
3
C
Figure 30-41 X-ray structure of yeast DNA polymerase ␦ (pol
␦) in complex with primer–template DNA and dCTP. The
protein is drawn in ribbon form with its five domains differently
colored as indicated.The DNA, whose primer and template
strands consist of 12 and 16 nt, together with the incoming dCTP,
is drawn in stick form with template C green, primer C cyan,
dCTP C magenta, N blue, O red, and P orange and with
successive P atoms in each DNA strand connected by orange
rods.The Ca
2⫹
ions at the polymerase (Pol) active site are
represented by dark green spheres as is the Ca
2⫹
ion at the
exonuclease (Exo) active site. [Based on an X-ray structure by
Aneel Aggarwal, Mount Sinai School of Medicine, New York,
New York. PDBid 3IAY.]
␣␥␦ε
Location Nucleus Mitochondrion Nucleus Nucleus
Subunit masses (kD)
a
167, 79, 62, 48 144 125, 55, 40 256, 78, 23, 22
(166, 66, 59, 50) (140, 55) (124, 51, 51) (262, 60, 17, 12)
Family B A B B
Table 30-5 Properties of Eukaryotic DNA Polymerases That Participate
in DNA Replication
a
Yeast S. cerevisiae (human cells).
Source: Mainly Johnson, A. and O’Donnell, M., Annu. Rev. Biochem. 74, 283 (2005).
JWCL281_c30_1173-1259.qxd 8/10/10 9:11 PM Page 1203
1204 Chapter 30. DNA Replication, Repair, and Recombination
near the primer strand, following which pol ␦ binds to the
PCNA and processively extends the DNA strand.
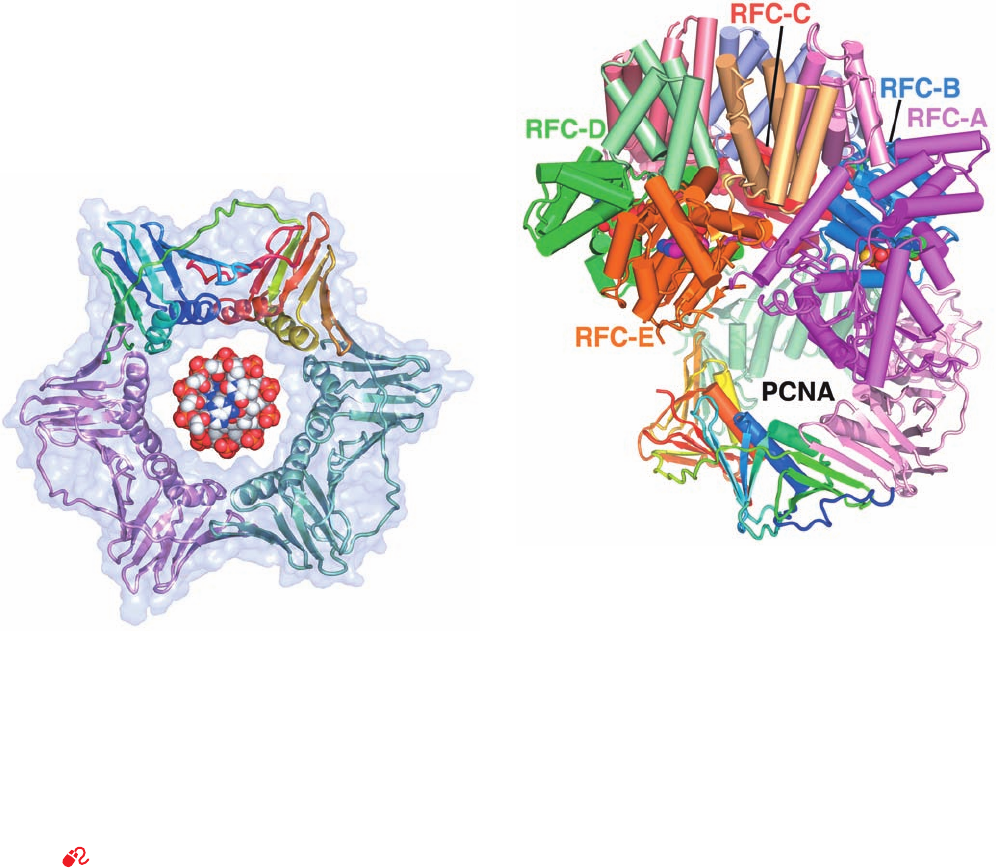
RFC, like the E. coli clamp loader, is a heteropentamer
of AAA⫹ family subunits, but whereas the clamp loader
contains three identical subunits (␥
3
␦␦¿), RFC consists of
five different subunits, RFC-A through RFC-E (alterna-
tively, RFC1–RFC5).The X-ray structure of yeast RFC in
complex with PCNA (Fig. 30-43), determined by O’Don-
nell and Kuriyan, reveals that RFC’s A, B, and C subunits
bind in conserved hydrophobic grooves on the face of
PCNA on which its C-terminal residues are located (since
PCNA’s three identical subunits are linked head-to-tail,
its two faces are different). In fact, this so-called C side of
PCNA, which faces away from the direction of poly-
merase motion, binds many of the proteins that partici-
pate in replicative processes, including most DNA poly-
merases, and hence PCNA plays a major role in
In contrast to pol ␣, which exhibits only moderate pro-
cessivity (⬃100 nucleotides), that of pol ␦ is essentially un-
limited (replicates the entire length of a template), but only
when it is in complex with a protein named proliferating
cell nuclear antigen (PCNA; so named because it occurs
only in the nuclei of proliferating cells and reacts with anti-
bodies produced by a subset of patients with the autoim-
mune disease systemic lupus erythematosus). The X-ray
structure of PCNA (Fig. 30-42), determined by Kuriyan, re-
veals that it forms a trimeric ring with almost identical
structure (and presumably function) as the E. coli 
2
slid-
ing clamp (Fig. 30-13).Thus, each PCNA subunit consists of
four rather than six of the structurally similar ␣ mo-
tifs from which the E. coli  subunit is constructed. Intrigu-
ingly, PCNA and the  subunit exhibit no significant se-
quence identity, even when their structurally similar
portions are aligned.Archaea also have sliding clamps with
pseudohexagonal symmetry.
Pol ␦ in complex with PCNA is required for lagging
strand synthesis. In contrast, pol ␣/primase functions to
synthesize ⬃12-nt RNA primers, which it extends by an ad-
ditional ⬃20 nt of DNA. Then, in a process called poly-
merase switching, the eukaryotic counterpart of the E. coli
clamp loader (Section 30-3Cc), replication factor C (RFC),
displaces the pol ␣ and loads PCNA on the template DNA
Figure 30-42 X-ray structure of human PCNA. Its three
subunits, which form a 3-fold symmetric ring, are drawn in ribbon
form embedded in their semitransparent surface diagram. One of
these subunits is colored in rainbow order from its N-terminus
(blue) to its C-terminus (red), another is pink, and the third is
light green.A space-filling model of B-DNA viewed along its
helix axis has been drawn in the center of the PCNA ring.
Compare this structure with that of the 
2
sliding clamp of the
E. coli Pol III holoenzyme (Fig. 30-13). [Based on an X-structure
by John Kuriyan, University of California at Berkeley. PDBid
1AXC.]
See Interactive Exercise 35
Figure 30-43 X-ray structure of yeast RFC in complex with
PCNA, ADP, and ATP␥S. Both proteins are drawn in tube-and-
arrow form with the RFC subunits colored as their homologs in
Fig. 30-34a (with the RFC-A, B, C, D, and E subunits
corresponding to the E. coli clamp loader’s ␦, ␥3, ␥2, ␥1, and ␦¿
subunits, respectively) and the PCNA colored as in Fig. 30-42.
The view is similar to that of the E. coli clamp loader in Fig. 30-34a.
ADP, which binds to only RFC-E, and ATP␥S, which binds to the
other four subunits, are shown in space-filling form with ATP␥S
C green,ADP C magenta, N blue, O red, P orange, and S yellow.
Note that all five RFC subunits have a nucleotide binding site,
whereas the E. coli ␦ and ␦¿ subunits do not. [Based on an
X-ray structure by Mike O’Donnell,The Rockefeller University,
and John Kuriyan, University of California at Berkeley. PDBid
1SXJ.]
JWCL281_c30_1173-1259.qxd 10/19/10 10:29 AM Page 1204
(replication units; DNA segments that are each served by
a replication origin) are activated simultaneously. New
replicons are activated throughout S phase until the en-
tire chromosome has been replicated. During this
process, replicons that have already been replicated are
distinguished from those that have not; that is, a cell’s
chromosomal DNA is replicated once and only once per
cell cycle.
c. The Assembly of the Eukaryotic Initiation Complex
Occurs in Two Stages
The once-and-only-once replication of eukaryotic DNA
per cell cycle is conferred by a type of binary switch.A pre-
replicative complex (pre-RC) is assembled at each replica-
tion origin during the G
1
phase of the cell cycle. This is the
only period of the cell cycle during which the pre-RC can
form and hence this process is known as licensing. How-
ever, a licensed pre-RC cannot initiate DNA replication.
Rather, it must be activated to do so, a process that occurs
only during S phase. This temporal separation of pre-RC
assembly and origin activation ensures that a new pre-RC
cannot assemble on an origin that has already “fired” (com-
menced replication) so that an origin can only fire once per
cell cycle. How does this occur?
The licensing process and how the pre-RC is activated
to form an initiation complex are still incompletely under-
stood. Thus, although it appears that most of the proteins
forming these complexes have been identified, their struc-
tures, interactions, and, in many cases, their functions are
largely unknown. Keeping this in mind, let us consider
what is known about these processes.
Replication origins are surprisingly variable among
species, often within the same organism, and even vary with
a given organism’s developmental stage. Thus, whereas S.
cerevisiae origins, which are known as autonomously repli-
cating sequences (ARS), contain a highly conserved 11-bp
AT-rich sequence within a less well defined ⬃125-bp re-
gion, some metazoan origins are dispersed over 10 to 50 kb
“initiation zones” that contain multiple origins and, in
some cases, require no specific DNA sequence at all. De-
spite this disparity, the proteins that participate in eukary-
otic DNA replication are highly conserved from yeast to
humans.
Section 30-4. Eukaryotic Replication 1205
recruiting the components of the various types of replica-
tion forks. The C-terminal domains of each RFC subunit
associate to form a ring-shaped collar, as do the C-termi-
nal domains of the E. coli clamp loader (Fig. 30-34). Like-
wise, RFC’s AAA⫹ domains are arranged in a right-
handed spiral that matches the helical path of the
sugar–phosphate backbone of B-DNA. Presumably,
prokaryotic and eukaryotic clamp loaders interact with
primer–template DNA and their corresponding sliding
clamps in similar ways.
Pol d, a heterotetrameric nuclear enzyme, is the most
enigmatic participant in DNA replication. Pol ε is highly
processive in the absence of PCNA and has a 3¿S5¿ ex-
onuclease activity that degrades single-stranded DNA to
6- or 7-residue oligonucleotides rather than to mononu-
cleotides, as does that of pol ␦. Although pol ε is necessary
for the viability of yeast, its essential function can be car-
ried out by only the noncatalytic C-terminal half of its
256-kD catalytic subunit, which is unique among B-family
DNA polymerases. This suggests that the C-terminal half
of the pol ε catalytic subunit is required for the assembly
of the replication complex. Nevertheless, Thomas Kunkel
has shown that pol ε is probably the leading strand repli-
case, although it may also contribute to lagging strand syn-
thesis. Moreover, pol ␦ may also participate in leading
strand synthesis.
Pol ␥, a monomer, occurs exclusively in the mitochon-
drion, where it presumably replicates the mitochondrial
DNA. Chloroplasts contain a similar enzyme.
Eukaryotic cells contain batteries of DNA polymerases.
These include the DNA polymerases that participate in
chromosomal DNA replication (pols ␣, ␦, and ε) and sev-
eral that take part in DNA repair processes (Section 30-5)
including pols , , , , and (alternatively, POLB, POLH,
POLI, POLK, and POLZ). Pol , an X-family enzyme, is
remarkable for its small size (a 335-residue monomer in
humans).
b. Eukaryotic Chromosomes Consist of
Numerous Replicons
Eukaryotic and prokaryotic DNA replication systems
differ most obviously in that eukaryotic chromosomes
have multiple replication origins in contrast to the single
replication origin of prokaryotic chromosomes. Eukary-
otic cells replicate DNA at the rate of ⬃50 nt/s (⬃20
times slower than does E. coli) as was determined by
autoradiographically measuring the lengths of pulse-
labeled sections of eukaryotic chromosomes. Since a eu-
karyotic chromosome typically contains 60 times more
DNA than those of prokaryotes, its bidirectional replica-
tion from a single origin would require ⬃1 month to
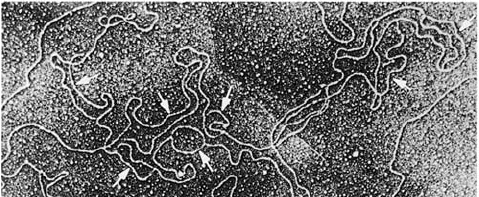
complete. Electron micrographs such as Fig. 30-44, how-
ever, reveal that eukaryotic chromosomes contain multi-
ple origins, one every 3 to 300 kb depending on both the
species and the tissue, so that S phase usually occupies
only a few hours.
Cytological observations indicate that the various
chromosomal regions are not all replicated simultane-
ously; rather, clusters of 20 to 80 adjacent replicons
Figure 30-44 Electron micrograph of a fragment of replicating
Drosophila DNA. The arrows indicate its multiple replication
eyes. [From Kreigstein, H.J. and Hogness, D.S., Proc. Natl. Acad.
Sci. 71, 136 (1974).]
JWCL281_c30_1173-1259.qxd 8/10/10 9:11 PM Page 1205
1206 Chapter 30. DNA Replication, Repair, and Recombination
The conversion of a licensed pre-RC to an active initia-
tion complex requires the addition of pol ␣/primase, pol ε,
and several accessory proteins, which only occurs at the on-
set of S phase. This process begins with addition of Mcm10
protein (which shares no sequence similarity with any of
the subunits of the MCM complex) to the pre-RC, which
probably displaces Cdt1.This is followed by the addition of
at least two protein kinases, a Cdk and Ddk, the latter be-
ing a heterodimer of the protein kinase Cdc7 with its
activating subunit Dbf4 (Ddk stands for Dbf4-dependent
kinase). Ddk acts to phosphorylate five of the six MCM
subunits (all but Mcm2) so as to activate the MCM com-
plex as a helicase. In contrast, the way in which Cdks acti-
vate the pre-RC is poorly understood, although several
ORC and MCM proteins as well as Cdc6/Cdc18 are phos-
phorylated by Cdks. Ddk together with a Cdk also recruits
Cdc45 to the growing initiation complex. Cdc45, in turn, is
required for the assembly of the initiating synthetic ma-
chinery at the replication fork, including pol ␣/primase, pol
ε, PCNA, and replication protein A (RPA), the het-
erotrimeric eukaryotic counterpart of SSB, thereby form-
ing an active initiation complex.
d. Re-Replication Is Prevented through the Actions
of Cdks and Geminin
Once initiation (priming) has occurred, the initiation
complex is joined by RFC and pol ␦ and, as is described
above, is converted to an active replicative complex by
polymerase switching. DNA replication then proceeds
bidirectionally until each replication fork has collided with
an oppositely traveling replication fork, thereby complet-
ing the replication of the replicon. An active replication
fork will destroy any licensed pre-RCs and unfired initia-
tion complexes in its path, thereby preventing the DNA at
such sites from being replicated twice. Eukaryotes appear
to lack termination sequences and proteins analogous to
the Ter sites and Tus protein in E. coli.
Several redundant mechanisms ensure that a pre-RC
can initiate DNA synthesis only once. Cdks are active
from late G
1
phase through late M phase. These elevated
Cdk levels, which are required to activate initiation, also
prevent reinitiation. The Cdk-mediated phosphorylation of
Cdc6/Cdc18, which occurs late in G
1
after the pre-RCs have
formed, causes Cdc6/Cdc18 to be proteolytically degraded
in yeast and exported from the nucleus in mammalian cells.
Evidently, Cdc6/Cdc18 is only required for the assembly of
the pre-RC, not its activation. The helicase activity of the
MCM complex is inhibited by phosphorylation, at least in
vitro. Moreover, MCM proteins are exported from the nu-
cleus in G
2
and M phases, a process that is interrupted by
Cdk inactivation. However, the function of Cdk-mediated
phosphorylation of ORC proteins is unclear.
Metazoan cells have yet another mechanism to prevent
the assembly of a licensed pre-RC on already replicated
DNA. High levels of a protein named geminin appear in S
phase and continue to accumulate until late M phase, when
geminin is degraded. Geminin associates with Cdt1 (which
together with Cdc6/Cdc18 loads the MCM complex onto
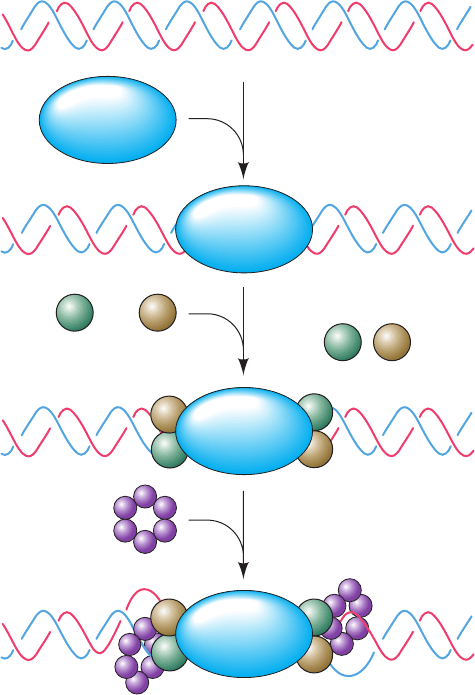
The assembly of the pre-RC (Fig. 30-45) begins late in
M phase or early in G
1
phase with the binding of the ori-
gin recognition complex (ORC), a hexamer of related
proteins (Orc1 through Orc6), to the origin, where it re-
mains bound during most or all of the cell cycle. ORC, the
functional analog of DnaA protein in E. coli replication
initiation (Section 30-3Ca), then recruits two proteins,
Cdc6 in S. cerevisiae (Cdc18 in S. pombe; Cdc for cell divi-
sion cycle) and Cdt1. These proteins then cooperate with
the ORC to load the MCM complex [named for its
minichromosome (plasmid) maintenance functions], a
hexamer of related subunits (Mcm2 through Mcm7), onto
the DNA to yield the licensed pre-RC. The MCM com-
plex, a ring-shaped ATP-driven helicase, is the analog of
E. coli DnaB helicase, whereas Cdc6/Cdc18 together with
Cdt1 appears to be an analog of E. coli DnaC (which fa-
cilitates DnaB loading).With the exception of Cdt1, all of
these proteins, Orc1 through Orc6, Cdc6/Cdc18, Mcm2
through Mcm7, as well as E. coli DnaA, DnaB, and DnaC,
are AAA⫹ ATPases.
Figure 30-45 Schematic diagram for the assembly of the
eukaryotic pre-replicative complex (pre-RC). The actual
stoichiometries, positions, and interactions of its various
components are largely unknown.The pre-RC only forms during
the G
1
phase of the cell cycle.
+
Origin-containing DNA
Licensed pre-RC
ORC
Cdc6/Cdc18 Cdt1
MCM Complex
JWCL281_c30_1173-1259.qxd 8/10/10 9:11 PM Page 1206
C. Reverse Transcriptase
The retroviruses, which are RNA-containing eukaryotic
viruses such as certain tumor viruses and human immuno-
deficiency virus (HIV), contain an RNA-directed DNA
polymerase (reverse transcriptase). This enzyme, which
was independently discovered in 1970 by Howard Temin
and David Baltimore, acts much like Pol I in that it synthe-
sizes DNA in the 5¿S3¿ direction from primed templates.
In the case of reverse transcriptase, however, RNA is the
template. The discovery of reverse transcriptase caused a
mild sensation in the biochemical community because it
was perceived by some as being heretical to the central
dogma of molecular biology (Section 5-4). There is, how-
ever, no thermodynamic prohibition to the reverse tran-
scriptase reaction; in fact, under certain conditions, Pol I
can likewise copy RNA templates.
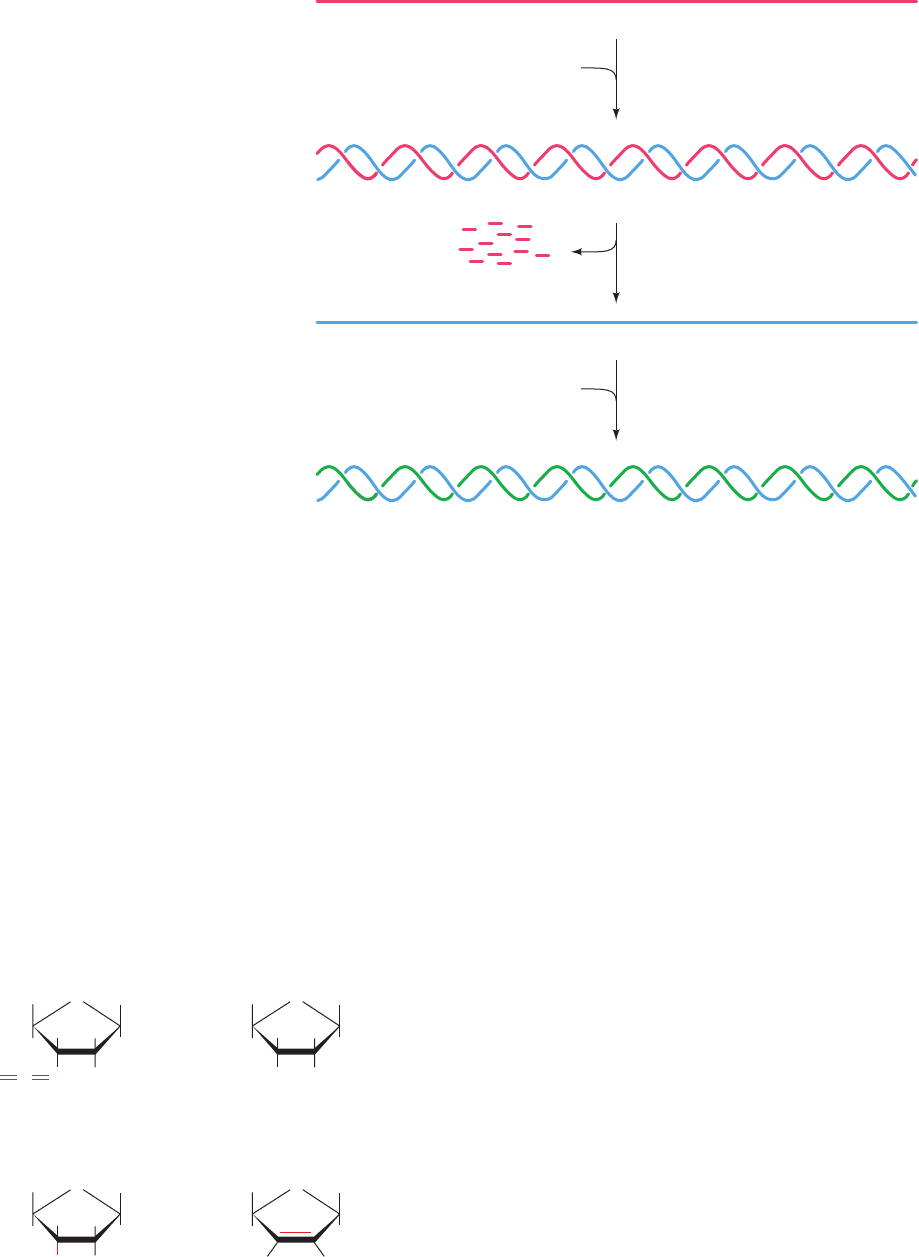
Reverse transcriptase transcribes the retrovirus’s single-
stranded RNA genome to a double-stranded DNA as fol-
lows (Fig. 30-47):
1. The retroviral RNA acts as a template for the synthe-
sis of its complementary DNA (RNA-directed DNA poly-
merase activity), yielding an RNA–DNA hybrid helix. The
DNA synthesis is primed by a host cell tRNA whose 3¿ end
partially unfolds to base pair with a complementary seg-
ment of the viral RNA.
2. The RNA strand is then nucleolytically degraded
(RNase H activity; H for hybrid).
3. The DNA strand acts as a template for the synthesis
of its complementary DNA (DNA-directed DNA poly-
merase activity), yielding double-stranded DNA.
The DNA is then integrated into a host cell chromosome.
Reverse transcriptase has been a particularly useful tool
in genetic engineering because of its ability to transcribe
mRNAs to complementary strands of DNA (cDNA). In
transcribing eukaryotic mRNAs, which have poly(A) tails
(Section 31-4Ab), the primer can be oligo(dT). cDNAs
have been used, for example, as probes in Southern blot-
ting (Section 5-5D) to identify the genes coding for their
Section 30-4. Eukaryotic Replication 1207
the ORC) so as to inhibit the assembly of the pre-RC.This
inhibition can be reversed by the addition of excess Cdt1. It
therefore seems likely that the presence of geminin pro-
vides protection against DNA re-replication under condi-
tions when Cdks are inhibited by checkpoint activation. In
addition, re-replication is also prevented by the degrada-
tion of Cdt1 after the origin with which it is associated has
fired.The requirement for DNA replication in this process
is indicated by its blockage by the DNA polymerase in-
hibitor aphidicolin.
Finally, cells that have shifted to the G
0
(quiescent)
phase of the cell cycle (Fig. 30-40)—the majority of cells in
the human body—cease making DNA. Such cells are char-
acterized by the absence of Cdk activity. In proliferating
cells, this would permit the re-replication of DNA. How-
ever,cells in G
0
also lack the proteins of the MCM complex
and are therefore incapable of assembling licensed pre-
RCs. Since cancerous cells are characterized by being in a
state of rapid proliferation (Section 19-3B), the presence of
MCM complex proteins in what should be quiescent cells is
a promising diagnostic marker for cancer.
e. Primers Are Removed by RNase H1 and Flap
Endonuclease-1
In lagging strand synthesis, when pol ␦ reaches the pre-
viously synthesized Okazaki fragment, it partially displaces
its RNA primer through DNA synthesis, thereby generat-
ing an RNA flap. The primer is then removed through the
actions of two enzymes: RNase H1 removes most of the
RNA leaving only a 5¿ ribonucleotide adjacent to the DNA,
which is then removed by flap endonuclease-1 (FEN1).
However, as we have seen, pol ␣/primase extends the RNA
primers it has made by ⬃20 nt of DNA before it is dis-
placed by pol ␦. Since pol ␣ lacks proofreading ability, this
primer extension is more likely to contain errors than the
DNA synthesized by pol ␦. However, FEN1, which is re-
cruited to the replication fork by its binding to the C side of
PCNA, provides what is, in effect, pol ␣’s proofreading
function: It is also an endonuclease that excises mismatch-
containing oligonucleotides up to 15 nt long from the 5¿
end of an annealed DNA strand. Moreover, FEN1 can
make several such excisions in succession to remove more
distant mismatches. The excised segment is later replaced
by pol ␦ as it synthesizes the succeeding Okazaki fragment.
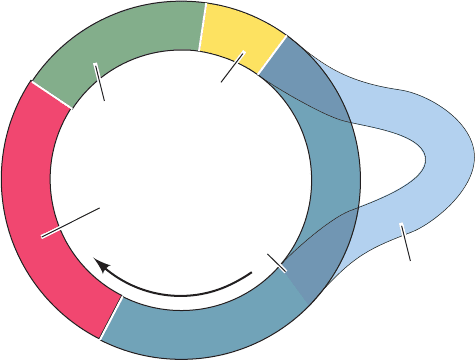
f. Mitochondrial DNA Is Replicated via RNA
Okazaki Fragments
Mammalian mitochondria contain two to ten copies of
their ⬃16-kb circular chromosome. The pol ␥–mediated
replication of this chromosome occurs unidirectionally
from a single origin. In this process, as Ian Holt showed, the
lagging strand Okazaki fragments are entirely synthesized
as RNA (Fig. 30-46). The RNA is then replaced by DNA,
although the way this occurs is poorly understood. One
possibility is that this RNA is excised in much the same
way as the primers in the nucleus, that is, through the ac-
tions of RNase H1 and FEN1, followed by the synthesis of
lagging strand DNA by pol ␥.
Leading strand DNA
Lagging strand RNA
Okazaki fragments
Motion of
replication fork
Origin
Mitochondrial chromosome
5'
3'
3'
5'
Figure 30-46 Replication of the mammalian mitochondrial
chromosome. This circular chromosome is unidirectionally
replicated from a single origin by a process in which the Okazaki
fragments are entirely RNA.
JWCL281_c30_1173-1259.qxd 8/26/10 8:20 PM Page 1207
1208 Chapter 30. DNA Replication, Repair, and Recombination
O
H
HH
NNN H
H
HOCH
2
T
⫺⫹
3'-Azido-3'-deoxythymidine
(AZT; zidovudine)
O
H
HH
H H
H
HOCH
2
Hypoxanthine
2',3'-Dideoxyinosine,
(ddI; didanosine)
O
H
HH
H H
H
HOCH
2
C
2',3'-Dideoxycytidine
(ddC; zalcitabine)
O
H
HH
H
HOCH
2
T
2',3'-Didehydro-3'-
deoxythymidine
(stavudine)
are RT inhibitors. Unfortunately, resistant strains of HIV-1
arise quite rapidly because RT lacks a proofreading exonu-
clease function and hence is highly error prone.Thus, as we
have seen (Section 15-4Cd), effective long-term anti-HIV
therapy requires the concurrent administration of at least
one RT inhibitor and an HIV protease inhibitor.
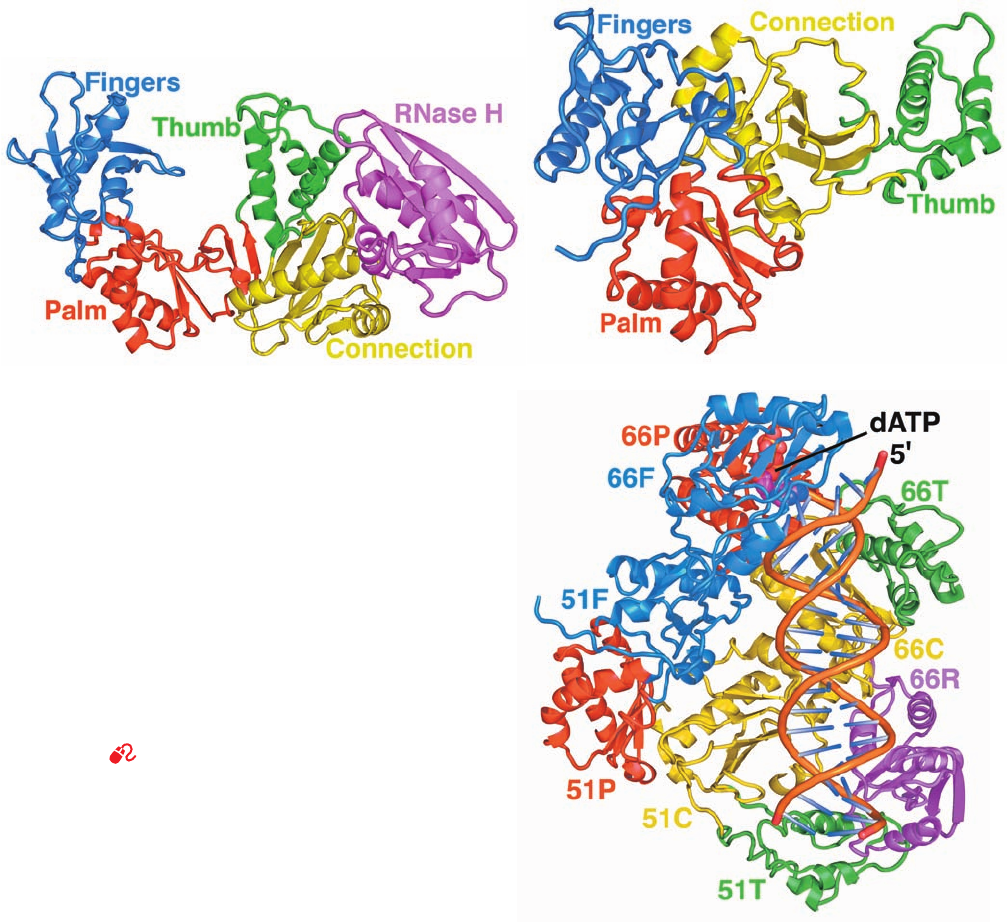
Edward Arnold determined the X-ray structure of RT
complexed to a 21-bp primer–template DNA with a 5-nt
overhang at the 5¿ end of its template strand, a dideoxy-dG
(ddG) residue at the 3¿ end of its primer strand (which pre-
vents its further extension), and in complex with dATP
(Fig. 30-48). The polymerase domains of p66 and p51 each
contain four subdomains, which, because of their collective
resemblance in p66 to DNA polymerases, are named, from
N- to C-terminus, fingers, palm, thumb, and connection. In-
deed, reverse transcriptases form a separate family of DNA
polymerases, family RT. In p66, the RNase H domain fol-
lows the connection.
p51 has undergone a remarkable conformational
change relative to p66: The connection has rotated by 155°
and translated by 17 Å to bring it from a position in p66 in
which it contacts only the RNase H domain (Fig. 30-48a) to
one in p51 in which it contacts all three other polymerase
subdomains (Fig. 30-48b). This permits p66 and p51 to
bring different surfaces of their connections into juxtaposi-
tion to form, in part, RT’s DNA-binding groove. Thus, the
chemically identical polymerase domains of p66 and p51
are not related by 2-fold molecular symmetry (a rare but
not unprecedented phenomenon), but instead, associate in
a sort of head-to-head and tail-to-tail arrangement (Fig. 30-
48c). Consequently, RT has only one polymerase active site
and one RNase H active site.This is an example of viral ge-
netic economy: HIV-1, with its limited genome size, has
corresponding mRNAs. An RNA’s base sequence can be
easily determined by sequencing its cDNA (Section 7-2A).
a. X-Ray Structure of HIV-1 Reverse Transcriptase
HIV-1 reverse transcriptase (RT) is a dimeric protein
whose subunits are synthesized as identical 66-kD
polypeptides, known as p66 (p for protein), that each con-
tain a polymerase domain and an RNase H domain. How-
ever, the RNase H domain of one of the two subunits is
proteolytically excised, thereby yielding a 51-kD polypep-
tide named p51. Thus, RT is dimer of p66 and p51.
The first drugs to be clinically approved to treat AIDS,
3ⴕ-azido-3ⴕ-deoxythymidine (AZT; zidovudine), 2ⴕ,3ⴕ-
dideoxyinosine (ddI; didanosine), 2ⴕ,3ⴕ-dideoxycytidine
(ddC; zalcitabine), and 2ⴕ,3ⴕ-didehydro-3ⴕ-deoxythymidine
(stavudine),
3'
5'
5'
3'5'
3'
5'3'
RNA
.
DNA hybrid
RNase H
2
3'
5'
5'
3'
Double-stranded DNA
Single-stranded RNA
RNA-directed DNA polymerasedNTP
NMP
1
Single-stranded DNA
DNA-directed DNA polymerasedNTP
3
Figure 30-47 The reactions catalyzed by
reverse transcriptase.
JWCL281_c30_1173-1259.qxd 8/10/10 9:11 PM Page 1208
succeeded in using a single polypeptide for what are essen-
tially two different functions.
The dATP binds at the 3¿ end of the primer strand, near
p66’s three catalytically essential Asp side chains, where it
pairs with a template dT base.The DNA assumes a confor-
mation that, near the polymerase active site, resembles A-
DNA (note the A-DNA-like tilt of the bases with respect
to the helix axis below the dATP in Fig. 30-48c), but else-
where more closely resembles B-DNA (in which the bases
are nearly perpendicular to the helix axis), a phenomenon
that also has been observed in several structures of A-
family DNA polymerases in their complexes with DNA
(Section 30-2Ae). Most of the protein–DNA interactions
involve the DNA’s sugar–phosphate backbone.
The RT active site region contains the few sequence
motifs that are conserved among the various polymerases.
Indeed, this region of p66 has a striking structural resem-
blance to DNA polymerases of known structure (Sections
30-2A and 30-4Ba). This suggests that other DNA poly-
merases are likely to bind DNA in a similar manner.
D. Telomeres and Telomerase
The ends of linear chromosomes cannot be replicated by
any of the mechanisms we have yet considered. This is be-
cause the RNA primer at the 5¿ end of a completed lagging
strand cannot be replaced with DNA; the primer required
to do this would have no place to bind. How, then, are the
DNA sequences at the ends of eukaryotic chromosomes,
the telomeres (Greek: telos, end), replicated?
Telomeric DNA has an unusual sequence: It consists of
up to several thousand tandem repeats of a simple, species-
dependent, G-rich sequence concluding the 3¿-ending
strand of each chromosomal terminus. For example, the cil-
iated protozoan Tetrahymena has the repeating telomeric
sequence TTGGGG, whereas in all vertebrates it is
Section 30-4. Eukaryotic Replication 1209
Figure 30-48 X-ray structure of HIV-1 reverse transcriptase in
complex with primer–template DNA and dATP. (a) A ribbon
diagram of the p66 subunit in which the N-terminal fingers
domain is blue, the palm is red, the thumb is green, the
connection is yellow, and the RNase H domain is magenta.
(b) The p51 subunit with its palm subunit oriented similarly to
that of p66. Note the different orientations of its four domains
relative to p66. (c) A ribbon diagram of the HIV-1 RT p66/p51
heterodimer in complex with DNA and dATP.The domains of
p66 and p51 are colored as in Parts a and b (the labels indicate
subunit and domain; e.g., 51F and 66R denote the p51 finger
domain and the p66 RNase H domain).The DNA is drawn in
ladder form.The complex is oriented with its p66 polymerase
domain toward the top of the figure and viewed toward the
protein’s template–primer binding cleft (whose floor is largely
composed of the connection domains of p66 and p51). [Based on
an X-ray structure by Edward Arnold, Rutgers University.
PDBid 3JYT.]
See Interactive Exercise 36
(a)
(b)
(c)
JWCL281_c30_1173-1259.qxd 10/19/10 10:30 AM Page 1209
1210 Chapter 30. DNA Replication, Repair, and Recombination
TTAGGG. Moreover, this strand ends with an overhang
that varies from ⬃20 nt in yeast to ⬃200 bp in humans.
Elizabeth Blackburn, Carol Greider, and Jack Szostak
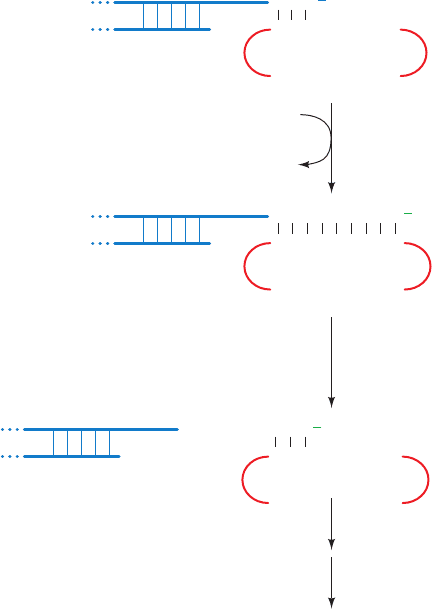
have shown that telomeric DNA is synthesized by a novel
mechanism.The enzyme that synthesizes the G-rich strand
of telomeric DNA is named telomerase. Tetrahymena
telomerase, for example, adds tandem repeats of the telo-
meric sequence TTGGGG to the 3¿ end of any G-rich
telomeric oligonucleotide independently of any exoge-
nously added template. A clue as to how this occurs came
from the discovery that telomerases are ribonucleopro-
teins whose RNA components contain a segment that is
complementary to the repeating telomeric sequence. This
sequence apparently acts as a template in a kind of reverse
transcriptase reaction that synthesizes the telomeric se-
quence, translocates to the DNA’s new 3¿ end, and repeats
the process (Fig. 30-49).This hypothesis is confirmed by the
observation that mutationally altering the telomerase
RNA gene segment complementary to telomere DNA re-
sults in telomere DNA with the corresponding altered se-
quence. In fact, telomerase’s highly conserved protein com-
ponent, which is named TERT, is homologous to known
reverse transcriptases (its RNA component is called TER).
The DNA strand complementary to the telomere’s G-rich
strand is apparently synthesized by the normal cellular ma-
chinery for lagging strand synthesis, thereby accounting for
the 3¿ overhang of the G-rich strand.
a. TERT Resembles Other DNA Polymerases
The X-ray structure of the 596-residue TERT from the
red flour beetle Tribolium castaneum, determined by Em-
manuel Skordalakes, reveals that this subunit contains the
familiar fingers–palm–thumb domain organization of
other DNA polymerases (Fig. 30-50) and, in particular, re-
sembles the corresponding domains of the HIV-1 reverse
transcriptase p66 subunit (Fig. 30-48). In addition, TERT
has an N-terminal RNA-binding domain named TRBD
(for telomere repeat binding domain). TRBD closes the
gap between the thumb and fingers, thereby yielding a
ringlike protein with a hole that is ⬃26 Å wide and ⬃21 Å
deep. This is sufficient to accommodate an ⬃8-bp segment
of double-stranded nucleic acid.
b. Telomeres Must Be Capped
Without the action of telomerase, a chromosome
would be shortened at both ends by 50 to 100 nt with
every cycle of DNA replication and cell division. It was
therefore initially assumed that, in the absence of active
telomerase, essential genes located near the ends of chro-
mosomes would eventually be lost, thereby killing the de-
scendents of the originally affected cells. However, it is
now evident that telomeres serve a vital chromosomal
function that is compromised before this can happen.
Free DNA ends, which are subject to nuclease degrada-
tion, trigger DNA damage repair systems that normally
function to rejoin the ends of broken chromosomes (as
well as cell cycle arrest until this has happened).Thus ex-
posed telomeric DNA would result in the end-to-end fu-
sion of chromosomes, a process that leads to chromoso-
mal instability and eventual cell death [fused
chromosomes often break in mitosis (their two cen-
tromeres may cause them to be pulled in opposite direc-
tions), activating DNA damage checkpoints]. However, in
a process known as capping, telomeric DNA is specifi-
cally bound by proteins that sequester the DNA ends.
There is mounting evidence that capping is a dynamic
process in which the probability of a telomere sponta-
neously upcapping increases as telomere length de-
creases. Since most somatic cells in multicellular organ-
isms have very low levels of telomerase activity, this
explains why such cells in culture can only undergo a lim-
ited number of doublings (20–60) before they reach
senescence (a stage in which they cease dividing) and
eventually die (Section 19-3B). Indeed, otherwise immor-
tal Tetrahymena cultures with mutationally impaired
telomerases exhibit characteristics reminiscent of senes-
cent mammalian cells before dying off. Apparently, the
loss of telomerase function in somatic cells is a basis for ag-
ing in multicellular organisms.
c. Telomere Length Correlates with Aging
There is strong experimental evidence in support of this
theory of aging.The analysis of cultured human fibroblasts
Figure 30-49 Proposed mechanism for the synthesis of
telomeric DNA by Tetrahymena telomerase. The telomere’s 5¿-
ending strand is later extended by normal lagging strand
synthesis. [After Greider, C.W. and Blackburn, E.H., Nature 337,
336 (1989).]
5′
3′ 5′
3′
5′3′
T T G
A A C C C C A A C
OH
Telomeric DNA
Telomerase RNA
polymerize
5′
3′ 5′
5′3′
T
A
T
A
G
C
G
C
G
C
G
C
T
A
T
A
G
C
5′
3′ 5′
5′3′
TTGGGG
CCCAAC
T
A
T
A
G
C
translocate
dGTP + dTTP
PP
i
3′
OH
3′
OH
JWCL281_c30_1173-1259.qxd 8/10/10 9:11 PM Page 1210
