Voet D., Voet Ju.G. Biochemistry
Подождите немного. Документ загружается.

Problems 1171
General
Bloomfield,V.A., Crothers, D.M., and Tinoco, I., Jr., Nucleic Acids:
Structures, Properties, and Functions, University Science Books
(2000).
Creighton, T.E., The Biophyical Chemistry of Nucleic Acids &
Proteins, Chapters 2–6, Helvetian Press (2010).
Neidle, S. (Ed.), Oxford Handbook of Nucleic Acid Structure,
Oxford University Press (1999).
Saenger,W., Principles of Nucleic Acid Structure, Springer-Verlag
(1984).
The double helix—50 years, Nature 421, 395–453 (2003). [A sup-
plement containing a series of articles on the historical, cul-
tural, and scientific influences of the DNA double helix cele-
brating the fiftieth anniversary of its discovery.]
Structures and Stabilities of Nucleic Acids
Arnott, S., Historical article: DNA polymorphism and the early his-
tory of the double helix, Trends Biochem. Sci. 31, 349–364 (2006).
Bacolla,A. and Wells, R.D., Non-B DNA conformations, genomic
rearrangements, and human disease, J. Biol. Chem. 46,
47411–47414 (2004).
Dickerson, R.E., Sequence-dependent B-DNA conformation in
crystals and in protein complexes, in Sarma, R.H. and Sarma,
M.H. (Eds.), Structure, Motion, Interaction and Expression in
Biological Molecules, pp. 17–35, Adenine Press (1998); and
DNA bending: the prevalence of kinkiness and the virtues of
normality, Nucleic Acids Res. 26, 1906–1926 (1998).
Fairhead, H., Setlow, B., and Setlow, P., Prevention of DNA dam-
age in spores and in vitro by small, acid-soluble proteins from
Bacillus species, J. Bacteriol. 175, 1367–1374 (1993).
Ha, C.H., Lowenhaupt, K., Rich, A., Kim, Y.-G., and Kim, K.K.,
Crystal structure of a junction between B-DNA and Z-DNA
reveals two extruded bases, Nature 437, 1183–1186 (2005).
Horton, N.C. and Finzel, B.C.,The structure of an RNA/DNA hy-
brid: A substrate of the ribonuclease activity of HIV-1 reverse
transcriptase, J. Mol. Biol. 264, 521–533 (1996).
Rich,A.,The double helix: tale of two puckers, Nature Struct.Biol.
10, 247–249 (2003). [A historical review.]
Schwartz,T., Rould, M.A.,Lowenhaupt, K., Herbert,A., and Rich,
A., Crystal structure of the Z domain of the human editing
enzyme ADAR1 bound to left-handed Z-DNA, Science 284,
1841–1845 (1999).
Wing, R., Drew, H.,Takano,T., Broka, C.,Tanaka, S., Itakura, K.,and
Dickerson, R.E., Crystal structure analysis of a complete turn of
B-DNA, Nature 287, 755–758 (1980); and Shui, X., McFail-Isom,
L., Hu, G.G., and Williams, L.D., The B-DNA decamer at high
resolution reveals a spine of sodium, Biochemistry 37,
8341–8355 (1998). [The Dickerson dodecamer at its original
2.5 Å resolution and at its later-determined 1.4 Å resolution.]
Supercoiled DNA
Bates, A.D. and Maxwell, A., DNA Topology (2nd ed.) Oxford
University Press (2005).
Champoux, J.J., DNA topoisomerases: Structure, function, and
mechanism, Annu. Rev. Biochem. 70, 369–413 (2001).
Changela,A., DiGate, R., and Mondragón,A., Crystal structure of
a complex of a type IA DNA topoisomerase with a single-
stranded DNA, Nature 411, 1077–1081 (2001); and Mondragón,
A. and DiGate, R., The structure of Escherichia coli DNA
topoisomerase III, Structure 7, 1373–1383 (1999).
Classen, S., Olland, S., and Berger, J., Structure of the topoiso-
merase II ATPase region and its mechanism of inhibition by
the chemotherapeutic agent ICRF-187, Proc. Natl. Acad. Sci.
100, 10629–10634 (2003).
Deweese, J.E., Osheroff, M.A., and Osheroff, N., DNA topology
and topoisomerases, Biochem. Mol. Biol. Educ. 37, 2–10
(2009).
Dong, K.C. and Berger, J.M., Structural basis for the gate-DNA
recognition and bending by type IIA topoisomerases. Nature
450, 1201–1205 (2007). [X-ray structure of the yeast topoiso-
merase II breakage/reunion domain in complex with DNA.]
Dong, K.C. and Berger, J.M., Structure and function of DNA
topoisomerases, in Rice, P.A. and Correll, C.C. (Eds.),
Protein–Nucleic Acid Interactions, RSC Publishing (2008); and
Corbett, K.D. and Berger, J.M., Structure, molecular mecha-
nisms, and evolutionary relationships in DNA topoisomerases,
Annu. Rev. Biophys. Biomol. Struct. 33, 95–118 (2004).
Gadelle, D., Filée, J., Buhler, C., and Forterre, P., Phylogenomics of
type II DNA topoisomerases, Bioessays 25, 232–242 (2003).
Lebowitz, J., Through the looking glass: The discovery of super-
coiled DNA, Trends Biochem. Sci. 15, 202–207 (1990). [An in-
formative eyewitness account of how DNA supercoiling was
discovered.]
Li, T.-K. and Liu, L.F., Tumor cell death induced by topoiso-
merase-targeting drugs, Annu. Rev. Pharmacol. Toxicol. 41,
53–77 (2001).
Maxwell, A., DNA gyrase as a drug target, Biochem. Soc. Trans.
27, 48–53 (1999).
Redinbo, M.R., Stewart, L., Kuhn, P., Champoux, J.J., and Hol,
W.G.J., Crystal structures of human topoisomerase I in cova-
lent and noncovalent complexes with DNA, Science 279,
1504–1513 (1998); Stewart, L., Redinbo, M.R., Qiu, X., Hol,
W.G.J., and Champoux, J.J., A model for the mechanism of hu-
man topoisomerase I, Science 279, 1534–1541 (1998); and Red-
inbo, M.R., Champoux, J.J., and Hol, W.G.J., Structural insights
into the function of type IB topoisomerases, Curr. Opin. Struct.
Biol. 9, 29–36 (1999).
Taneja, B., Patel, A., Slesarev, A., and Mondraón, A., Structure of
the N-terminal fragment of topoisomerase V reveals a new
family of topoisomerases, EMBO J. 25, 398–408 (2006).
Wang, J.C., Moving one DNA double helix through another by a
type II DNA topoisomerase: The story of a simple molecular
machine, Q. Rev. Biophys. 31, 107–144 (1998).
Wang, J.C., Cellular roles of DNA topoisomerases: a molecular
perspective, Nature Rev. Mol. Cell Biol. 3, 430–440 (2002).
REFERENCES
1. A ⴢ T base pairs in DNA exhibit greater variability in their
propeller twisting than do G ⴢ C base pairs. Suggest the structural
basis of this phenomenon.
PROBLEMS
*2. At Na
concentrations 5M, the T
m
of DNA decreases
with increasing [Na
]. Explain this behavior. (Hint: Consider the
solvation requirements of Na
.)
JWCL281_c29_1143-1172.qxd 8/11/10 11:01 AM Page 1171
*3. Why are the most commonly observed conformations of
the ribose ring those in which either atom C2¿ or atom C3¿ is out
of the plane of the other four ring atoms? (Hint: In puckering a
planar ring such that one atom is out of the plane of the other
four, the substituents about the bond opposite the out-of-plane
atom remain eclipsed. This is best observed with a ball-and-stick
model.)
4. Polyomavirus DNA can be separated by sedimentation at
neutral pH into three components that have sedimentation coeffi-
cients of 20, 16, and 14.5S and that are known as Types I, II, and III
DNAs, respectively. These DNAs all have identical base se-
quences and molecular masses. In 0.15M NaCl, both Types II and
III DNA have melting curves of normal cooperativity and a T
m
of
88°C. Type I DNA, however, exhibits a very broad melting curve
and a T
m
of 107°C.At pH 13,Types I and III DNAs have sedimen-
tation coefficients of 53 and 16S, respectively, and Type II sepa-
rates into two components with sedimentation coefficients of 16
and 18S. How do Types I, II, and III DNAs differ from one an-
other? Explain their different physical properties.
5. When the helix axis of a closed circular duplex DNA of
2310 bp is constrained to lie in a plane, the DNA has a twist (T) of
207. When released, the DNA takes up its normal twist of 10.5 bp
per turn. Indicate the values of the linking number (L), writhing
number (W), and twist for both the constrained and uncon-
strained conformational states of this DNA circle.What is the su-
perhelix density,
, of both the constrained and unconstrained
DNA circles?
6. A covalently closed circular duplex DNA has a 100-bp seg-
ment of alternating C and G residues. On transfer to a solution
containing a high salt concentration, this segment undergoes a
transition from the B conformation to the Z conformation. What
is the accompanying change in its linking number, writhing num-
ber, and twist?
7. You have discovered an enzyme secreted by a particularly
virulent bacterium that cleaves the C2¿
¬C3¿ bond in the deoxyri-
bose residues of duplex DNA.What is the effect of this enzyme on
supercoiled DNA?
8. A bacterial chromosome consists of a protein–DNA com-
plex in which its single DNA molecule appears to be supercoiled,
as demonstrated by ethidium bromide titration. However, in con-
trast to the case with naked circular duplex DNA, the light single-
strand nicking of chromosomal DNA does not abolish this super-
coiling.What does this indicate about the structure of the bacterial
chromosome, that is, how do its proteins constrain its DNA?
9. Although types IA and IIA topoisomerases exhibit no sig-
nificant sequence similarity, it has been suggested that they are
distantly related based on the similarities of certain aspects of
their enzymatic mechanisms.What are these similarities?
10. Draw the mechanism of DNA strand cleavage and rejoin-
ing mediated by topoisomerase IA.
1172 Chapter 29. Nucleic Acid Structures
JWCL281_c29_1143-1172.qxd 8/2/10 8:06 PM Page 1172
CHAPTER 30
DNA Replication,
Repair, and
Recombination
1 DNA Replication: An Overview
A. Replication Forks
B. Role of DNA Gyrase
C. Semidiscontinuous Replication
D. RNA Primers
2 Enzymes of Replication
A. DNA Polymerase I
B. DNA Polymerase III
C. Unwinding DNA: Helicases and Single-Strand Binding Protein
D. DNA Ligase
E. Primase
3 Prokaryotic Replication
A. Bacteriophage M13
B. Bacteriophage X174
C. Escherichia coli
D. Fidelity of Replication
4 Eukaryotic Replication
A. The Cell Cycle
B. Eukaryotic Replication Mechanisms
C. Reverse Transcriptase
D. Telomeres and Telomerase
5 Repair of DNA
A. Direct Reversal of Damage
B. Excision Repair
C. Mismatch Repair
D. The SOS Response
E. Double-Strand Break Repair
F. Identification of Carcinogens
6 Recombination and Mobile Genetic Elements
A. Homologous Recombination
B. Transposition and Site-Specific Recombination
7 DNA Methylation and Trinucleotide Repeat
Expansions
Here we begin a three-chapter series on the basic processes
of gene expression: DNA replication (this chapter), tran-
scription (Chapter 31), and translation (Chapter 32).These
processes have been outlined in Section 5-4. We shall now
discuss them in greater depth with an emphasis on how we
have come to know what we know.
1 DNA REPLICATION: AN OVERVIEW
Watson and Crick’s seminal paper describing the DNA
double helix ended with the statement:“It has not escaped
our notice that the specific pairing we have postulated
immediately suggests a possible copying mechanism for
the genetic material.” In a succeeding paper they expanded
on this rather cryptic remark by pointing out that a DNA
strand could act as a template to direct the synthesis of its
complementary strand. Although Meselson and Stahl
demonstrated, in 1958, that DNA is, in fact, semiconserva-
tively replicated (Section 5-3B),it was not until some 20 years
later that the mechanism of DNA replication in prokary-
otes was understood in reasonable detail. This is because,
as we shall see in this chapter, the DNA replication process
rivals translation in its complexity but is mediated by often
loosely associated protein assemblies that are present in
only a few copies per cell. The surprising intricacy of DNA
replication compared to the chemically similar transcription
process (Section 31-2) arises from the need for extreme
accuracy in DNA replication so as to preserve the integrity
of the genome from generation to generation.
A. Replication Forks
DNA is replicated by enzymes known as DNA-directed
DNA polymerases or simply DNA polymerases.These en-
zymes utilize single-stranded DNA as templates on which
to catalyze the synthesis of the complementary strand from
the appropriate deoxynucleoside triphosphates (Fig. 30-1).
The incoming nucleotides are selected by their ability to
form Watson–Crick base pairs with the template DNA so
that the newly synthesized DNA strand forms a double
helix with the template strand. Nearly all known DNA
polymerases can only add a nucleotide donated by a nucle-
oside triphosphate to the free 3¿-OH group of a base paired
polynucleotide so that DNA chains are extended only in the
5¿S3¿ direction. DNA polymerases are discussed further
in Sections 30-2A, 30-2B, and 30-4B.
a. Duplex DNA Replicates Semiconservatively
at Replication Forks
John Cairns obtained the earliest indications of how
chromosomes replicate through the autoradiography of
replicating DNA. Autoradiograms of circular chromo-
somes grown in a medium containing [
3
H]thymidine show
JWCL281_c30_1173-1259.qxd 8/10/10 9:10 PM Page 1173
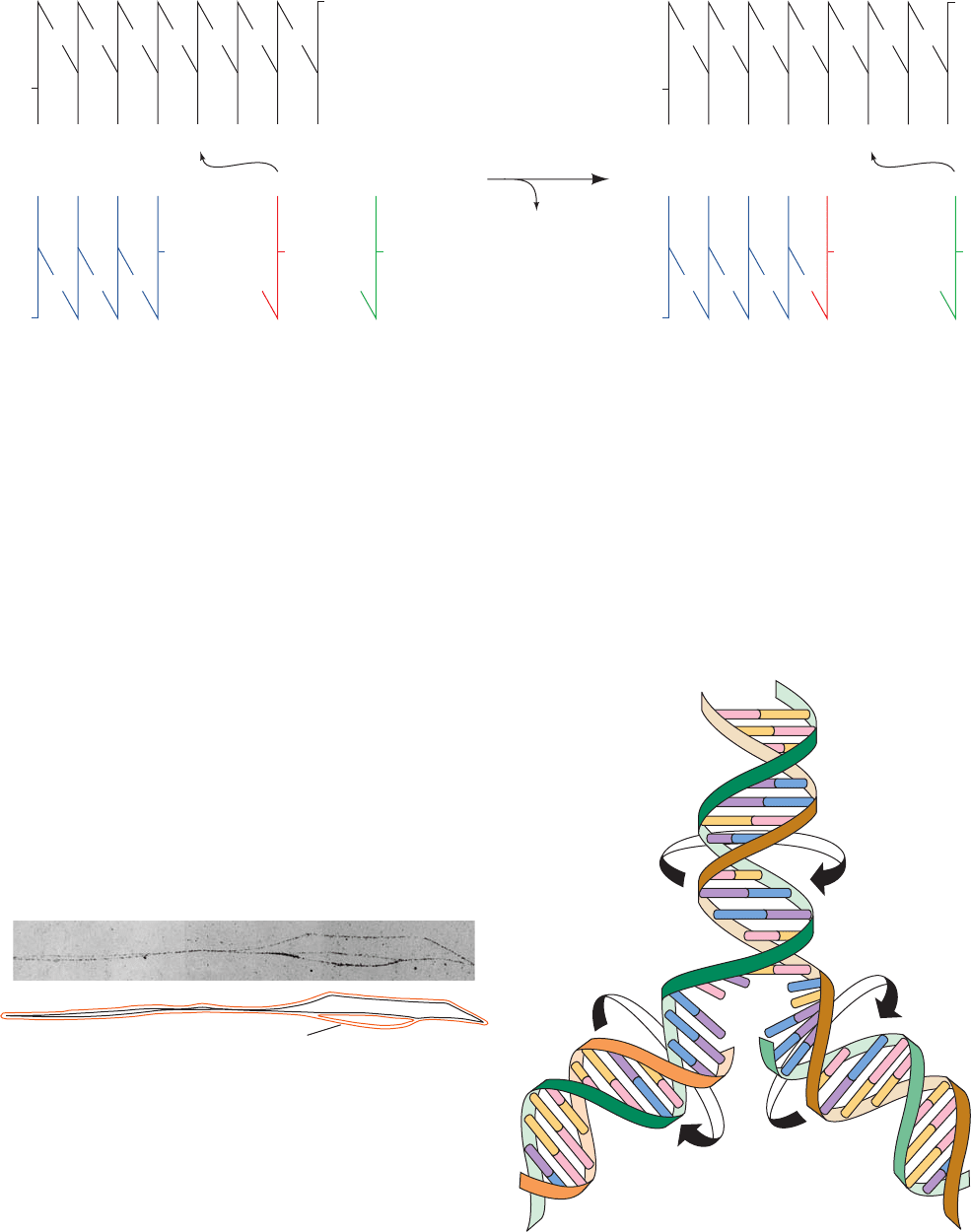
Replica DNAs
Parent DNA
Replication eye
1174 Chapter 30. DNA Replication, Repair, and Recombination
the presence of replication “eyes” or “bubbles” (Fig. 30-2).
These so-called structures (after their resemblance to the
Greek letter theta) indicate that double-stranded DNA
(dsDNA) replicates by the progressive separation of its two
parental strands accompanied by the synthesis of their com-
plementary strands to yield two semiconservatively repli-
cated duplex daughter strands (Fig. 30-3). DNA replication
involving structures is known as replication.
A branch point in a replication eye at which DNA syn-
thesis occurs is called a replication fork. A replication bub-
ble may contain one or two replication forks (unidirec-
tional or bidirectional replication). Autoradiographic
studies have demonstrated that replication is almost
always bidirectional (Fig. 30-4). Moreover, such experi-
ments, together with genetic evidence, have established
that prokaryotic and bacteriophage DNAs have but one
replication origin (point where DNA synthesis is initiated).
B. Role of DNA Gyrase
The requirement that the parent DNA unwind at the repli-
cation fork (Fig. 30-3) presents a formidable topological
obstacle. For instance, E. coli DNA is replicated at a rate of
⬃1000 nucleotides/s. If its 1300-m-long chromosome
were linear, it would have to flail around within the con-
fines of a 3-m-long E. coli cell at ⬃100 revolutions/s
Figure 30-1 Action of DNA polymerase. DNA polymerases
assemble incoming deoxynucleoside triphosphates on
Figure 30-2 Autoradiogram and its interpretive drawing of a
replicating E. coli chromosome. The bacterium had been grown
for somewhat more than one generation in a medium containing
[
3
H]thymidine, thereby labeling the subsequently synthesized
DNA so that it appears as a line of dark grains in the
photographic emulsion (red lines in the interpretive drawing).The
size of the replication eye indicates that the circular chromosome
is about one-sixth duplicated in the present round of replication.
[Courtesy of John Cairns, Cold Spring Harbor Laboratory,
New York.]
Figure 30-3 Replication of DNA.
p p p p
pp p
p p p
C
3⬘
5⬘ 3⬘Primer dCTP
DNA
polymerase
G
...
...
5⬘
...
...
A
T
...
G
C
OH
...
TGA
Template
AT
A
...
ppp
PP
i
OH
C
dTTP
ppp
OH
T
+++etc.
p p p p
p
p p p
p p p
C
G
...
...
...
...
A
T
...
G
C
...
...
TGAAT
A
...
OH
C
ppp
OH
T
++etc.
single-stranded DNA templates such that the growing strand is
elongated in its 5¿S3¿ direction.
JWCL281_c30_1173-1259.qxd 8/10/10 9:10 PM Page 1174
(recall that B-DNA has ⬃10 bp per turn). But since the
E. coli chromosome is, in fact, circular, even this could not
occur. Rather, the DNA molecule would accumulate ⫹100
supercoils/s (see Section 29-3A for a discussion of super-
coiling) until it became too tightly coiled to permit further
unwinding. Naturally occurring DNA’s negative supercoil-
ing promotes DNA unwinding but only to the extent of
⬃5% of its duplex turns (recall that naturally occurring
DNAs are typically underwound by one supercoil per ⬃19
duplex turns; Section 29-3Bb). In prokaryotes, however,
negative supercoils may be introduced into DNA through
the action of a type IIA topoisomerase (DNA gyrase; Sec-
tion 29-3Cd) at the expense of ATP hydrolysis.This process
is essential for prokaryotic DNA replication as is demon-
strated by the observation that DNA gyrase inhibitors,
such as novobiocin, arrest DNA replication except in mu-
tants whose DNA gyrase does not bind these antibiotics.
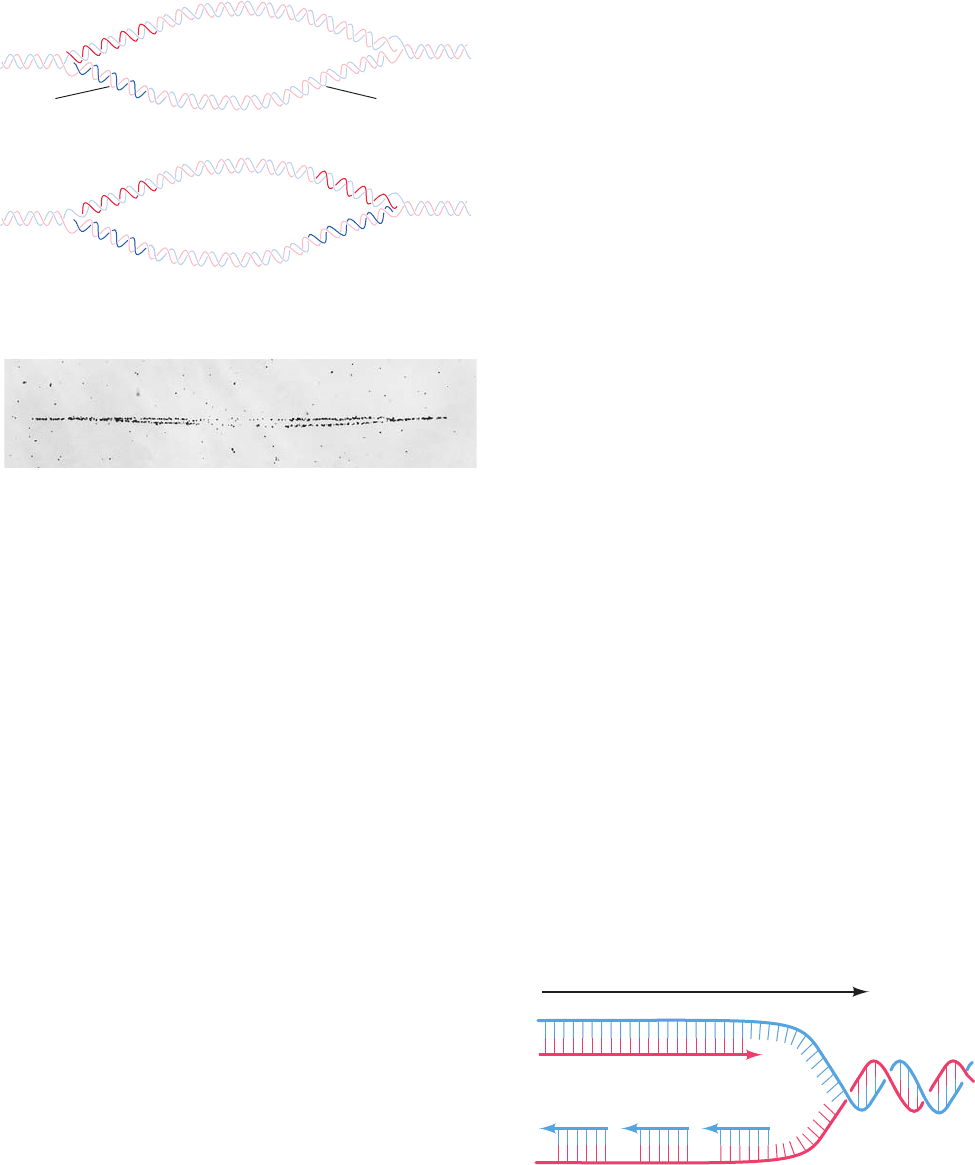
C. Semidiscontinuous Replication
The low-resolution images provided by autoradiograms
such as Figs. 30-2 and 30-4b suggest that dsDNA’s two
antiparallel strands are simultaneously replicated at an ad-
vancing replication fork.Yet, all known DNA polymerases
can only extend DNA strands in the 5¿S3¿ direction.How,
then, does DNA polymerase copy the parent strand that
extends in the 5¿S3¿ direction past the replication fork?
This question was answered in 1968 by Reiji Okazaki
through the following experiments. If a growing E. coli cul-
ture is pulse-labeled for 30 s with [
3
H]thymidine, much of
the radioactive and hence newly synthesized DNA has a
sedimentation coefficient in alkali of 7S to 11S. These so-
called Okazaki fragments evidently consist of only 1000 to
2000 nucleotides (nt; 100–200 nt in eukaryotes). If, how-
ever, following the 30 s [
3
H]thymidine pulse, the E. coli are
transferred to an unlabeled medium (a pulse–chase exper-
iment), the resulting radioactively labeled DNA sediments
at a rate that increases with the time that the cells had
grown in the unlabeled medium. The Okazaki fragments
must therefore become covalently incorporated into larger
DNA molecules.
Okazaki interpreted his experimental results in terms of
the semidiscontinuous replication model (Fig. 30-5). The
two parent strands are replicated in different ways. The
newly synthesized DNA strand that extends 5¿S3¿ in the di-
rection of replication fork movement, the so-called leading
strand, is essentially continuously synthesized in its 5¿S3¿
direction as the replication fork advances. The other newly
synthesized strand, the lagging strand, is also synthesized in
its 5¿S3¿ direction but discontinuously as Okazaki frag-
ments.The Okazaki fragments are only covalently joined to-
gether sometime after their synthesis in a reaction catalyzed
by the enzyme DNA ligase (Section 30-2D).
The semidiscontinuous model of DNA replication is
corroborated by electron micrographs of replicating
DNA showing single-stranded regions on one side of the
Section 30-1. DNA Replication: An Overview 1175
Figure 30-4 Autoradiographic differentiation of unidirectional
and bidirectional replication of DNA. (a) An organism is
grown for several generations in a medium that is lightly labeled
with [
3
H]thymidine so that all of its DNA will be visible in an
autoradiogram.A large amount of [
3
H]thymidine is then added
to the medium for a few seconds before the DNA is isolated
(pulse labeling) in order to label only those bases near the
replication fork(s). Unidirectional DNA replication will exhibit
only one heavily labeled branch point (above), whereas
bidirectional DNA replication will exhibit two such branch
points (below). (b) An autoradiogram of E. coli DNA so treated,
demonstrating that it is bidirectionally replicated. [Courtesy of
David M. Prescott, University of Colorado.]
Unidirectional
replication
Heavily
labeled DNA
Bidirectional
replication
Lightly
labeled DNA
(a)
(b)
3′
5′
3′
5′
3′
5′
5′
3′
Leading strand
Lagging strand (Okazaki fragments)
Parental strands
Motion of
replication
fork
Figure 30-5 Semidiscontinuous DNA replication. In DNA
replication, both daughter strands (leading strand red, lagging
strand blue) are synthesized in their 5¿S3¿ directions.The
leading strand is synthesized continuously, whereas the lagging
strand is synthesized discontinuously.
JWCL281_c30_1173-1259.qxd 8/10/10 9:10 PM Page 1175

3′
3′
5′
5′
RNA primer
5′
3′
3′
5′
Okazaki fragment
5′
3′
3′
5′
5′
replication fork (Fig. 30-6). In bidirectionally replicating
DNA, moreover, the two single-stranded regions occur,
as expected, on diagonally opposite sides of the replica-
tion bubble.
D. RNA Primers
DNA polymerases’ all but universal requirement for a free
3¿-OH group to extend a DNA chain poses a question that
was emphasized by the establishment of the semidiscontin-
uous model of DNA replication: How is DNA synthesis
initiated? Careful analysis of Okazaki fragments revealed
that their 5¿ ends consist of RNA segments of 1 to 60 nt (a
length that is species dependent) that are complementary to
the template DNA chain (Fig. 30-7). E. coli has two enzymes
that can catalyze the formation of these RNA primers:
RNA polymerase, the ⬃459-kD multisubunit enzyme that
mediates transcription (Section 31-2), and the much
smaller primase (60 kD), the monomeric product of the
dnaG gene.
Primase is insensitive to the RNA polymerase inhibitor
rifampicin (Section 31-2Bb). The observation that ri-
fampicin inhibits only leading strand synthesis therefore
indicates that primase initiates the Okazaki fragment
primers. The initiation of leading strand synthesis in E. coli,
a much rarer event than that of Okazaki fragments, can be
mediated in vitro by either RNA polymerase or primase
alone but is greatly stimulated when both enzymes are
present. It is therefore thought that these enzymes act syn-
ergistically in vivo to prime leading strand synthesis.
Mature DNA does not contain RNA.The RNA primers
are eventually removed and the resulting single-strand
gaps are filled in with DNA by a mechanism described in
Section 30-2Aj.
2 ENZYMES OF REPLICATION
DNA replication is a complex process involving a great
variety of enzymes. It requires, to list only its major actors
in their order of appearance: (1) DNA topoisomerases,
(2) enzymes known as helicases that separate the DNA
strands at the replication fork, (3) proteins that prevent
them from reannealing before they are replicated, (4) en-
zymes that synthesize RNA primers, (5) a DNA poly-
merase, (6) an enzyme to remove the RNA primers, and
(7) an enzyme to covalently link successive Okazaki frag-
ments. In this section, we describe the properties and func-
tions of many of these proteins.
A. DNA Polymerase I
In 1957, Arthur Kornberg reported that he had discovered
an enzyme that catalyzes the synthesis of DNA in extracts
of E. coli through its ability to incorporate the radioactive
label from [
14
C]thymidine triphosphate into DNA. This en-
zyme, which has since become known as DNA polymerase
I or Pol I, consists of a monomeric 928-residue polypeptide.
Pol I couples deoxynucleoside triphosphates on DNA
templates (Fig. 30-1) in a reaction that occurs through the
nucleophilic attack of the growing DNA chain’s 3¿-OH
group on the ␣-phosphoryl of an incoming nucleoside
triphosphate.The reaction is driven by the resulting elimina-
tion of PP
i
and its subsequent hydrolysis by inorganic py-
rophosphatase. The overall reaction resembles that cat-
alyzed by RNA polymerase (Fig. 5-23) but differs from it
by the strict requirement that the incoming nucleoside be
linked to a free 3¿-OH group of a polynucleoside that is
base paired to the template (RNA polymerase initiates
1176 Chapter 30. DNA Replication, Repair, and Recombination
Figure 30-7 Priming of DNA synthesis by short RNA
segments.
Figure 30-6 Electron micrograph of a replication eye in
Drosophila melanogaster DNA. Note that the single-stranded
regions (arrows) near the replication forks have the trans
configuration consistent with the semidiscontinuous model of
DNA replication. [From Kreigstein, H.J. and Hogness, D.S., Proc.
Natl.Acad. Sci. 71, 173 (1974).]
JWCL281_c30_1173-1259.qxd 8/10/10 9:10 PM Page 1176

transcription by linking together two ribonucleoside
triphosphates on a DNA template; Section 31-2C). The
complementarity between the product DNA and the tem-
plate was at first inferred through base composition and
hybridization studies but was eventually directly estab-
lished by base sequence determinations. The error rate of
Pol I in copying the template is extremely low, as was first
demonstrated by its in vitro replication of the 5386-nt
DNA from bacteriophage X174 to yield fully infective
phage DNA. In fact, its measured error rate is around one
wrong base per 10 million.
Pol I is said to be processive in that it catalyzes a series
of successive polymerization steps, typically 20 or more,
without releasing the template (the opposite of “proces-
sive” is “distributive”). Pol I can, of course, work in reverse
by degrading DNA through pyrophosphorolysis. This re-
verse reaction, however, probably has no physiological sig-
nificance because of the low in vivo concentration of PP
i
resulting from the action of inorganic pyrophosphatase.
a. Pol I Recognizes the Incoming dNTP According
to the Shape of the Base Pair It Forms with the
Template DNA
The specificity of Pol I for an incoming base arises
from the requirement that it form a Watson–Crick base
pair with the template rather than direct recognition of
the incoming base (recall that the four base pairs, A ⴢ T,
T ⴢ A, G ⴢ C, and C ⴢ G, have nearly identical shapes; Fig.
5-12). Thus, as Eric Kool demonstrated, when the “base”
2,4-difluorotoluene (F),
which is isosteric with (has the same shape as) thymine but
does not accept hydrogen bonds, is synthetically inserted
into a template DNA, Pol I incorporates A opposite the F
with a similar rate of mismatches as it incorporates A
opposite T. Likewise, dFTP is incorporated opposite tem-
plate A with a similar fidelity as is dTTP. Yet the incorpo-
ration of F opposite an A in DNA destabilizes the double
helix by 15 kJ/mol relative to T opposite this A. Evidently,
Pol I selects an incoming dNTP largely according to its
ability to form a Watson–Crick-shaped pair with the tem-
plate base but with little regard for its hydrogen bonding
properties. Indeed, the NMR structure of a 12-bp DNA
containing a centrally located F opposite an A reveals that
it assumes a B-DNA conformation in which the F–A pair
closely resembles a T ⴢ A base pair in the same position of
an otherwise identical DNA.
F
O
H
H
2,4-Difluorotoluene base (F) Thymine (T)
F
H
CH
3
H
O
CH
3
N
N
b. Pol I Can Edit Its Mistakes
In addition to its polymerase activity, Pol I has two inde-
pendent hydrolytic activities:
1. It can act as a 3¿S5¿ exonuclease.
2. It can act as a 5¿S3¿ exonuclease.
The 3¿S5¿ exonuclease reaction differs chemically from
the pyrophosphorolysis reaction (the reverse of the poly-
merase reaction) only in that H
2
O rather than PP
i
is the
nucleotide acceptor. Kinetic and crystallographic studies,
however, indicate that these two catalytic activities oc-
cupy separate active sites (see below).The 3¿S5¿ exonu-
clease function is activated by an unpaired 3¿-terminal
nucleotide with a free OH group. If Pol I erroneously in-
corporates a wrong (unpaired) nucleotide at the end of a
growing DNA chain, the polymerase activity is inhibited
and the 3¿S5¿ exonuclease excises the offending nu-
cleotide (Fig. 5-36). The polymerase activity then re-
sumes DNA replication. Pol I therefore has the ability to
proofread or edit a DNA chain as it is synthesized so as to
correct its mistakes. This explains the great fidelity of
DNA replication by Pol I: The overall fraction of bases
that the enzyme misincorporates, ⬃10
⫺7
, is the product of
the fraction of bases that its polymerase activity misin-
corporates and the fraction of misincorporated bases
that its 3¿S5¿ exonuclease activity fails to excise. The
price of this high fidelity is that ⬃3% of correctly incor-
porated nucleotides are also excised.
The Pol I 5¿S3¿ exonuclease binds to dsDNA at single-
strand nicks with little regard to the character of the 5¿ nu-
cleotide (5¿-OH or phosphate group; base paired or not). It
cleaves the DNA in a base paired region beyond the nick
such that the DNA is excised as either mononucleotides or
oligonucleotides of up to 10 residues (Fig. 5-33). In con-
trast, the 3¿S5¿ exonuclease removes only unpaired
mononucleotides with 3¿-OH groups.
c. Pol I’s Polymerase and Two Exonuclease
Functions Each Occupy Separate Active Sites
The 5¿S3¿ exonuclease activity of Pol I is independent
of both its 3¿S5¿ exonuclease and its polymerase activi-
ties. In fact, as we saw in Section 7-2A, proteases such as
subtilisin or trypsin cleave Pol I into two fragments: a larger
C-terminal or Klenow fragment (residues 324–928), which
contains both the polymerase and the 3¿S5¿ exonuclease
activities; and a smaller, N-terminal fragment (residues
1–323), which contains the 5¿S3¿ exonuclease activity.
Thus Pol I contains three active sites on a single polypep-
tide chain.
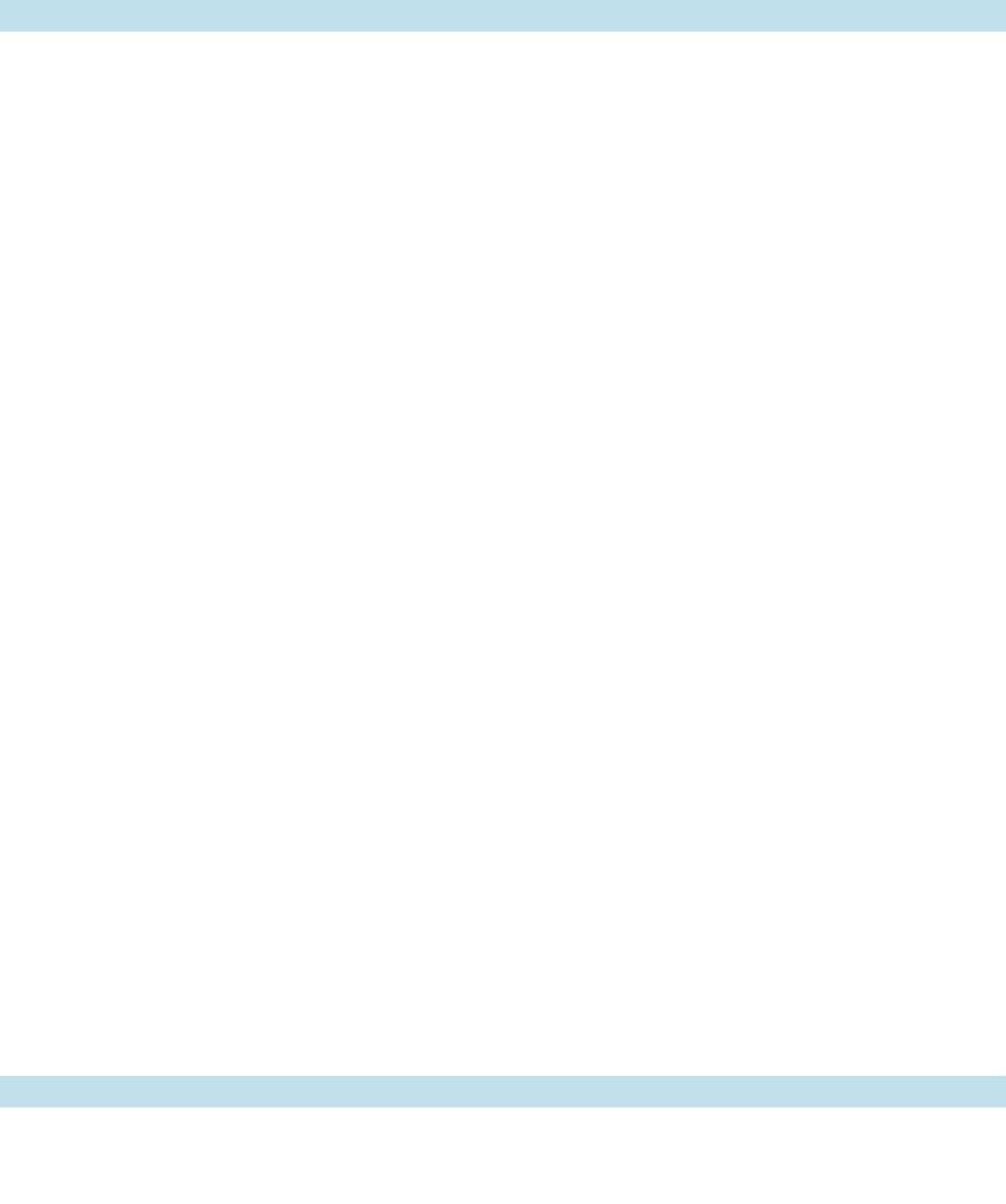
d. The X-Ray Structure of Klenow Fragment
Indicates How It Binds DNA
The X-ray structure of Klenow fragment in complex
with dsDNA, determined by Thomas Steitz, reveals that
Section 30-2. Enzymes of Replication 1177
JWCL281_c30_1173-1259.qxd 8/10/10 9:10 PM Page 1177
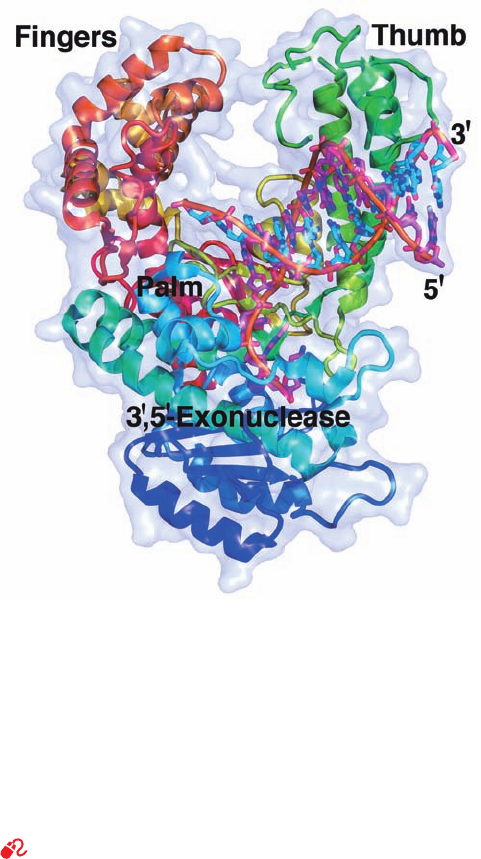
the enzyme consists of two domains (Fig. 30-8).The smaller
domain (residues 324–517) contains the 3¿S5¿ exonucle-
ase site, as was demonstrated by the absence of this func-
tion but not polymerase activity in a genetically engineered
Klenow fragment mutant that lacks the divalent metal
ion–binding sites known to be essential for 3¿S5¿ exonu-
clease activity but which otherwise has a normal structure.
The larger domain (residues 521–928) contains the poly-
merase active site at the bottom of a prominent cleft, a sur-
prisingly large distance (⬃25 Å) from the 3¿S5¿ exonucle-
ase site. The cleft, which is lined with positively charged
residues, has the appropriate size (⬃22 Å wide by ⬃30 Å
deep) and shape to bind a B-DNA molecule in a manner
resembling a right hand grasping a rod. Consequently,
Klenow fragment’s polymerase domains are known as its
“fingers,”“thumb,” and “palm” (Fig. 30-8). Indeed, all DNA
polymerases of known structure have similar shapes with a
fingers domain that binds the incoming nucleotide and the
templating base, a thumb domain that guides the newly
formed dsDNA as it leaves the active site, and a palm do-
main that contains the active site at the bottom of the cleft
between the fingers and thumb domains (Sections 30-2Ba,
30-4Ba, and 30-4Ca).
e. DNA Polymerase Distinguishes Watson–Crick
Base Pairs via Sequence-Independent Interactions
That Induce Domain Movements
DNA polymerase I from the thermophile Thermus
aquaticus (Taq), which is 51% identical to its E. coli ho-
molog, lacks a functional 3¿S5¿ exonuclease. Its X-ray
structure (Fig.30-9a), also determined by Steitz, reveals that
its C-terminal domain closely resembles Klenow fragment
(Fig. 30-8), although the metal ion–binding carboxylate
residues that are essential for the 3¿S5¿ exonuclease func-
tion of Klenow fragment are absent in Taq polymerase. Its
N-terminal 5¿S3¿ exonuclease domain, which appears to
be only loosely tethered to the C-terminal polymerase do-
main, contains a conserved cluster of metal ions at the bot-
tom of a cleft that is ⬃70 Å from the polymerase active site.
It is therefore is unclear how the polymerase and 5¿S3¿
exonuclease functions work in concert, as we shall see (Sec-
tion 30-2Ai), to produce dsDNA with a single nick.
Gabriel Waksman crystallized the C-terminal domain of
Taq DNA polymerase I [Klentaq1; which is often used in
PCR experiments (Section 5-5F)] in complex with an 11-bp
dsDNA that has a GGAAA-5¿ overhang at the 5¿ end of its
template strand, and the crystals were incubated with 2¿,3¿-
dideoxy-CTP (ddCTP; which lacks a 3¿-OH group). The X-
ray structure of these crystals (Fig. 30-9b) reveals that a
ddC residue had been covalently linked to the 3¿ end of the
primer and formed a Watson–Crick pair with the template
overhang’s 3¿ G. Moreover, a second ddCTP molecule (to
which the primer’s new 3¿-terminal ddC residue is inca-
pable of forming a covalent bond) occupies the enzyme’s
active site where it forms a Watson–Crick pair with the
template’s next G that is largely out of contact with the sur-
rounding aqueous solution. Clearly, Klentaq1 retains its
catalytic activity in this crystal.
A DNA polymerase must distinguish correctly paired
bases from mismatches and yet do so via sequence-inde-
pendent interactions with the incoming dNTP. The forego-
ing X-ray structure reveals that this occurs through an ac-
tive site pocket that is complementary in shape to
Watson–Crick base pairs. The pocket is formed by the
stacking of a conserved Tyr side chain on the template
base, as well as by van der Waals interactions with the pro-
tein and with the preceding base pair. In addition, although
the dsDNA is mainly in the B conformation, the 3 base
pairs nearest the active site assume the A conformation, as
has also been observed in the X-ray structures of several
other DNA polymerases in their complexes with DNA.
The resulting wider and shallower minor groove (Section
29-1Ba) permits protein side chains to form hydrogen
bonds with the otherwise inaccessible N3 atoms of the
purine bases and O2 atoms of the pyrimidine bases. The
positions of these hydrogen bond acceptors are sequence-
1178 Chapter 30. DNA Replication, Repair, and Recombination
Figure 30-8 X-ray structure of E. coli DNA polymerase I
Klenow fragment in complex with a dsDNA. The protein is
drawn in ribbon form colored in rainbow order from its
N-terminus (blue) to its C-terminus (red) and embedded in its
semitransparent molecular surface. The DNA is shown in stick
form with the C atoms of its 10-nt template strand cyan, the
C atoms of its 13-nt primer strand magenta, N blue, O red, and
P orange, and with successive P atoms in each polynucleotide
strand connected by orange rods.A Zn
2⫹
ion (purple sphere)
marks the 3¿S5¿-exonuclease active site. [Based on an X-ray
structure by Thomas Steitz, Yale University. PDBid 1KLN.]
See Interactive Exercise 33
JWCL281_c30_1173-1259.qxd 10/19/10 10:28 AM Page 1178
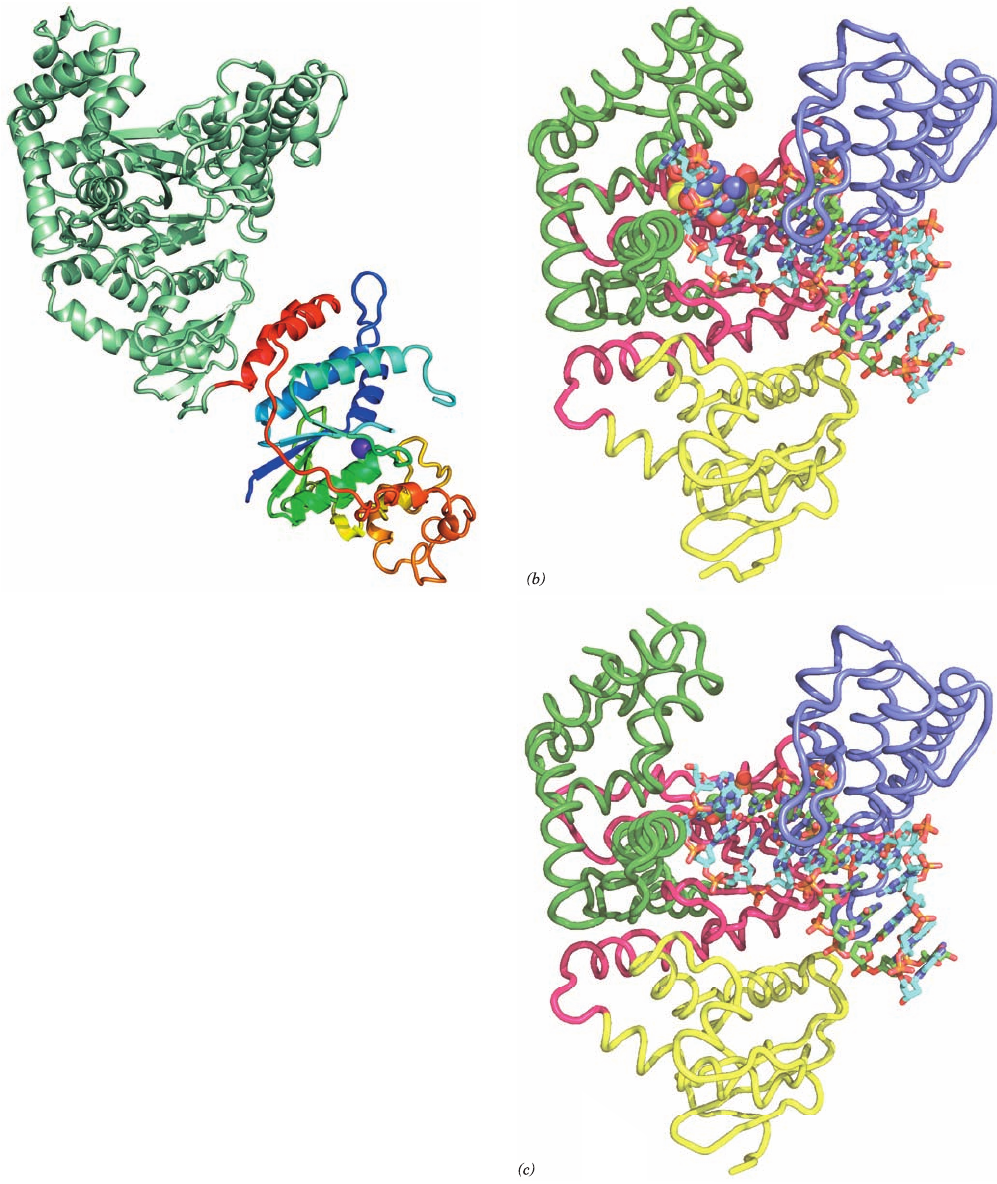
independent as can be seen from an inspection of Fig. 5-12
[in contrast, the positions of the hydrogen bonding accep-
tors in the major groove vary with both the identity (A ⴢ T
vs G ⴢ C) and the orientation (e.g., A ⴢ T vs T ⴢ A) of the
base pair]. However, with a non-Watson–Crick pairing,
these hydrogen bonds would be greatly distorted if not
completely disrupted.The protein also makes extensive se-
quence-independent hydrogen bonding and van der Waals
interactions with the DNA’s sugar–phosphate backbone.
The above Klentaq1 ⴢ DNA ⴢ ddCTP crystals were par-
tially depleted of ddCTP by soaking them in a stabilizing
solution that lacks ddCTP. The X-ray structure of the
ddCTP-depleted crystals (Fig. 30-9c) reveals that Klentaq1
Section 30-2. Enzymes of Replication 1179
(a)
Figure 30-9 X-ray structures of T. thermophilus DNA
polymerase I. (a) X-ray structure of the entire protein drawn in
ribbon form.The N-terminal 5¿S3¿ exonuclease domain is
colored in rainbow order from its N-terminus (blue) to its
C-terminus (red).The Zn
2⫹
ion at its active site is represented by
a purple sphere. The C-terminal Klentaq1 domain, which is
oriented similarly to the Klenow fragment in Fig. 30-8, is light
green. (b) The X-ray structures of Klentaq1 in its closed
conformation (with bound ddCTP) and (c) in its open
conformation (ddCTP depleted).The protein, which is viewed
similarly to Fig. 30-8, is drawn in worm form with its N-terminal,
palm, fingers, and thumb domains colored yellow, magenta,
green, and blue, respectively. The DNA is shown in stick form,
whereas the primer strand’s 3¿-terminal ddC residue is drawn in
space-filling form as is the ddCTP in Part b that is base paired
with a template G residue in the enzyme’s active site pocket.The
atoms are colored according to type with template strand C cyan,
primer strand C green, ddCTP C yellow, N blue, O red, and P
orange. [Part a based on an X-ray structure by Thomas Steitz,
Yale University. PDBid 1TAQ. Parts b and c based on X-ray
structures by Gabriel Waksman, Washington University School
of Medicine. PDBids 3KTQ and 2KTQ.]
JWCL281_c30_1173-1259.qxd 8/10/10 9:10 PM Page 1179
assumes a so-called open conformation, which differs sig-
nificantly from that in the so-called closed conformation
described above by a 46° hingelike motion of the fingers
domain away from the polymerase active site (Fig. 30-9c).
Evidently, the formation of a Watson–Crick base pair at the
polymerase active site triggers the formation of a produc-
tive ternary complex that buries the incoming nucleotide.
In particular, the foregoing Tyr side chain, which extends
from the rightmost helix of the fingers domain (Fig. 30-9b),
stacks on the incoming base, an interaction that is absent in
the open conformation. Moreover, a Lys and an Arg side
chain, that also extend from the rightmost helix of the fin-
gers domain, form salt bridges with the ␣- and -phosphate
groups of the incoming dNTP. These observations are con-
sistent with kinetic measurements on Pol I indicating that
the binding of the correct dNTP to the enzyme induces a
rate-limiting conformational change that yields a tight ter-
nary complex. It therefore appears that the enzyme rapidly
samples the available dNTPs in its open conformation but
only when it binds the correct dNTP in a Watson–Crick
pairing with the template base does it form the catalytically
competent closed conformation. The subsequent reaction
steps then rapidly yield the product complex which, follow-
ing a second conformational change, releases the product
PP
i
. Finally, the DNA is translocated in the active site,
probably via a linear diffusion mechanism, so as to position
it for the next reaction cycle.
The comparison of the above X-ray structures with that
of Klentaq1 alone indicates that on binding DNA, the
thumb domain moves to wrap around the DNA. It is likely
that this conformational change is largely responsible for
Pol I’s processivity. In both Klentaq1 ⴢ DNA structures, nei-
ther the dsDNA nor the single-stranded DNA (ssDNA)
passes through the cleft between the thumb and fingers do-
main as the shape and position of the cleft suggest. Rather,
the template strand makes a sharp bend at the first unpaired
base, thereby unstacking this base and positioning this
ssDNA on the same side of the cleft as the dsDNA. Similar
arrangements have been observed in X-ray structures of
other DNA polymerases in their complexes with DNA.
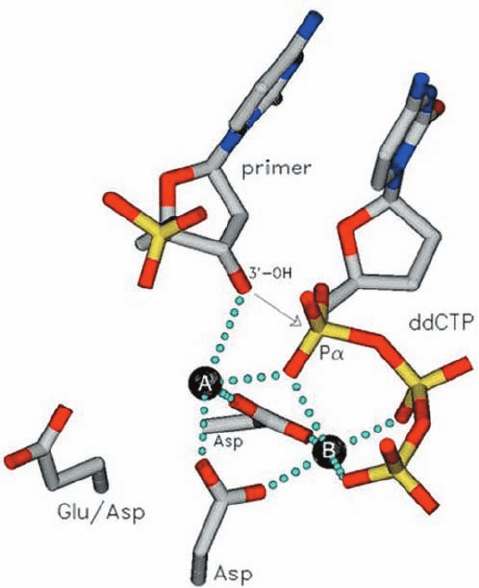
f. The DNA Polymerase Catalytic Mechanism
Involves Two Metal Ions
The X-ray structures of a variety of DNA polymerases
suggest that they share a common catalytic mechanism for
nucleotidyl transfer (Fig. 30-10). Their active sites all con-
tain two metal ions, usually Mg
2⫹
, that are liganded by two
invariant Asp side chains in the palm domain. Metal ion B
in Fig. 30-10 is liganded by all three phosphate groups of the
bound dNTP, whereas metal ion A bridges the ␣-phosphate
group of this dNTP and the primer’s 3¿-OH group. Metal
ion A presumably activates the primer’s 3¿-OH group for an
in-line nucleophilic attack on the ␣-phosphate group
(Fig. 16-6b), whereas metal ion B functions to orient its
bound triphosphate group and to electrostatically shield
their negative charges as well as the additional negative
charge on the transition state leading to the release of the
PP
i
ion (Section 16-2B).
g. Editing Complexes Contain the Primer Strand
in the 3ⴕ S 5ⴕ Exonuclease Site
The complex of Klenow fragment with DNA shown in
Fig. 30-8 contains a 13-nt primer strand [d(GCCTCGCG-
GCGGC)] and a 10-nt template strand [d(GCCGC-
GAGGC)] that is complementary to the 10-nt segment at
the 5¿ end of the primer strand. The 3¿-terminal nucleotide
of the primer strand (the last one that an active polymerase
would have added) is bound at the 3¿S5¿ exonuclease ac-
tive site.This arrangement is made possible by the opening
up of the G ⴢ C base pair that would otherwise be formed
by the 5¿ nucleotide of the template strand, which remains
bound near the entrance of the polymerase active site. Ev-
idently, the Klenow fragment has bound the primer strand
in an “editing” complex rather than in the polymerase cleft.
In E. coli Pol I, how does the 3¿ end of the primer strand
transfer between the polymerase active site and the 3¿S5¿
exonuclease active site? This appears to occur through the
competition of these sites for the 3¿ end of the primer
strand, which base pairs to form dsDNA in the polymerase
1180 Chapter 30. DNA Replication, Repair, and Recombination
Figure 30-10 Schematic diagram for the nucleotidyl
transferase mechanism of DNA polymerases. A and B represent
enzyme-bound metal ions that usually are Mg
2⫹
.Atoms
are colored according to type (C gray, N blue, O red, and
P yellow) and metal ion coordination is represented by dotted
lines. Metal ion A activates the primer’s 3¿-OH group for in-line
nucleophilic attack on the incoming dNTP’s ␣-phosphate group
(arrow), whereas metal ion B acts to orient and electrostatically
stabilize the negatively charged triphosphate group. [Courtesy of
Tom Ellenberger, Harvard Medical School.]
JWCL281_c30_1173-1259.qxd 8/10/10 9:10 PM Page 1180
