Voet D., Voet Ju.G. Biochemistry
Подождите немного. Документ загружается.

catalytic triad and is a structural paradigm for two enzyme
families, Nature Struct. Biol. 3, 74–86 (1996).
Thoden, J.B., Kappock, T.J., Stubbe, J.A., and Holden, H.M.,
Three-dimensional structure of N
5
-carboxyaminoimidazole ri-
bonucleotide synthetase: A member of the ATP grasp protein
superfamily, Biochemistry 38, 15480–15492 (1999). [X-ray
structure of PurK.]
Toth, E.A. and Yeates, T.O., The structure of adenylosuccinate
lyase, an enzyme with dual activity in the de novo purine
biosynthetic pathway, Structure 8, 163–174 (2000).
Wang,W., Kappock,T.J., Stubbe,J.A., and Ealick, S.E., X-ray struc-
ture of glycinamide ribonucleotide synthetase from Es-
cherichia coli, Biochemistry 37, 15647–15662 (1998).
Zalkin, H. and Dixon, J.E., De novo purine nucleotide biosynthe-
sis, Prog. Nucleic Acid Res. Mol. Biol. 42, 259–285 (1992).
Pyrimidine Nucleotide Biosynthesis
Begley, T.P., Appleby, T.C., and Ealick, S.E., The structural basis for
the remarkable catalytic proficiency of orotidine 5¿-monophos-
phate decarboxylase, Curr. Opin. Struct. Biol. 10, 711–718 (2000).
Evans, D.R. and Guy, H.I., Mammalian pyrimidine biosynthesis:
Fresh insights into an ancient pathway, J. Biol. Chem. 279,
33035–33038 (2004).
Jones, M.E., Orotidylate decarboxylase of yeast and man, Curr.
Top. Cell Regul. 33, 331–342 (1992).
Liu, S., Neidhardt, E.A., Grossman, T.H., Ocain, T., and Clardy, J.,
Structures of human dihydroorotate dehydrogenase in com-
plex with antiproliferative agents, Structure 8, 25–33 (1999).
Miller,B.G.and Wolfenden,R.,Catalytic proficiency:The unusual case
of OMP decarboxylase, Annu. Rev. Biochem. 71, 847–885 (2002).
Scapin, G., Ozturk, D.H., Grubmeyer, C., and Sacchettini,J.C.,The
crystal structure of the orotate phosphoribosyltransferase
complexed with orotate and -
D-5-phosphoribosyl-1-
pyrophosphate, Biochemistry 34, 10744–10754 (1995).
Thoden, J.B., Phillips, G.N., Jr., Neal, T.M., Raushel, F.M., and
Holden, H.M., Molecular structure of dihydroorotase:A para-
digm for catalysis through the use of a binuclear center, Bio-
chemistry 40, 6989–6997 (2001).
Traut, T.W. and Jones, M.E., Uracil metabolism—UMP synthesis
from orotic acid or uridine and conversion of uracil to -alanine:
Enzymes and cDNAs, Prog. Nucleic Acid Res. Mol. Biol. 53,
1–78 (1996).
Synthesis of Deoxynucleotides
Carreras,C.W.and Santi,D.V.,The catalytic mechanism and structure
of thymidylate synthase, Annu. Rev. Biochem. 64, 721–762 (1995).
Eriksson, M., Uhlin, U., Ramaswamy, S., Ekberg, M., Regnström,
K., Sjöberg, B.-M., and Eklund, H., Binding of allosteric effec-
tors to ribonucleotide reductase protein R1: Reduction of ac-
tive site cysteines promotes substrate binding, Structure 5,
1077–1092 (1997).
Finer-Moore, J.S., Santi, D.V., and Stroud, R.M., Lessons and
conclusions from dissecting the mechanism of a bisubstrate
enzyme: thymidylate synthase mutagenesis, function, and
structure, Biochemistry 42, 248–256 (2003).
Kashlan, O.B., Scott, C.P., Lear, J.D., and Cooperman, B.S.,A com-
prehensive model for the allosteric regulation of mammalian
ribonuclease reductase. Functional consequences of ATP- and
dATP-induced oligomerization of the large subunit, Biochem-
istry 41, 462–474 (2002).
Knighton, D.R., Kan, C.-C., Howland, E., Janson, C.A., Hostom-
ska, Z., Welsh, K.M., and Matthews, D.A., Structure of and
kinetic channeling in bifunctional dihydrofolate reductase-
thymidylate synthase, Nature Struct. Biol. 1, 186–194 (1994).
Lennon, B.W.,Williams, J.R., Jr., and Ludwig, M.L.,Twists in catal-
ysis:Alternating conformations in Escherichia coli thioredoxin
reductase, Science 289, 1190–1194 (2000).
Logan, D.T.,Andersson, J., Sjöberg,B.-M., and Nordlund, P.,A gly-
cyl radical site in the crystal structure of a Class III ribonu-
cleotide reductase, Science 283, 1499–1504 (1999).
Matthews, D.A., Villafranca, J.E., Janson, C.A., Smith, W.W.,
Welsh, K., and Freer, S., Stereochemical mechanisms of action
for thymidylate synthase based on the X-ray structure of
the covalent inhibitory ternary complex with 5-fluoro-2¿-
deoxyuridylate and 5,10-methylenetetrahydrofolate, J. Mol.
Biol. 214, 937–948 (1990); and Hyatt, D.C., Maley, F., and
Montfort, W.R., Use of strain in a stereospecific catalytic
mechanism: Crystal structure of Escherichia coli thymidylate
synthase bound to FdUMP and methylenetetrahydrofolate,
Biochemistry 36, 4585–4594 (1997).
Mol, C.D., Harris, J.M., McIntosh, E.M., and Tainer, J.A., Human
dUTP pyrophosphatase: Uracil recognition by a hairpin and
active sites formed by three separate subunits, Structure 4,
1077–1092 (1996).
Nordlund, P. and Eklund, H., Structure and function of the
Escherichia coli ribonucleotide reductase protein R2, J. Mol.
Biol. 232, 123–164 (1993).
Nordlund, P. and Reichard, P., Ribonucleotide reductases, Annu.
Rev. Biochem. 75, 681–706 (2006).
Powis, G. and Montfort, W.R., Properties and biological activities
of thioredoxins, Annu. Rev. Biophys. Biomol. Struct. 30,
421–455 (2001).
Sintchak, M.D., Arjara, G., Kellog, B.A., Stubbe, J., and Drennan,
C.L.,The crystal structure of class II ribonucleotide reductase
reveals how an allosterically regulated monomer mimics a
dimer, Nature Struct. Biol. 9, 293–300 (2002).
Stubbe, J. and Riggs-Gelasco, P., Harnessing free radicals: Forma-
tion and function of the tyrosyl radical in ribonucleotide
reductase, Trends Biochem. Sci. 23, 438–443 (1998).
Stubbe, J., Ge, J., and Yee, C.S.,The evolution of ribonucleotide re-
duction revisited, Trends Biochem. Sci. 26, 93–99 (2001); and
Stubbe, J., Ribonucleotide reductases: The link between an
RNA and a DNA world, Curr. Opin. Struct. Biol. 10, 731–736
(2000).
Uhlin, U. and Eklund, H., Structure of ribonucleotide reductase
protein R1, Nature 370, 533–539 (1994).
Nucleotide Degradation
Enroth, C., Eger, B.T., Okamoto, K., Nishino, T., Nishino, T., and
Pai, E., Crystal structure of bovine milk xanthine dehydroge-
nase and xanthine oxidase: Structure based mechanism of con-
version, Proc. Natl.Acad. Sci. 97, 10723–10728 (2000).
Parkman, R., Weinberg, K., Crooks, G., Nolta, I., Kapoor, N., and
Kohn, D., Gene therapy for adenosine deaminase deficiency,
Annu. Rev. Med. 51, 33–47 (2000).
Wilson, D.K., Rudolph, F.B., and Quiocho, F.A., Atomic structure
of adenosine deaminase complexed with a transition-state ana-
log: Understanding catalysis and immunodeficiency mutations,
Science 252, 1278–1284 (1991); Wilson, D.K. and Quiocho, F.A.,
A pre-transition-state mimic of an enzyme: X-ray structure of
adenosine deaminase with bound 1-deazaadenosine and zinc-
activated water, Biochemistry 32, 1689–1694 (1993); and Crys-
tallographic observation of a trapped tetrahedral intermediate
in a metalloenzyme, Nature Struct. Biol. 1, 691–694 (1994).
Biosynthesis of Nucleotide Coenzymes
Belenky, P., Bogan, K.L., and Brenner, C., NAD
metabolism in
health and disease, Trends Biochem. Sci. 32, 12–19 (2007).
References 1141
JWCL281_c28_1107-1142.qxd 4/22/10 9:17 AM Page 1141
1142 Chapter 28. Nucleotide Metabolism
1. Azaserine (O-diazoacetyl-L-serine) and 6-diazo-5-oxo-L-
norleucine (DON)
are glutamine analogs. They form covalent bonds to nucleophiles
at the active sites of enzymes that bind glutamine, thereby irre-
versibly inactivating these enzymes. Identify the nucleotide
biosynthesis intermediates that accumulate in the presence of ei-
ther of these glutamine antagonists.
2. Suggest a mechanism for the AIR synthetase reaction
(Fig. 28-2, Reaction 6).
*3. What is the energetic price, in ATPs, of synthesizing the hy-
poxanthine residue of IMP from CO
2
and NH
4
⫹
?
4. Why is deoxyadenosine toxic to mammalian cells?
5. Indicate which of the following substances are mechanism-
based inhibitors and explain your reasoning. (a) Tosyl-
L-phenyl-
alanine chloromethylketone with chymotrypsin (Section 15-3Ab).
(b) Trimethoprim with bacterial dihydrofolate reductase. (c) The
␦-lactone analog of (NAG)
4
with lysozyme (Section 15-2Cb). (d)
Allopurinol with xanthine oxidase.
6. Why do individuals who are undergoing chemotherapy
with cytotoxic (cell killing) agents such as FdUMP or methotrex-
ate temporarily go bald?
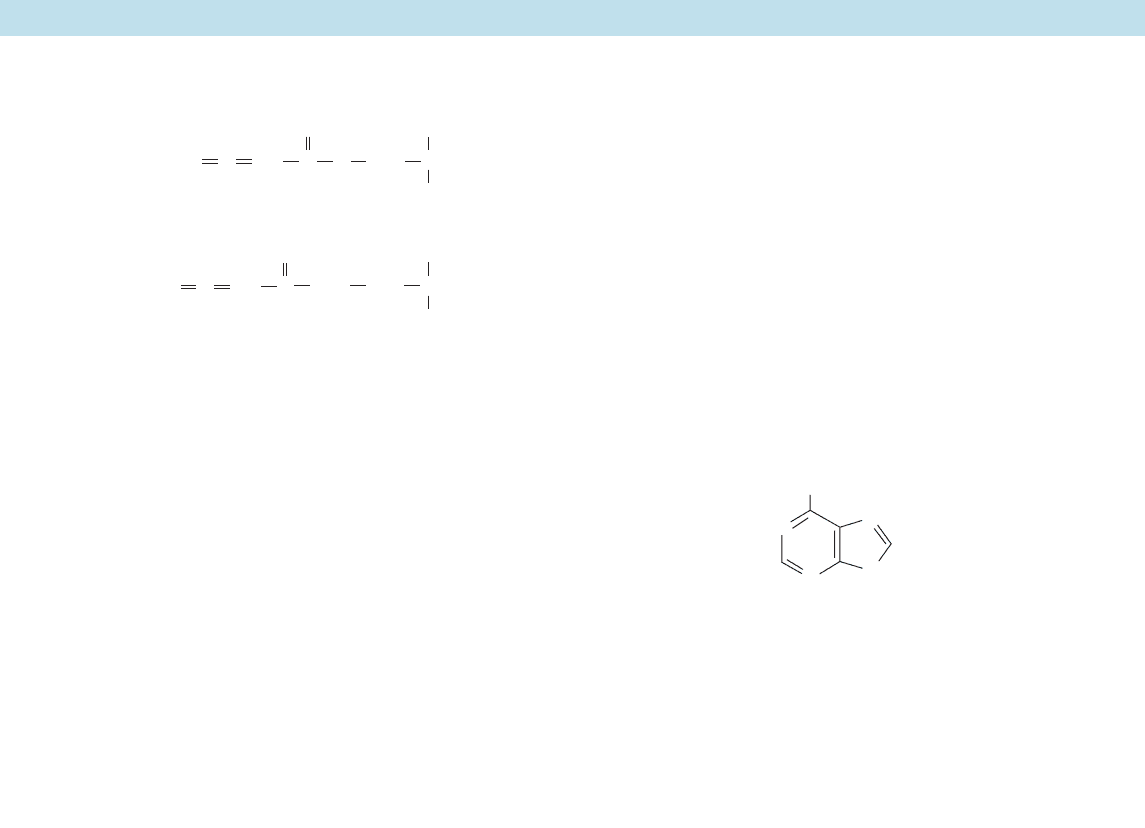
6-Diazo-5-oxo-L-norleucine (DON)
Azaserine
NNCH
CH
CH
COO
_
COO
_
NH
3
+
NH
3
+
H
2
C
CH
2
CH
2
CH
CO
O
C
O
_
+
NN
_
+
7. Normal cells die in a nutrient medium containing thymidine
and methotrexate that supports the growth of mutant cells defec-
tive in thymidylate synthase. Explain.
8. FdUMP and methotrexate, when taken together, are less ef-
fective chemotherapeutic agents than when either drug is taken
alone. Explain.
9. Some microorganisms lack DHFR activity, but their
thymidylate synthase has an FAD cofactor.What is the function of
the FAD?
10. Why is gout more prevalent in populations that eat rela-
tively large amounts of meat?
11. Gout resulting from the de novo overproduction of
purines can be distinguished from gout caused by impaired excre-
tion of uric acid by feeding a patient
15
N-labeled glycine and de-
termining the distribution of
15
N in his or her excreted uric acid.
What isotopic distributions are expected for each type of defect?
12. 6-Mercaptopurine,
after its conversion to the corresponding nucleotide through sal-
vage reactions, is a potent competitive inhibitor of IMP in the
pathways for AMP and GMP biosynthesis. It is therefore a clini-
cally useful anticancer agent.The chemotherapeutic effectiveness
of 6-mercaptopurine is enhanced when it is administered with al-
lopurinol. Explain the mechanism of this enhancement.
SH
N
N
N
N
H
6-Mercaptopurine
PROBLEMS
JWCL281_c28_1107-1142.qxd 7/21/10 7:10 PM Page 1142
PART V
EXPRESSION AND
TRANSMISSION
OF GENETIC
INFORMATION
Schematic diagram of the
eukaryotic preinitiation
complex that is required
for the transcription of
DNA to messenger RNA.
The TATA-box binding
protein is shown in orange.
JWCL281_c29_1143-1172.qxd 7/9/10 12:46 AM Page 1143
This page intentionally left blank
1145
CHAPTER 29
Nucleic Acid
Structures
1 Double Helical Structures
A. B-DNA
B. Other Nucleic Acid Helices
2 Forces Stabilizing Nucleic Acid Structures
A. Sugar–Phosphate Chain Conformations
B. Base Pairing
C. Base Stacking and Hydrophobic Interactions
D. Ionic Interactions
3 Supercoiled DNA
A. Superhelix Topology
B. Measurements of Supercoiling
C. Topoisomerases
The structure and properties of DNA are introduced in
Section 5-3. In this chapter we extend this discussion with
emphasis on DNA; the structures of RNAs are detailed in
Sections 31-4A and 32-2B. Methods of purifying, sequenc-
ing, and chemically synthesizing nucleic acids are discussed
in Sections 6-6, 7-2, and 7-6, and recombinant DNA tech-
niques are discussed in Section 5-5. Bioinformatics, as it
concerns nucleic acids, is outlined in Section 7-4, and the
Nucleic Acid Database is described in Section 8-3Cb.
1 DOUBLE HELICAL STRUCTURES
See Guided Exploration 23: DNA structures Double helical
DNA has three major helical forms, B-DNA,A-DNA, and
Z-DNA, whose structures are depicted in Fig. 29-1. In this
section we discuss the major characteristics of each of these
helical forms as well as those of double helical RNA and
DNA–RNA hybrid helices.
A. B-DNA
The structure of B-DNA (Fig. 29-1, middle panels), the bi-
ologically predominant form of DNA, is described in Sec-
tion 5-3A.To recapitulate (Table 29-1), B-DNA consists of
a right-handed double helix whose two antiparallel
sugar–phosphate chains wrap around the periphery of the
helix. Its aromatic bases (A, T, G, and C), which occupy
the core of the helix, form complementary A ⴢ T and G ⴢ C
Watson–Crick base pairs (Fig. 5-12), whose planes are
nearly perpendicular to the axis of the double helix. Neigh-
boring base pairs, whose aromatic rings are 3.4 Å thick, are
stacked in van der Waals contact, with the helix axis passing
through the middle of each base pair. B-DNA is ⬃20 Å in
diameter and has two deep grooves between its
sugar–phosphate chains: the relatively narrow minor
groove, which exposes that edge of the base pairs from
which the glycosidic bonds (the bonds from the base N to
the ribose C1¿) extend (toward the bottom of Fig. 5-12), and
the relatively wide major groove, which exposes the oppo-
site edge of each base pair (toward the top of Fig. 5-12).
Canonical (ideal) DNA has a helical twist of 10 base pairs
(bp) per turn and hence a pitch (rise per turn) of 34 Å.
The Watson–Crick base pairs in either orientation are
structurally interchangeable, that is, A ⴢ T, T ⴢ A, G ⴢ C, and
1145
There are two classes of nucleic acids, deoxyribonucleic
acid (DNA) and ribonucleic acid (RNA). DNA is the
hereditary molecule in all cellular life-forms, as well as in
many viruses. It has but two functions:
1. To direct its own replication during cell division.
2. To direct the transcription of complementary mole-
cules of RNA.
RNA, in contrast, has more varied biological functions:
1. The RNA transcripts of DNA sequences that specify
polypeptides, messenger RNAs (mRNAs), direct the ribo-
somal synthesis of these polypeptides in a process known
as translation.
2. The RNAs of ribosomes, which are about two-thirds
RNA and one-third protein, have functional as well as
structural roles.
3. During protein synthesis, amino acids are delivered
to the ribosome by molecules of transfer RNA (tRNA).
4. Certain RNAs are associated with specific proteins
to form ribonucleoproteins that participate in the post-
transcriptional processing of other RNAs.
5. A variety of short RNAs participate in the control of
eukaryotic gene expression and in protection against
viruses, a phenomenon known as RNA interference
(RNAi).
6. In many viruses, RNA, not DNA, is the carrier of
hereditary information.
JWCL281_c29_1143-1172.qxd 7/9/10 12:46 AM Page 1145
C ⴢ G can replace each other in the double helix without al-
tering the positions of the sugar–phosphate backbones’ C1¿
atoms. In contrast, any other combination of bases would
significantly distort the double helix since the formation of
a non-Watson–Crick base pair would require considerable
reorientation of the DNA’s sugar–phosphate backbones.
a. Real DNA Deviates from the Ideal
Watson–Crick Structure
The DNA samples that were available when James Wat-
son and Francis Crick formulated the Watson–Crick struc-
ture in 1953 were extracted from cells and hence consisted
of molecules of heterogeneous lengths and base sequences.
1146 Chapter 29. Nucleic Acid Structures
Minor
groove
Major
groove
A-DNA
(a)
Minor
groove
Major
groove
B-DNA
Minor
groove
Z-DNA
Major
groove
(b)
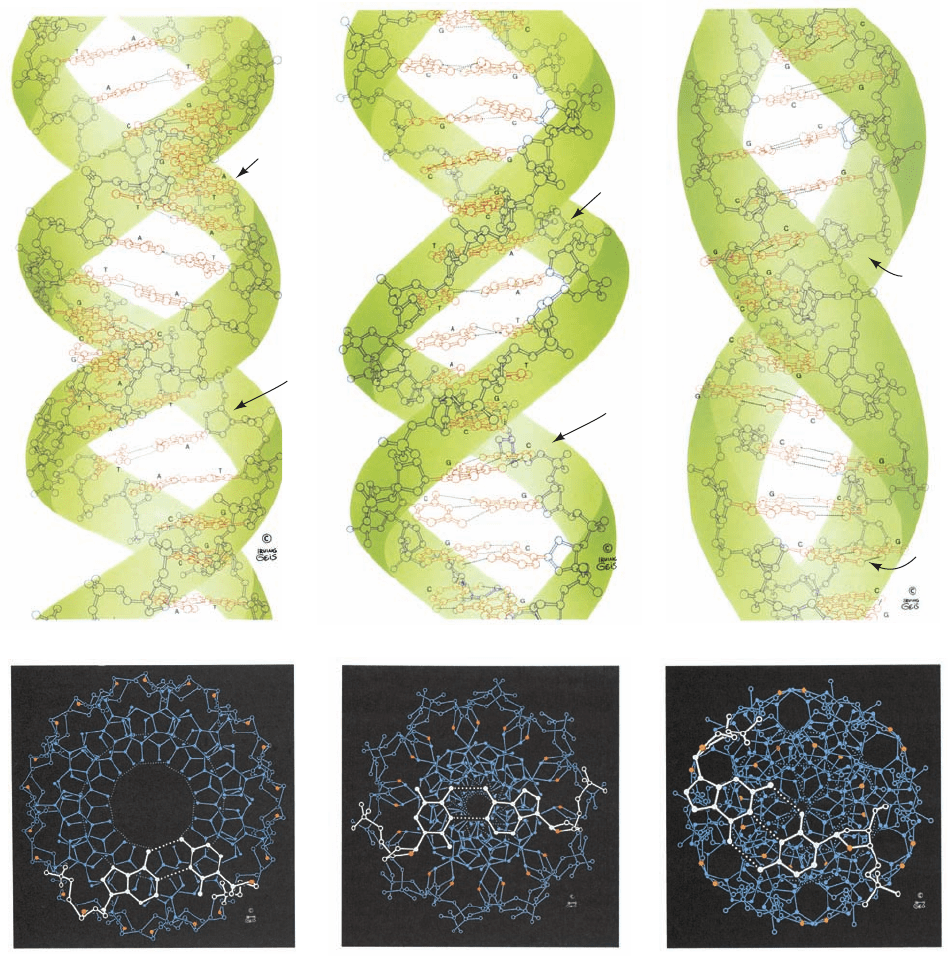
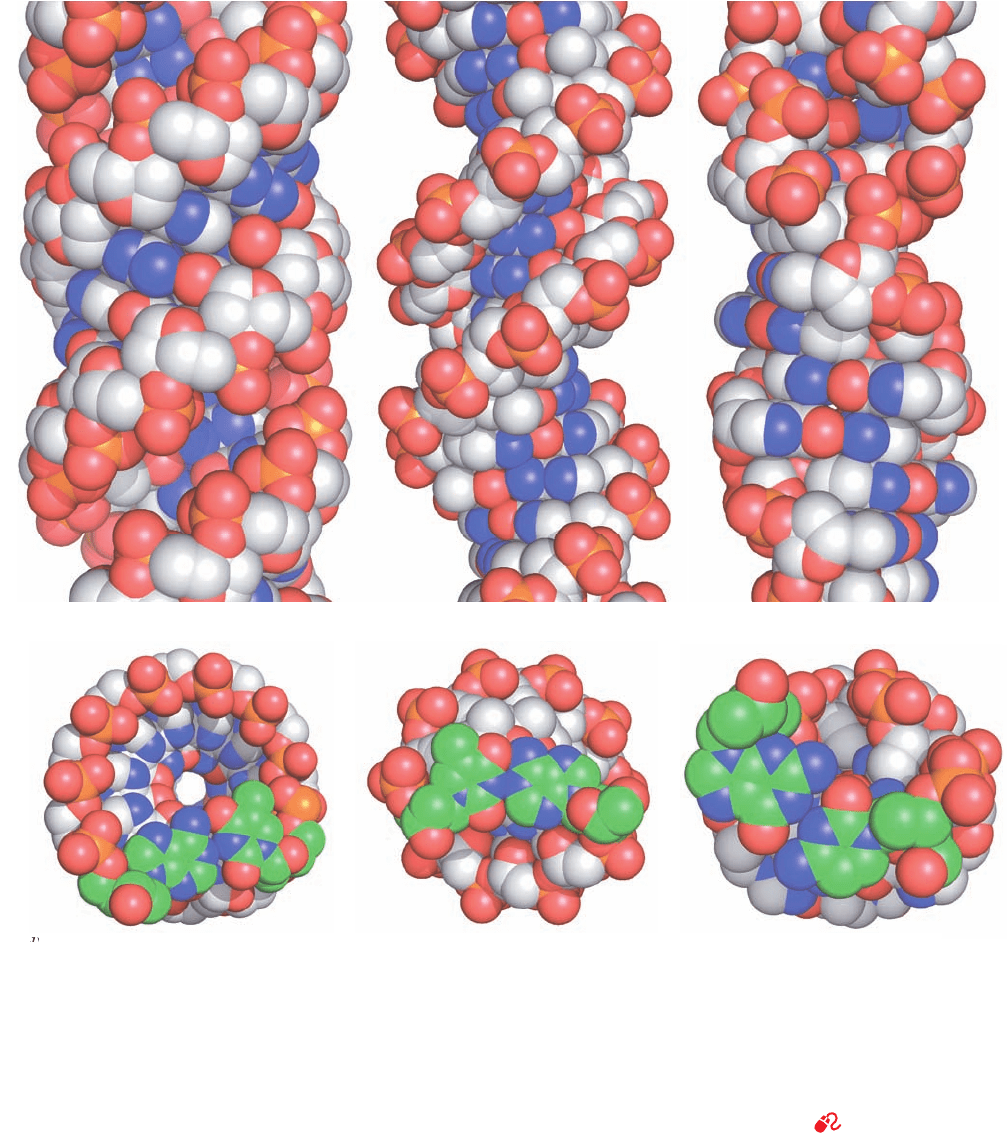
Figure 29-1 Structures of A-, B-, and Z-DNAs. (a) Ball-and
stick drawings viewed perpendicular to the helix axis.The
sugar–phosphate backbones, which wind about the periphery of
each molecule, are outlined by green ribbons, and the bases,
which occupy its core, are red. Note that the two sugar–phosphate
chains in each helix run in opposite directions so as to form right-
handed double helices in A- and B-DNAs and a left-handed
double helix in Z-DNA. (b) Views along the helix axis. The ribose
ring O atoms are red and the nucleotide pair closest to the viewer
is white. Note that the helix axis passes far “above” the major
groove of A-DNA, through the base pairs of B-DNA, and
through the edge of the minor groove of Z-DNA. Consequently,
A-DNA has a hollow core whereas B- and Z-DNAs have solid
cores.Also note that the deoxyribose residues in A- and B-DNAs
have the same conformation in each helix, but those in Z-DNA
have two different conformations so that alternate ribose residues
JWCL281_c29_1143-1172.qxd 8/2/10 8:06 PM Page 1146
A
-DN
A
B-DN
A
Z
-DN
A
(c)
(d)
Such elongated molecules do not crystallize, but can be
drawn into threadlike fibers in which the helix axes of the
DNA molecules are all approximately parallel to the fiber
axis but are poorly aligned, if at all, in any other way. The
X-ray diffraction patterns of such fibers provide only crude,
low-resolution images in which the base pair electron den-
sity is the average electron density of all the base pairs in
the fiber.The Watson–Crick structure was based, in part, on
the X-ray fiber diffraction pattern of B-DNA (Fig. 5-10).
By the late 1970s, advances in nucleic acid chemistry
permitted the synthesis and crystallization of ever longer
oligonucleotides of defined sequences (Section 7-6A),
Section 29-1. Double Helical Structures 1147
lie at different radii. (c) Space-filling models viewed perpendicular
to the helix axis and colored according to atom type (C white,
N blue, O red, and P orange). (d) Space-filling models viewed
along the helix axis and colored as in Part c but with the C atoms
of the nucleotide pair closest to the viewer green. H atoms in
Parts b, c, and d have been omitted for clarity. [Based on X-ray
structures by the following:A-DNA, Olga Kennard, Dov
Rabinovitch, Zippora Shakked, and Mysore Viswamitra,
Cambridge University, U.K. Nucleic Acid Database ID ADH010;
B-DNA, Richard Dickerson and Horace Drew, Caltech. PDBid
1BNA; and Z-DNA,Andrew Wang and Alexander Rich, MIT.
PDBid 2DCG. Illustration, Irving Geis. Image from the Irving
Geis Collection, Howard Hughes Medical Institute. Reprinted
with permission. Model coordinates for Parts c and d generated
by Helen Berman, Rutgers University.]
See Kinemage
Exercises 17-1, 17-4, 17-5, and 17-6
JWCL281_c29_1143-1172.qxd 8/2/10 8:06 PM Page 1147
many of which could be crystallized. Consequently,
some 25 years after the Watson–Crick structure was
formulated, its X-ray crystal structure was clearly vis-
ualized for the first time when Richard Dickerson and
Horace Drew determined the first X-ray crystal struc-
ture of a B-DNA, that of the self-complementary dode-
camer d(CGCGAATTCGCG), at near-atomic (1.9 Å) res-
olution. This molecule, whose structure was subsequently
determined at significantly higher (1.4 Å) resolution by
Loren Williams, has an average rise per residue of 3.3 Å
and has 10.1 bp per turn (a helical twist of 35.5° per bp),
values that are nearly equal to those of canonical B-DNA.
However, individual residues depart significantly from
this average conformation (Fig. 29-1a, middle panel). For
example, the helical twist per base pair in this dodecamer
ranges from 26° to 43°. Each base pair further deviates
from its ideal conformation by such distortions as pro-
peller twisting (the opposite rotation of paired bases
about the base pair’s long axis; in the 1.4-Å resolution
structure, this quantity ranges from 23° to 7°) and base
pair roll (the tilting of a base pair as a whole about its long
axis; this quantity ranges from 14° to 17°).
X-ray and NMR studies of numerous other double heli-
cal DNA oligomers have amply demonstrated that the con-
formation of DNA, particularly B-DNA, is irregular in a
sequence-specific manner, although the rules specifying
how sequence governs conformation have proved to be
surprisingly elusive. This is because base sequence does not
so much confer a fixed conformation on a double helix as it
establishes the deformability of the helix. Thus, 5¿-R–Y-3¿
steps (where R and Y are the abbreviations for purines and
pyrimidines, respectively) in B-DNA are easily bent
because they exhibit relatively little ring–ring overlap be-
tween adjacent base pairs. In contrast, both Y–R steps and
R–R steps (the latter, due to base pairing, are equivalent to
Y–Y steps), and most notably A–A steps, are more rigid
because the extensive ring–ring overlap between their ad-
jacent base pairs tends to keep these base pairs parallel.
This phenomenon, as we shall see, is important for the
sequence-specific binding of DNA to proteins that process
genetic information. This is because many of these proteins
wrap their target DNAs around them, in many cases by
bending them by well over 90°. DNAs with different
sequences than the target DNA would not bind so readily
to the protein because they would resist deformation to the
required conformation more than the target DNA.
B. Other Nucleic Acid Helices
X-ray fiber diffraction studies, starting in the mid-1940s, re-
vealed that nucleic acids are conformationally variable
molecules. Indeed, double helical DNA and RNA can as-
sume several distinct structures that vary with such factors
as the humidity and the identities of the cations present, as
well as with base sequence. For example, fibers of B-DNA
form in the presence of alkali metal ions such as Na
when
the relative humidity is 92%. In this subsection, we de-
scribe the other major conformational states of double-
stranded DNA as well as those of double-stranded RNA
and RNA–DNA hybrid helices.
a. A-DNA’s Base Pairs Are Inclined to the Helix Axis
When the relative humidity is reduced to 75%, B-DNA
undergoes a reversible conformational change to the so-
called A form. Fiber X-ray studies indicate that A-DNA
forms a wider and flatter right-handed helix than does B-
DNA (Fig. 29-1, left panels; Table 29-1). A-DNA has 11.6
bp per turn and a pitch of 34 Å, which gives A-DNA an ax-
ial hole (Fig. 29-1b, d, left panels). A-DNA’s most striking
feature, however, is that the planes of its base pairs are
1148 Chapter 29. Nucleic Acid Structures
Table 29-1 Structural Features of Ideal A-, B-, and Z-DNA
A-DNA B-DNA Z-DNA
Helical sense Right-handed Right-handed Left-handed
Diameter ⬃26 Å ⬃20 Å ⬃18 Å
Base pairs per 11.6 10 12 (6 dimers)
helical turn
Helical twist 31° 36° 9° for pyrimidine–purine steps;
per base pair 51° for purine–pyrimidine steps
Helix pitch 34 Å 34 Å 44 Å
(rise per turn)
Helix rise per 2.9 Å 3.4 Å 7.4 Å per dimer
base pair
Base tilt normal 20° 6° 7°
to the helix axis
Major groove Narrow and deep Wide and deep Flat
Minor groove Wide and shallow Narrow and deep Narrow and deep
Sugar pucker C3¿-endo C2¿-endo C2¿-endo for pyrimidines; C3¿-endo for purines
Glycosidic bond Anti Anti Anti for pyrimidines; syn for purines
Source: Mainly Arnott, S., in Neidle, S. (Ed.), Oxford Handbook of Nucleic Acid Structure, p. 35, Oxford University Press (1999).
JWCL281_c29_1143-1172.qxd 7/8/10 8:39 PM Page 1148
tilted 20° with respect to the helix axis. Since its helix axis
passes “above” the major groove side of the base pairs (Fig.
29-1b, d, left panels) rather than through them as in B-
DNA,A-DNA has a deep major groove and a very shallow
minor groove; it can be described as a flat ribbon wound
around a 6-Å-diameter cylindrical hole. Most self-comple-
mentary oligonucleotides of
10 base pairs, for example,
d(GGCCGGCC) and d(GGTATACC), crystallize in the
A-DNA conformation. Like B-DNA, these molecules ex-
hibit considerable sequence-specific conformational varia-
tion although the degree of variation is less than that in
B-DNA.
A-DNA has, so far, been observed in only three biolog-
ical contexts: at the cleavage center of topoisomerase II
(Section 29-3Cd), at the active site of DNA polymerase
(Section 30-2Ae), and in certain Gram-positive bacteria
that have undergone sporulation (the formation, under en-
vironmental stress, of resistant although dormant cell types
known as spores; a sort of biological lifeboat). Such spores
contain a high proportion (20%) of small acid-soluble
spore proteins (SASPs). Some of these SASPs induce B-
DNA to assume the A form, at least in vitro. The DNA in
bacterial spores exhibits a resistance to UV-induced dam-
age that is abolished in mutants that lack these SASPs.This
occurs because the B S A conformation change inhibits
the UV-induced covalent cross-linking of pyrimidine bases
(Section 30-5Aa), in part by increasing the distance be-
tween successive pyrimidines.
b. Z-DNA Forms a Left-Handed Helix
Occasionally, a seemingly well-understood or at least fa-
miliar system exhibits quite unexpected properties. Over
25 years after the discovery of the Watson–Crick structure,
the crystal structure determination of the self-complemen-
tary hexanucleotide d(CGCGCG) by Andrew Wang and
Alexander Rich revealed, quite surprisingly, a left-handed
double helix (Fig. 29-1, right panels; Table 29-1). A similar
helix is formed by d(CGCATGCG). This helix, which has
been dubbed Z-DNA, has 12 Watson–Crick base pairs per
turn, a pitch of 44 Å, and, in contrast to A-DNA, a deep mi-
nor groove and no discernible major groove (its helix axis
passes “below” the minor groove side of its base pairs; Fig.
29-1b,d, right panels). Z-DNA therefore resembles a left-
handed drill bit in appearance. The base pairs in Z-DNA
are flipped 180° relative to those in B-DNA (Fig. 29-2)
through conformational changes discussed in Section 29-
2A. As a consequence, the repeating unit of Z-DNA is a
dinucleotide, d(XpYp), rather than a single nucleotide as it
is in the other DNA helices. The line joining successive
phosphorus atoms on a polynucleotide strand of Z-DNA
therefore follows a zigzag path around the helix (Fig.29-1a,c,
right panels; hence the name Z-DNA) rather than a
smooth curve as it does in A- and B-DNAs (Fig. 29-1a,c,
left and middle panels).
Fiber diffraction and NMR studies have shown that
complementary polynucleotides with alternating purines
and pyrimidines, such as poly d(GC) ⴢ poly d(GC) and poly
Section 29-1. Double Helical Structures 1149
B-DNA
G
C
Z-DNA
B-DNA
B-DNA
(or
A-DNA)
G
C
3' 5' 3' 5'
5' 3' 5' 3'
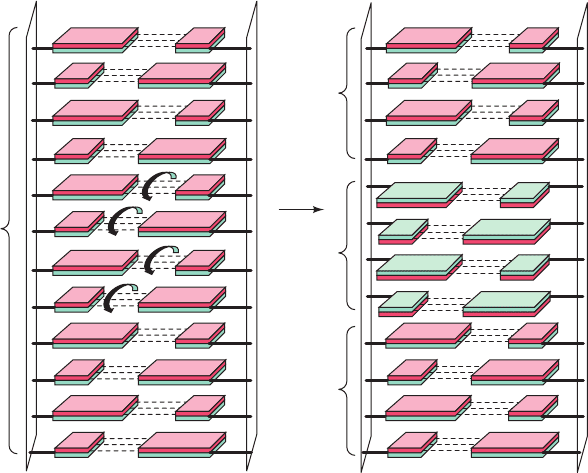
Figure 29-2 Conversion of B-DNA to Z-DNA. The conversion,
here represented by a 4-bp DNA segment, involves a 180°
flip of each base pair (curved arrows) relative to the sugar–
phosphate chains (compare the base pair orientations of B- and
Z-DNAs in Fig. 29-1b, d). Here, the different faces of the base pairs
are colored red and green. Note that if the drawing on the left is
taken as looking into the minor groove of unwound A- or B-DNA,
then in the drawing on the right, we are looking into the major
groove of the unwound Z-DNA segment. [After Rich, A., Nord-
heim,A., and Wang,A.H.-J., Annu. Rev. Biochem. 53, 799 (1984).]
JWCL281_c29_1143-1172.qxd 8/2/10 8:06 PM Page 1149
d(AC) ⴢ poly d(GT), take up the Z-DNA conformation at
high salt concentrations. Evidently, the Z-DNA conforma-
tion is most readily assumed by DNA segments with alter-
nating purine–pyrimidine base sequences (for structural
reasons explained in Section 29-2A). A high salt concentra-
tion stabilizes Z-DNA relative to B-DNA by reducing the
otherwise increased electrostatic repulsions between clos-
est approaching phosphate groups on opposite strands
(8 Å in Z-DNA vs 12 Å in B-DNA). The methylation of
cytosine residues at C5, a common biological modification
(Section 30-7), also promotes Z-DNA formation since a
hydrophobic methyl group in this position is less exposed
to solvent in Z-DNA than it is in B-DNA.
Does Z-DNA have any biological function? Rich has
proposed that the reversible conversion of specific seg-
ments of B-DNA to Z-DNA under appropriate circum-
stances acts as a kind of switch in regulating genetic expres-
sion, and there are indications that it transiently forms
behind actively transcribing RNA polymerase (Section 31-
4As). It was nevertheless surprisingly difficult to prove the
in vivo existence of Z-DNA.A major difficulty was demon-
strating that a particular probe for detecting Z-DNA, for
example, a Z-DNA-specific antibody, does not in itself
cause what would otherwise be B-DNA to assume the Z
conformation—a kind of biological uncertainty principle
(the act of measurement inevitably disturbs the system be-
ing measured). However, Rich has discovered several pro-
teins that specifically bind Z-DNA, including a family of Z-
DNA-binding protein domains named Z. The existence
of these proteins strongly suggests that Z-DNA does, in
fact, exist in vivo.
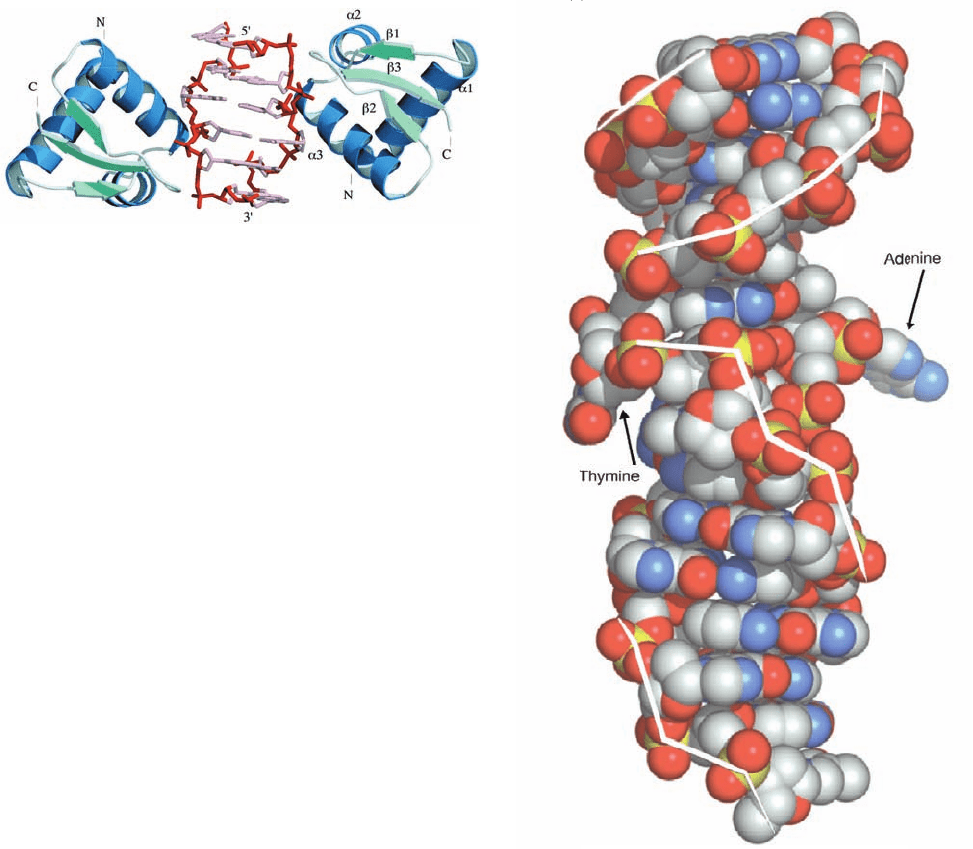
The X-ray structure of the 81-residue Z domain from
the RNA editing enzyme ADAR1 (Section 31-4As) in com-
plex with d(TCGCGCG) has been determined (Fig. 29-3a).
The CGCGCG segment of this heptanucleotide is self-com-
plementary, and therefore forms a 2-fold symmetric, 6-bp
segment of Z-DNA with an overhanging dT at the 5¿ end
of each strand (although these dT’s are disordered in the
1150 Chapter 29. Nucleic Acid Structures
(a)
(
b
)
Figure 29-3 Structures of DNA in complex with ADAR1 Z␣.
(a) X-ray structure of the complex of two Z domains with
Z-DNA consisting of a duplex of the self-complementary
hexamer d(CGCGCG) as viewed along its 2-fold axis of
symmetry. The Z-DNA is shown in stick form with its backbones
red and its remaining portions pink.The Z domains are drawn
in ribbon form with helices blue and sheets light green. Note that
each Z domain contacts only one strand of the Z-DNA.
(b) X-ray structure of a junction between Z-DNA and B-DNA.
The complex consists of d(GTCGCGCGCCATAAACC) and
d(ACGGTTTATGGCGCGCG) in complex with four Z
domains (not shown here for the sake of clarity).The DNA
forms a duplex with a 2-nucleotide overhang at each end in
which the lower 8 nucleotide pairs form Z-DNA, the upper 6
nucleotide pairs form B-DNA, and the A and T nucleotides at
the interface between these two DNA segments have been
extruded from the double helix.The DNA is drawn in space-
filling form with C gray, N blue, O red, and P yellow. The white
lines connect adjacent P atoms in the same polynucleotide,
thereby showing the zigzag pattern of the left-handed Z-DNA
and the smoother pattern of the right-handed B-DNA. [Courtesy
of Alexander Rich, MIT. PDBids 1QBJ and 2ACJ.]
JWCL281_c29_1143-1172.qxd 8/2/10 8:06 PM Page 1150
