Voet D., Voet Ju.G. Biochemistry
Подождите немного. Документ загружается.

pathways among these various intermediates (including ade-
nine), but all of these pathways lead to uric acid. Of course,
the intermediates in these processes may instead be reused to
form nucleotides via salvage reactions. In addition, ribose-1-
phosphate, a product of the reaction catalyzed by purine
nucleoside phosphorylase (PNP), is isomerized by phospho-
ribomutase to the PRPP precursor ribose-5-phosphate.
Adenosine and deoxyadenosine are not degraded by
mammalian PNP. Rather, adenine nucleosides and nu-
cleotides are deaminated by adenosine deaminase (ADA)
and AMP deaminase to their corresponding inosine deriv-
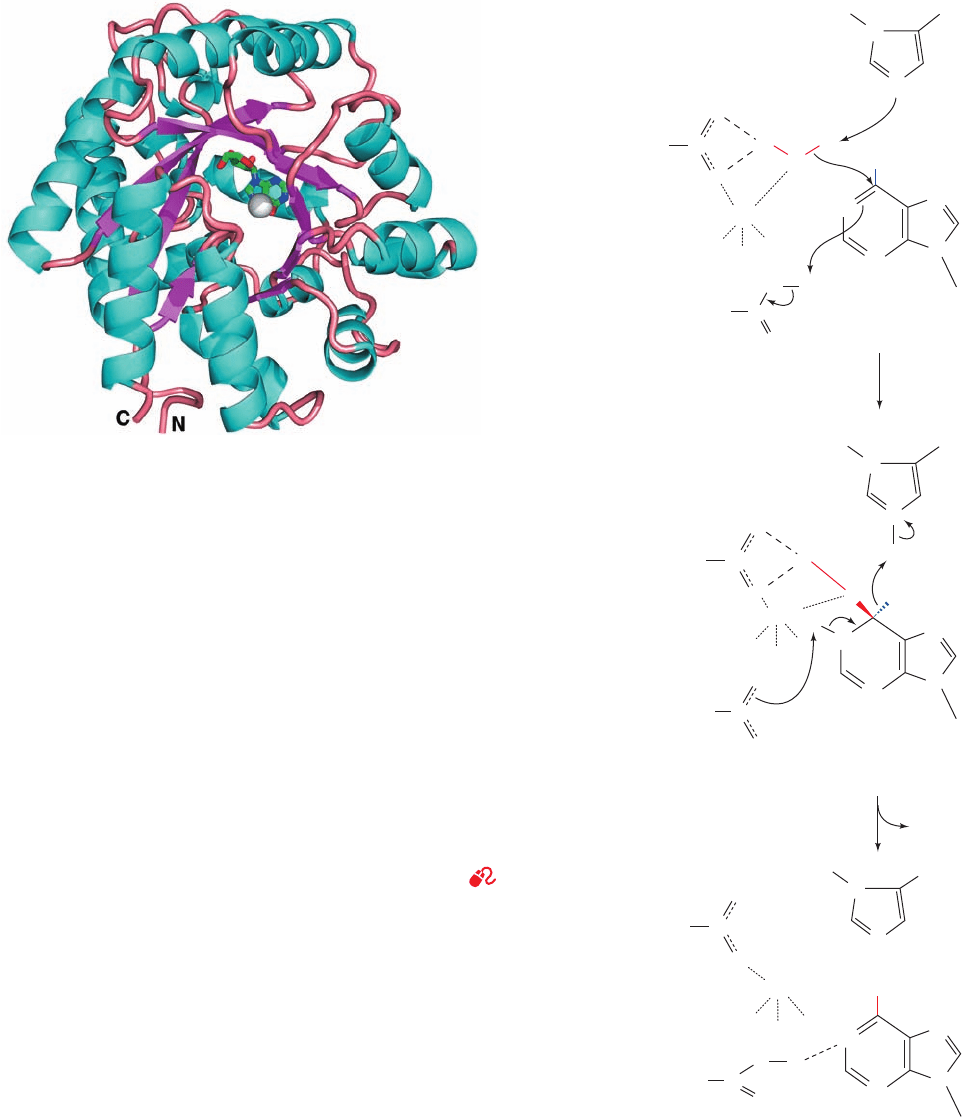
atives, which, in turn, may be further degraded. The X-ray
structure of murine ADA that was crystallized in the pres-
ence of its inhibitor purine ribonucleoside was determined
by Florante Quiocho (Fig. 28-24a). The enzyme forms an
eight-stranded ␣/ barrel with its active site in a pocket at the
C-terminal end of the  barrel, as occurs in nearly all known
␣/ barrel enzymes (Section 8-3Bh). Purine ribonucleoside
Section 28-4. Nucleotide Degradation 1131
Figure 28-24 X-ray structure and mechanism of adenosine deaminase. (a) A
ribbon diagram of murine adenosine deaminase in complex with its transition
state analog 6-hydroxy-1,6-dihydropurine ribonucleoside (HDPR).The
polypeptide is drawn in ribbon form colored according to its secondary structure
(helices cyan,  strands magenta, and loops salmon) and viewed approximately
down the axis of the enzyme’s ␣/ barrel from the N-terminal ends of its 
strands.The HDPR is shown in stick form with its C, N, and O atoms green, blue,
and red.The enzyme-bound Zn
2⫹
ion, which is coordinated by HDPR’s
6-hydroxyl group, is represented by a silver sphere. [Based on an X-ray structure
by Florante Quiocho, Baylor College of Medicine. PDBid 1ADA.] (b) The
proposed catalytic mechanism of adenosine deaminase. A Zn
2⫹
-polarized H
2
O
molecule (Section 15-1Cb) nucleophilically attacks C6 of the enzyme-bound
adenosine molecule in a process that is facilitated by His 238 acting as a general
base, Glu 217 acting as a general acid, and Asp 295 acting to orient the water
molecule via hydrogen bonding.The resulting tetrahedral intermediate
decomposes by the elimination of ammonia in a reaction that is aided by the now
imidazolium and carboxyl side chains of His 238 and Glu 217 acting as a general
acid and a general base, respectively. This yields inosine in its enol tautomeric
form, which, on its release from the enzyme, largely assumes its dominant keto
form.The Zn
2⫹
is coordinated by three His side chains that are not shown. [After
Wilson, D.K. and Quiocho, F.A., Biochemistry 32, 1692 (1993).]
See
Interactive Exercise 30
.
.
Inosine (enol tautomer)
Glu 217
Asp 295
C
O
O
O
O
C
–
C
–
O
O
H
N
N
OH
N
N
H His 238
Ribose
Glu 217
C
–
O
O
O
Asp 295
C
–
A
sp 295
C
O
OH
H
H
His 238
H
N
O
O
O
N
N
N
N
Glu 217
NH
3
NH
2
NH
2
N
N
Ribose
Zn
2+
Zn
2+
Zn
2+
Tetrahedral intermediate
N
H
H
N
N
N
N
N
H His 238
H
+
Ribose
Adenosine
.
.
N
(a) (b)
JWCL281_c28_1107-1142.qxd 10/19/10 9:59 AM Page 1131

binds to ADA in a normally rare hydrated form, 6-hydroxy-1,
6-dihydropurine ribonucleoside (HDPR),
a nearly ideal transition state analog of the ADA reaction.
Although it had been previously reported that ADA does
not require a cofactor, its X-ray structure clearly reveals
that a zinc ion is bound in the deepest part of the active site
pocket, where it is pentacoordinated by three His side
chains, a carboxyl oxygen of Asp 295, and the O6 atom of
HDPR. ADA’s active site complex suggests a catalytic
mechanism (Fig. 28-24b) reminiscent of that of carbonic
anhydrase (Section 15-1Cb): His 238, which is properly
positioned to act as a general base, abstracts a proton from
a bound Zn
2
-activated water molecule, which nucleophili-
cally attacks the adenine C6 atom to form a tetrahedral
intermediate. Products are then formed by the elimination
of ammonia.
a. Genetic Defects in ADA Result in Severe
Combined Immunodeficiency Disease
Abnormalities in purine nucleoside metabolism arising
from rare genetic defects in ADA selectively kill lympho-
cytes (a type of white blood cell). Since lymphocytes medi-
ate much of the immune response (Section 35-2A), ADA
deficiency results in severe combined immunodeficiency
disease (SCID) that, without special protective measures, is
invariably fatal in infancy due to overwhelming infection.
The mutations in all eight known ADA variants obtained
from SCID patients appear to structurally perturb the
active site of ADA.
Biochemical considerations provide a plausible expla-
nation of SCID’s etiology (causes). In the absence of active
ADA, deoxyadenosine is phosphorylated to yield levels of
dATP that are 50-fold greater than normal. This high con-
centration of dATP inhibits ribonucleotide reductase (Sec-
tion 28-3Ad), thereby preventing the synthesis of the other
dNTPs, choking off DNA synthesis and thus cell prolifera-
tion. The tissue-specific effect of ADA deficiency on the
immune system may be explained by the observation that
lymphoid tissue is particularly active in deoxyadenosine
phosphorylation.
SCID caused by ADA defects does not respond to treat-
ment by the intravenous injection of ADA because the
liver clears this enzyme from the bloodstream within min-
utes. If, however, several molecules of the biologically inert
polymer polyethylene glycol (PEG)
Polyethylene glycol
HO [¬CH
2
¬CH
2
¬O¬]
n
H
H
N
N
N
N
Ribose
H
N
H
HO
N
N
N
Ribose
6-Hydroxy-1,6-dihydropurine
ribonucleoside (HDPR)
Purine ribonucleoside
are covalently linked to surface groups on ADA,the resulting
PEG–ADA remains in the blood for 1 to 2 weeks, thereby
largely resuscitating the SCID victim’s immune system. The
protein-linked PEG only reduces the catalytic activity of
ADA by ⬃40% but, evidently, masks it from the receptors
that filter it out of the blood. SCID can therefore be treated
effectively by PEG–ADA.This treatment, however,is expen-
sive and not entirely satisfactory. Consequently, ADA defi-
ciency was selected as one of the first genetic diseases to be
treated by gene therapy (Section 5-5Hb): Lymphocytes were
extracted from the blood of an ADA-deficient child and
grown in the laboratory, had a normal ADA gene inserted
into them via genetic engineering techniques (Section 5-5),
and were then returned to the child.After 12 years, 20 to 25%
of the patient’s lymphocytes contained the introduced ADA
gene. However, ethical considerations have mandated that
the patient continue receiving injections of PEG–ADA so
that the efficacy of this gene therapy protocol is unclear.
b. The Purine Nucleotide Cycle
The deamination of AMP to IMP, when combined with
the synthesis of AMP from IMP (Fig. 28-4,left), has the effect
of deaminating aspartate to yield fumarate (Fig. 28-25). John
Lowenstein demonstrated that this purine nucleotide cycle
has an important metabolic role in skeletal muscle. An in-
crease in muscle activity requires an increase in the activity
of the citric acid cycle. This process usually occurs through
the generation of additional citric acid cycle intermediates
(Section 21-4). Muscles, however, lack most of the enzymes
that catalyze these anaplerotic (filling up) reactions in other
tissues. Rather, muscle replenishes its citric acid cycle inter-
mediates as fumarate generated in the purine nucleotide cy-
cle.The importance of the purine nucleotide cycle in muscle
metabolism is indicated by the observation that the activities
of the three enzymes involved are all severalfold higher in
muscle than in other tissues. In fact, individuals with an in-
herited deficiency in muscle AMP deaminase (myoadeny-
late deaminase deficiency) are easily fatigued and usually
suffer from cramps after exercise.
c. Xanthine Oxidase Is a Mini-Electron-
Transport Protein
Xanthine oxidase (XO) converts hypoxanthine to xan-
thine, and xanthine to uric acid (Fig. 28-23, bottom). In
mammals, this enzyme occurs mainly in the liver and the
small intestinal mucosa. XO is a homodimer of ⬃1330-
residue subunits, each of which binds a variety of electron-
transfer agents: an FAD, two spectroscopically distinct
[2Fe–2S] clusters, and a molybdopterin complex (Mo-pt)
OH
2
CH
2
OPO
3
2
–
H
2
N
O
O
S
Molybdopterin complex (Mo-pt)
Mo
S
S
H
N
H
O
H
N
H
....
N
HN
1132 Chapter 28. Nucleotide Metabolism
JWCL281_c28_1107-1142.qxd 6/8/10 10:39 AM Page 1132
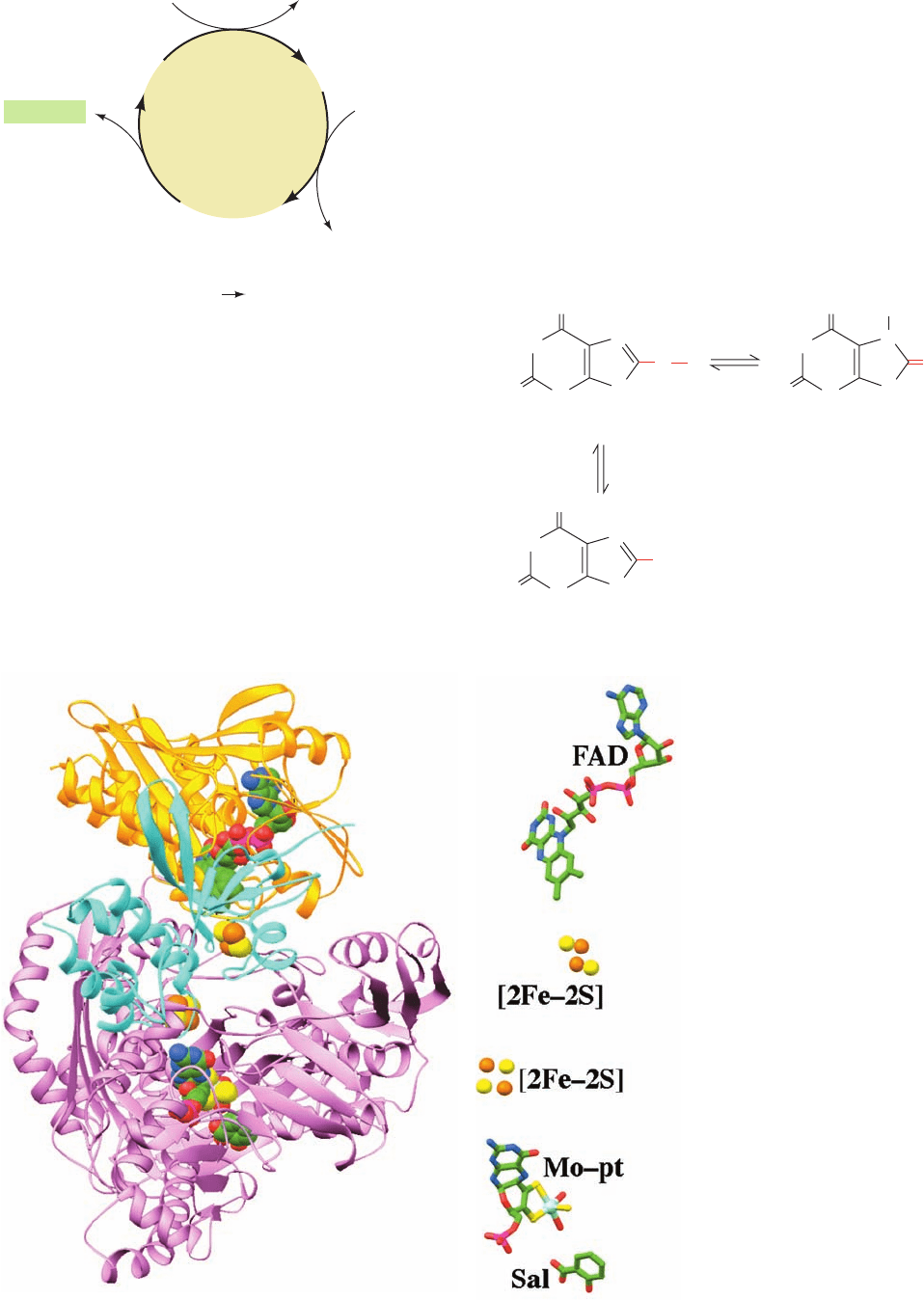
in which the Mo atom cycles between its Mo(VI) and
Mo(IV) oxidation states. The final electron acceptor is O
2
,
which is converted to H
2
O
2
, a potentially harmful oxidizing
agent that is subsequently disproportionated to H
2
O and
O
2
by catalase (Section 1-2Ad). In XO, the polypeptide has
been proteolytically cleaved into three segments (the un-
cleaved enzyme, which is known as xanthine dehydroge-
nase, preferably uses NAD
as its electron acceptor,
whereas XO does not react with NAD
).
The X-ray structure of XO from cow’s milk in complex
with the competitive inhibitor salicylic acid (Fig. 25-74),
determined by Emil Pai, reveals that the FAD and the molyb-
dopterin complex are interposed by the two [2Fe–2S] clusters
to form a mini-electron-transport chain (Fig. 28-26). Each of
its three peptide segments forms a separate domain with the
N-terminal domain binding the two [2Fe–2S] clusters, the cen-
tral domain binding the FAD, and the C-terminal domain
binding the Mo-pt complex. Although the salicylic acid does
not contact the Mo-pt complex, it binds to XO in a way that
blocks the approach of substrates to the metal center.
XO hydroxylates xanthine at its C8 position (and hy-
poxanthine at its C2 position), yielding uric acid in its enol
form that tautomerizes to the more stable keto form:
Uric acid (enol tautomer) Uric acid (keto tautomer)
Urate
pK = 5.4
HN
O
N
H
N
H
N
H
N
H
N
H
N
H
HNHN
N
O
O
_
H
+
+
OO
O
OH
O
O
H
NN
6
5
4
3
2
1
7
8
9
Section 28-4. Nucleotide Degradation 1133
Figure 28-25 The purine nucleotide cycle. This pathway
functions, in muscle, to prime the citric acid cycle by generating
fumarate.
Figure 28-26 X-ray structure of
xanthine oxidase from cow’s milk in
complex with salicylic acid.
(a) Ribbon diagram of its
1332-residue subunit in which the
N-terminal domain (residues 2–165)
is cyan, the central domain (residues
224–528) is gold, and the C-terminal
domain (residues 571–1315) is
lavender.The enzyme’s redox
cofactors and bound salicylic acid
are shown in space-filling form with
C green, N blue, O red, S yellow, P
magenta, Fe orange, and Mo light
blue. The ⬃50-residue peptide
segments spanning domains are
disordered and are apparently
highly flexible. (b) The enzyme’s
redox cofactors and salicylic acid
(Sal) drawn in stick form with their
S, Fe, and Mo atoms represented by
spheres.The atoms are colored as in
Part a and viewed from the same
direction but with greater
magnification. [Based on an X-ray
structure by Emil Pai, University of
Toronto,Toronto, Ontario, Canada.
PDBid 1FIQ.]
AMP
GTP
Aspartate
GDP P
i
Adenylosuccinate
4
NH
+
adenylosuccinate
lyase
adenylosuccinate
synthetase
AMP
deaminase
IMP
Fumarate
4
NH
+
Net:
H
2
O + Aspartate + GTP + GDP P
i
++fumarate
+
+
H
2
O
(a) (b)
JWCL281_c28_1107-1142.qxd 4/22/10 9:17 AM Page 1133
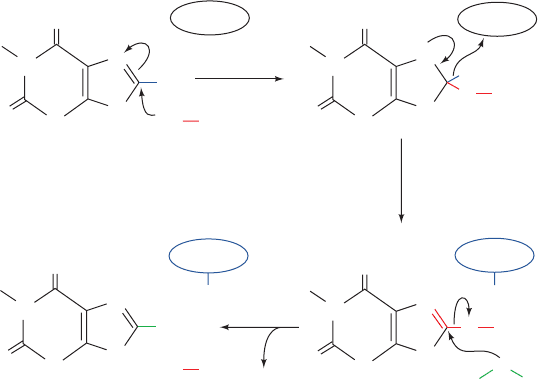
(its enol form ionizes with a pK of 5.4; hence, the name uric
acid).
18
O-labeling experiments have demonstrated that
the C8 keto oxygen of uric acid is derived from H
2
O,
whereas the oxygen atoms of H
2
O
2
come from O
2
. Chemi-
cal and spectroscopic studies suggest that the enzyme has
the following mechanism (Fig. 28-27):
1. The reaction is initiated by the attack of an enzyme
nucleophile, X, on the C8 position of xanthine.
2. The C8¬H atom is eliminated as a hydride ion that
combines with the Mo(VI) complex, thereby reducing it to
the Mo(IV) state.
3. Water displaces the enzyme nucleophile producing
uric acid.
In the second stage of the reaction, the now reduced en-
zyme is reoxidized to its original Mo(VI) state by reaction
with O
2
. This complex process, not surprisingly, is but
poorly understood. EPR measurements indicate that elec-
trons are funneled from the Mo(IV) through the two
[2Fe–2S] clusters to the flavin and ultimately to O
2
, yield-
ing H
2
O
2
and regenerated enzyme.
B. Fate of Uric Acid
In humans and other primates, the final product of purine
degradation is uric acid, which is excreted in the urine. The
same is true of birds, terrestrial reptiles, and many insects,
but these organisms, which do not excrete urea, also catab-
olize their excess amino acid nitrogen to uric acid via
purine biosynthesis. This complicated system of nitrogen
excretion has a straightforward function: It conserves water.
Uric acid is only sparingly soluble in water, so that its ex-
cretion as a paste of uric acid crystals is accompanied by
very little water. In contrast, the excretion of an equivalent
amount of the much more water-soluble urea osmotically
sequesters a significant amount of water.
In all other organisms, uric acid is further processed be-
fore excretion (Fig. 28-28). Mammals other than primates
oxidize it to their excretory product, allantoin, in a reaction
catalyzed by the Cu-containing enzyme urate oxidase. A
further degradation product, allantoic acid, is excreted by
teleost (bony) fish. Cartilaginous fish and amphibia further
degrade allantoic acid to urea prior to excretion. Finally,
marine invertebrates decompose urea to their nitrogen
excretory product, NH
4
.
a. Gout Is Caused by an Excess of Uric Acid
Gout is a disease characterized by elevated levels of uric
acid in body fluids. Its most common manifestation is ex-
crutiatingly painful arthritic joint inflammation of sudden
onset, most often in the big toe (Fig. 28-29), caused by
deposition of nearly insoluble crystals of sodium urate.
Sodium urate and/or uric acid may also precipitate in the
kidneys and ureters as stones, resulting in renal damage
and urinary tract obstruction. Gout, which affects ⬃3 per
1000 persons, predominantly males, has been tradition-
ally, although inaccurately, associated with overindulgent
eating and drinking. The probable origin of this associa-
tion is that in previous centuries, when wine was often
contaminated with lead during its manufacture and stor-
age, heavy drinking resulted in chronic lead poisoning,
which, among other things, decreases the kidney’s ability
to excrete uric acid.
1134 Chapter 28. Nucleotide Metabolism
Figure 28-27 Mechanism of xanthine oxidase. The reduced enzyme is subsequently reoxidized
by O
2
, yielding H
2
O
2
.
Uric acid
(enol tautomer)
Enzyme
nucleophile
Xanthine
Reduced
enzyme
Reduced
enzyme
Enzyme–Mo
complex
(fully oxidized form)
O
H
N
O
N
H
N
H
N
H
_
X
_
X
X
X
E
1
8
Mo(VI)
Mo(VI)
Mo(IV)
Mo(IV)
2
3
O
N
H
N
H
N
H
OH
N
H
O
E
H
+
H
N
O
O
N
H
N
H
N
E
H
HH
O
H
N
O
N
H
O
N
N
H
H
E
••
_
JWCL281_c28_1107-1142.qxd 4/22/10 9:17 AM Page 1134
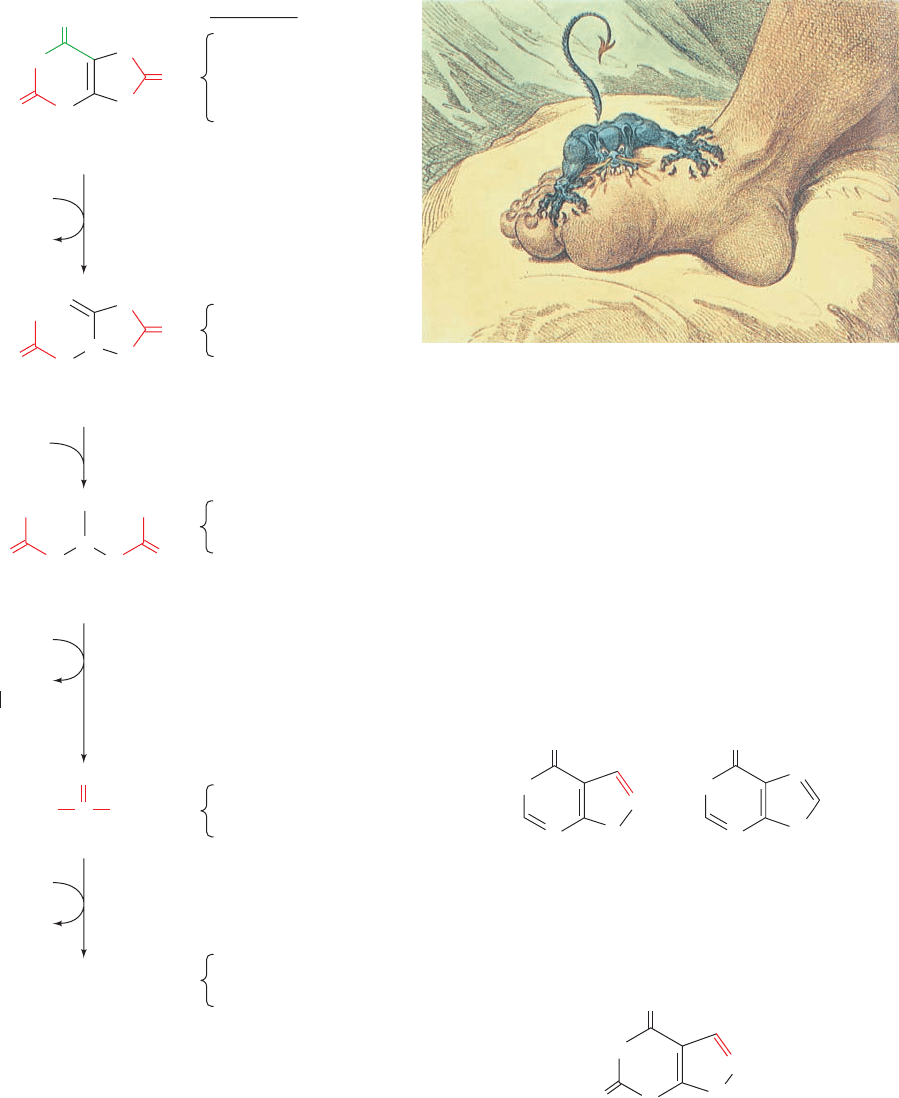
The most prevalent cause of gout is impaired uric acid
excretion (although usually for other reasons than lead
poisoning). Gout may also result from a number of meta-
bolic insufficiencies, most of which are not well character-
ized. One well-understood cause is HGPRT deficiency
(Lesch–Nyhan syndrome in severe cases), which leads to
excessive uric acid production through PRPP accumula-
tion (Section 28-1D). Uric acid overproduction is also
caused by glucose-6-phosphatase deficiency (von Gierke’s
glycogen storage disease; Section 18-4): The increased
availability of glucose-6-phosphate stimulates the pentose
phosphate pathway (Section 23-4), increasing the rate of
ribose-5-phosphate production and consequently that of
PRPP, which in turn stimulates purine biosynthesis.
Gout may be treated by administration of the xanthine
oxidase inhibitor allopurinol, a hypoxanthine analog with
interchanged N7 and C8 positions.
Xanthine oxidase hydroxylates allopurinol, as it does hy-
poxanthine, yielding alloxanthine,
which remains tightly bound to the reduced form of the en-
zyme, thereby inactivating it. Allopurinol consequently al-
leviates the symptoms of gout by decreasing the rate of uric
acid production while increasing the levels of the more sol-
uble hypoxanthine and xanthine. Although allopurinol
controls the gouty symptoms of Lesch–Nyhan syndrome, it
has no effect on its neurological symptoms.
HN
N
O
O
N
N
H
H
Alloxanthine
HN
Allopurinol Hypoxanthine
O
N
7
8
N
H
HN
N
O
N
N
H
N
Section 28-4. Nucleotide Degradation 1135
Figure 28-28 Degradation of uric acid to ammonia. The
process is arrested at different stages in the indicated species and
the resulting nitrogen-containing product is excreted.
Figure 28-29 The Gout, a cartoon by James Gillray (1799).
[Yale University Medical Historical Library.]
HN
O
O
O
N
H
N
H
H
N
Primates
Birds
Reptiles
Insects
Uric acid
2 H
2
O + O
2
CO
2
+ H
2
O
2
urate oxidase
H
2
O
allantoinase
Urea
H
2
O
COOH
CHO
Glyoxylic acid
allantoicase
O
O
O
N
H
N
H
H
N
2H
2
O
2CO
2
urease
Other mammals
Allantoin
Excreted by
C
H
OO
N
H
N
H
NH
2
NH
2
4 NH
4
H
2
N
H
2
NC
O
Teleost fish
Allantoic acid
Cartilaginous fish
Amphibia
Marine
invertebrates
C
H
COOH
2
+
H
2
N
JWCL281_c28_1107-1142.qxd 4/23/10 10:38 AM Page 1135
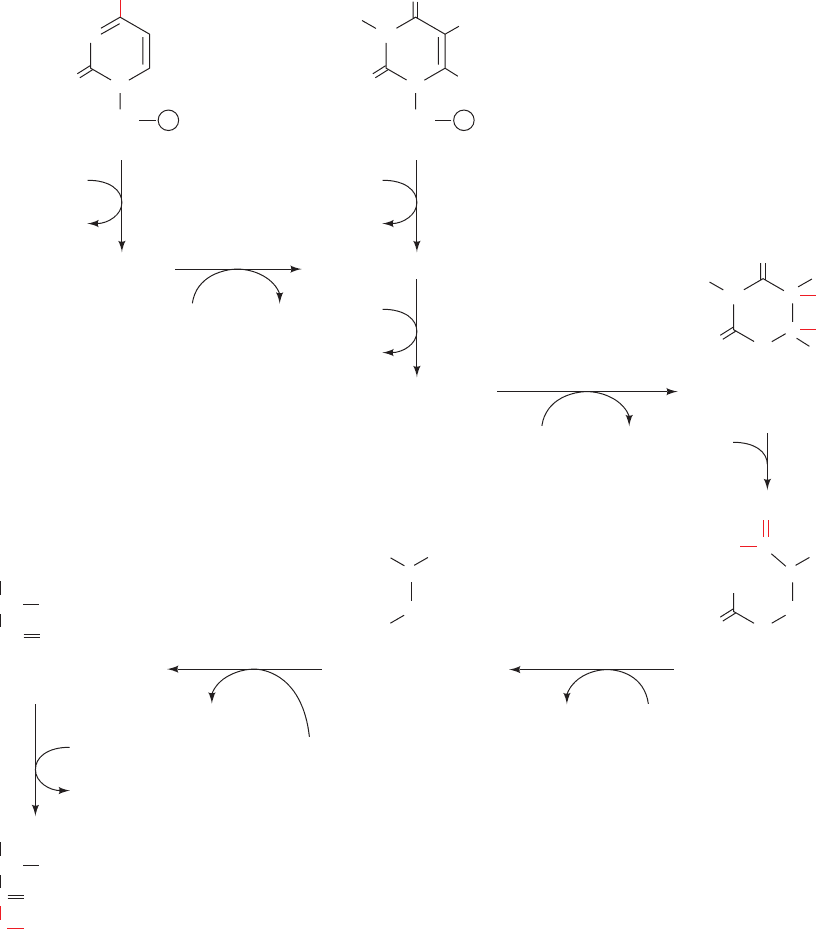
C. Catabolism of Pyrimidines
Animal cells degrade pyrimidine nucleotides to their compo-
nent bases (Fig. 28-30, top). These reactions, like those of
purine nucleotides, occur through dephosphorylation, deam-
ination, and glycosidic bond cleavages. The resulting uracil
and thymine are then broken down in the liver through
reduction (Fig. 28-30, middle) rather than by oxidation, as
occurs in purine catabolism. The end products of pyrimidine
catabolism, -alanine and -aminoisobutyrate, are amino
acids and are metabolized as such. They are converted,
through transamination and activation reactions, to malonyl-
CoA and methylmalonyl-CoA (Fig. 28-30, bottom left) for
further utilization (Sections 25-4A and 25-2Ea).
5 BIOSYNTHESIS OF NUCLEOTIDE
COENZYMES
In this section we outline the assembly, in animals, of the
nucleotide coenzymes NAD
and NADP
, FMN and
FAD, and coenzyme A, from their vitamin precursors.
These vitamins are synthesized de novo only by plants and
microorganisms.
A. Nicotinamide Coenzymes
The nicotinamide moiety of the nicotinamide coenzymes
(NAD
and NADP
) is derived, in humans, from dietary
nicotinamide, nicotinic acid,or the essential amino acid tryp-
tophan (Fig. 28-31). Nicotinate phosphoribosyltransferase,
1136 Chapter 28. Nucleotide Metabolism
Figure 28-30 Major pathways of pyrimidine catabolism in ani-
mals. The amino acid products of these reactions are taken up in
other metabolic processes. UMP and dTMP are degraded by the
same enzymes; the pathway for dTMP degradation is given in
parentheses.
CH
CH
CH
2
H
2
O
P
i
nucleotidase
H
2
O
P
i
nucleotidase
Cytidine Uridine (Deoxythymidine)
H
2
O
NH
4
+
cytidine
deaminase
P
i
uridine
phosphorylase
(d)Ribose-1-P
N
N
O
Rib P
CMP
H
N
O
N
O
H
(d)Rib
P
UMP (dTMP)
CH
3
NH
2
Uracil (Thymine)
dihydrouracil
dehydrogenase
NADP + H
+
NADP
+
()
H
N
O
N
H
O
H
Dihydrouracil
(Dihydrothymine)
CH
3
()
C
C
H
H
H
2
O
hydropyrimidine
hydratase
N
O
N
H
O
-Ureidopropionate
( -Ureidoisobutyrate)
CH
3
()
H
CO
–
β
β
H
2
ONH
4
+
+
CO
2
-ureidopropionaseβ
CH
CH
2
CH
3
()
OOC
-Alanine
( -Aminoisobutyrate)
–
β
β
Glutamate
-Ketoglutarate
α
Malonic semialdehyde
(Methylmalonic semialdehyde)
CH
O
CH
3
()
COO
–
NADH + H
+
NAD
+
+CoA
C
Malonyl-CoA
(Methylmalonyl-CoA)
CH
O
CH
3
()
COO
–
S
CoA
aminotransferase
H
2
NH
2
JWCL281_c28_1107-1142.qxd 4/22/10 9:17 AM Page 1136
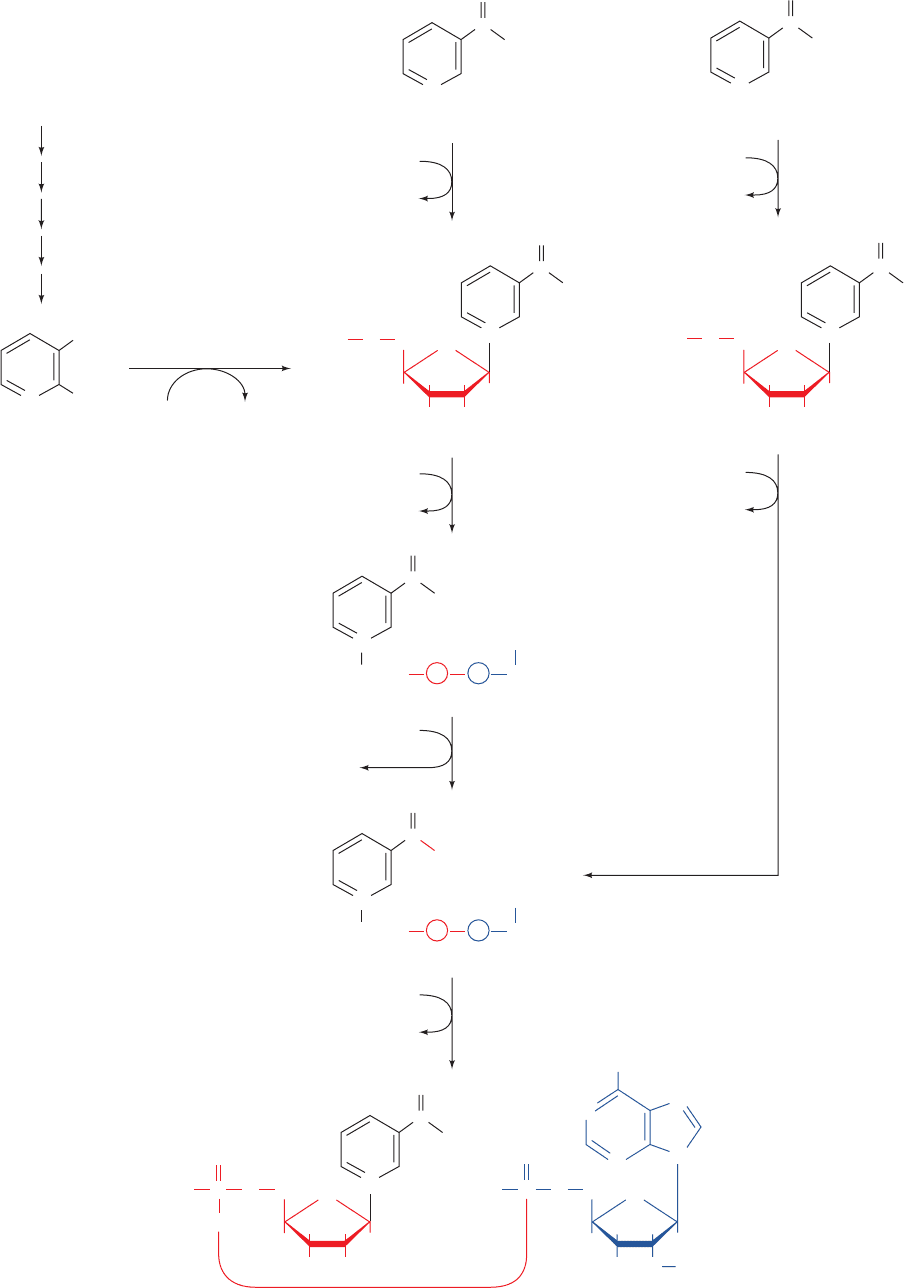
Section 28-5. Biosynthesis of Nucleotide Coenzymes 1137
Figure 28-31 Pathways for the biosynthesis of NAD
and NADP
. These nicotinamide coenzymes are
synthesized from their vitamin precursors,
nicotinate and nicotinamide, and from the
tryptophan degradation product, quinolinate.
Nicotinamide adenine dinucleotide phosphate (NADP
+
)
Nicotinamide adenine dinucleotide (NAD
+
)
Nicotinate adenine dinucleotide
Nicotinate mononucleotide
Nicotinamide mononucleotide (NMN)
Tryptophan
Quinolinate
Nicotinate
Nicotinamide
PRPP
nicotinate
phosphoribosyl
transferase
PP
i
ATP
pyrophosphorylase
PP
i
PRPP
nicotinamide
phosphoribosyl
transferase
PP
i
N
H
O
_
O
C
+
N
O
_
O
C
+
N
Ribose Ribose
Adenine
+
++
N
H
NH
2
NH
2
CH
2
CO
2
PP
i
PRPP
_
2
O
3
P
H
2
O
Glutamine
COO
_
COO
_
O
_
O
C
+
N
H
+
N
+
O
C
O
H
H
HH
OHOH
O
CH
2
_
2
O
3
P
O
HH
H
H
OHOH
O
quinolinate
phosphoribosyl
transferase
+
C
O
NAD
+ ATP
pyrophosphorylase
PP
i
NAD
+
ATP
NAD
+
synthetase
NAD
+
kinase
_
O
+
Glutamate
ATP
ATP
ADP
P P
N
Ribose Ribose
Adenine
+
NH
2
C
O
N
+
CH
2
CH
2
NH
2
PO
3
2
_
HH
HH
O
N
N
N
N
OH O
NH
2
OP
_
O
OP
O
O
O
C
O
HH
OHOH
HH
O
P P
JWCL281_c28_1107-1142.qxd 4/22/10 9:17 AM Page 1137
which occurs in most mammalian tissues, catalyzes the
formation of nicotinate mononucleotide from nicotinate
and PRPP. This intermediate may also be synthesized
from quinolinate, a degradation product of tryptophan
(Section 26-3G), in a reaction mediated by quinolinate
phosphoribosyltransferase, which occurs mainly in liver
and kidney. A poor diet, nevertheless, may result in pella-
gra (nicotinic acid deficiency; Section 13-3), since, under
such conditions, tryptophan will be almost entirely uti-
lized in protein biosynthesis. Nicotinate mononucleotide
is joined via a pyrophosphate linkage to an ATP-derived
AMP residue by NAD
⫹
pyrophosphorylase to yield
nicotinate adenine dinucleotide (desamido NAD
ⴙ
). Finally,
NAD
ⴙ
synthetase converts this intermediate to NAD
⫹
by
a transamidation reaction in which glutamine is the NH
2
donor.
NAD
⫹
may also be synthesized from nicotinamide. This
vitamin is converted to nicotinamide mononucleotide
(NMN) by nicotinamide phosphoribosyltransferase, a
widely occurring enzyme distinct from nicotinate phospho-
ribosyltransferase. However, NAD
⫹
is synthesized from
NMN and ATP by NAD pyrophosphorylase, the same
enzyme that synthesizes nicotinate adenine dinucleotide.
NADP
⫹
is formed via the ATP-dependent phosphoryla-
tion of the NAD
⫹
adenosine residue’s C2¿ OH group by
NAD
ⴙ
kinase.
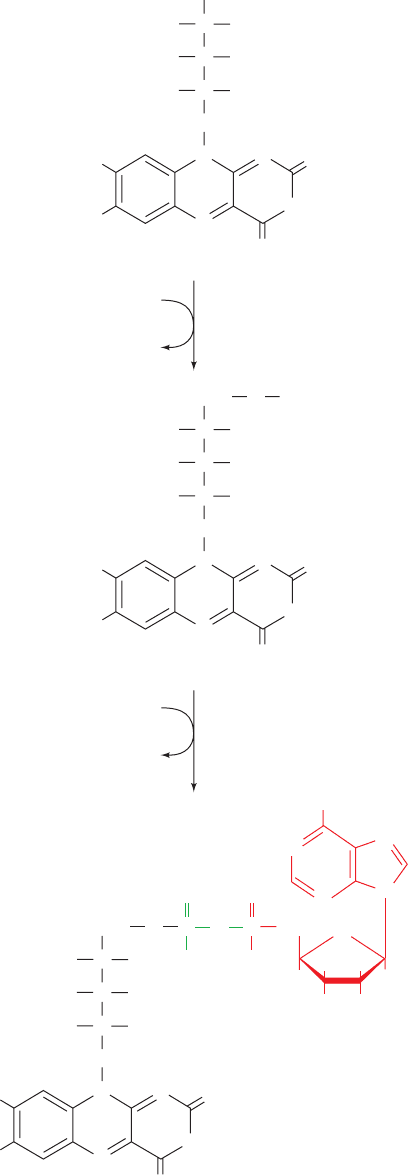
B. Flavin Coenzymes
FAD is synthesized from riboflavin in a two-reaction path-
way (Fig. 28-32). First, the 5¿-OH group of riboflavin’s
ribityl side chain is phosphorylated by flavokinase, yielding
flavin mononucleotide (FMN; not a true nucleotide since
its ribityl residue is not a true sugar). FAD may then be
formed by the coupling of FMN and ATP-derived AMP in
a pyrophosphate linkage in a reaction catalyzed by FAD
pyrophosphorylase. Both of these enzymes are widely
distributed in nature.
C. Coenzyme A
Coenzyme A is synthesized in mammalian cells according
to the pathway diagrammed in Fig. 28-33. Pantothenate,
an essential vitamin, is phosphorylated by pantothenate
kinase and then coupled to cysteine, the future business
end of CoA, by phosphopantothenoylcysteine synthetase.
After decarboxylation by phosphopantothenoylcysteine
decarboxylase, the resulting 4ⴕ-phosphopantethiene is
coupled to AMP in a pyrophosphate linkage by dephospho-
CoA pyrophosphorylase and then phosphorylated at its
adenosine 3¿ OH group by dephospho-CoA kinase to
form CoA. The latter two enzymatic activities occur on a
single protein.
1138 Chapter 28. Nucleotide Metabolism
Figure 28-32 Biosynthesis of FMN and FAD from the vitamin
precursor riboflavin.
FAD
pyrophosphorylase
ATP
PP
i
O
_
O
_
OCH
2
CH
2
CH
2
H
3
C
H
3
C
OHH
H
H
C
C
C
OH
OH
NH
2
P
P
O
O
O
N
N
N
N
O
HH
OHOH
HH
O
N
O
O
NN
NH
CH
2
CH
2
H
3
C
H
3
C
OHH
H
H
C
C
C
OH
OH
O
N
O
O
NN
NH
Flavin adenine dinucleotide (FAD)
Flavin mononucleotide (FMN)
flavokinase
ATP
ADP
CH
2
OH
CH
2
H
3
C
H
3
C
OHH
H
H
C
C
C
OH
OH
N
O
O
NN
NH
Riboflavin
PO
3
2
_
JWCL281_c28_1107-1142.qxd 8/13/10 6:46 PM Page 1138
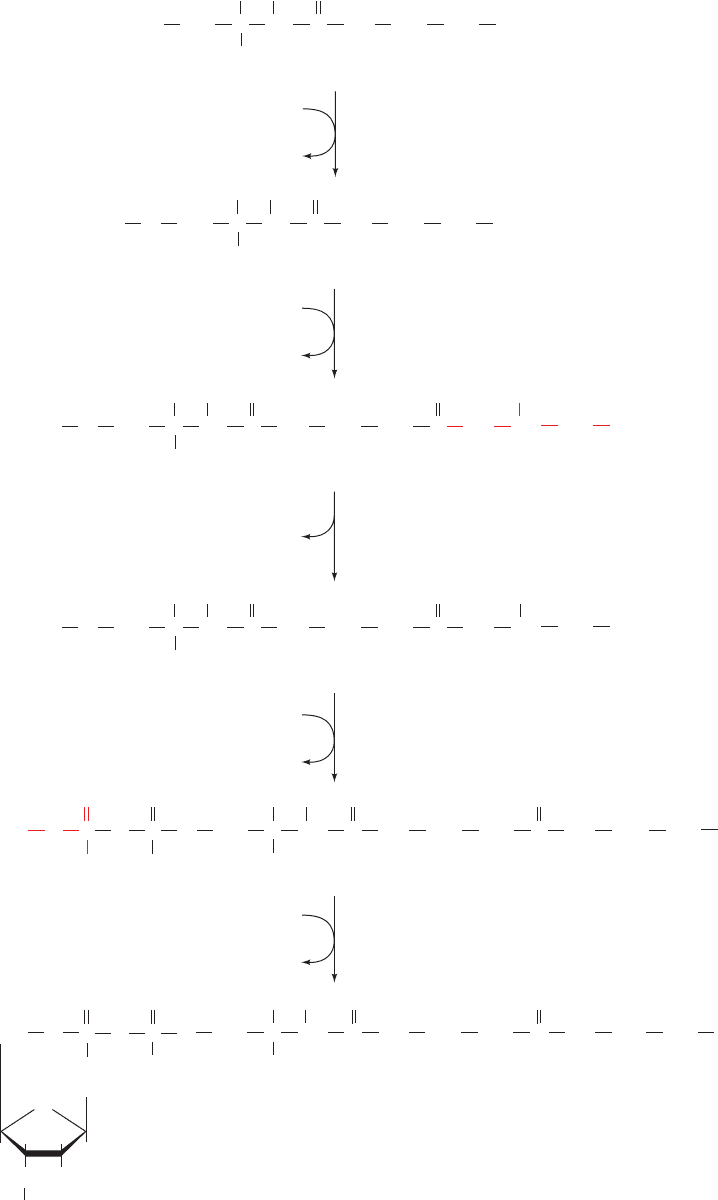
Section 28-5. Biosynthesis of Nucleotide Coenzymes 1139
Figure 28-33 Biosynthesis of coenzyme A from pantothenate, its vitamin precursor.
O
–
O
–
O
HH
OHO
H
Adenine
H
pantothenate kinase
ATP
ADP
H
3
C
H
3
C
CH
2
CH
2
CH C
O
C
HO
NH
CH
2
COO
–
Pantothenate
OH
phosphopantothenoylcysteine
synthetase
ATP Cysteine
P
i
ADP
H
3
C
H
3
C
CH
2
CH
2
CH C
O
C
O
–2
O
3
P
NH
CH
2
COO
–
4′-Phosphopantothenate
OH
phosphopantothenoylcysteine
decarboxylase
CO
2
H
3
C
H
3
C
CH
2
CH
2
CH C
O
C
O
–2
O
3
P
NH
CH
2
C
O
NH
CH
2
CH
SH
COO
–
4′-Phosphopantothenoylcysteine
OH
dephospho-CoA
pyrophosphorylase
PP
i
ATP
H
3
C
H
3
C
CH
2
CH
2
CH C
O
C
O
–2
O
3
P
NH
CH
2
C
O
NH
CH
2
CH
SH
H
4′-Phosphopantetheine
OH
dephospho-CoA
kinase
ADP
ATP
H
3
C
H
3
C
CH
2
CH
2
CH C
O
C
O
POPAdenosine O
NH
CH
2
C
O
OO
NH
CH
2
CH
2
SH
Dephosphocoenzyme A
OH
O
–
PO
3
2
–
O
–
H
3
C
H
3
C
CH
2
CH
2
C
CH
2
O
P
OP
O
OH
CH
NH
CH
2
NH
CH
2
SHCH
2
C
O
C
O
O
O
Coenzyme A (CoA)
JWCL281_c28_1107-1142.qxd 4/22/10 9:17 AM Page 1139

1140 Chapter 28. Nucleotide Metabolism
1 Synthesis of Purine Ribonucleotides Almost all cells
synthesize purine nucleotides de novo via similar metabolic
pathways.The purine ring is constructed in an 11-step reaction
sequence that yields IMP. AMP and GMP are then synthe-
sized from IMP in separate pathways. Nucleoside diphos-
phates and triphosphates are sequentially formed from these
products via phosphorylation reactions. The rates of synthesis
of these various nucleotides are interrelated through feedback
inhibition mechanisms that monitor their concentrations.
Purine nucleotides may also be synthesized from free purines
salvaged from nucleic acid degradation processes. The impor-
tance of these salvage reactions is demonstrated, for example,
by the devastating and bizarre consequences of Lesch–Nyhan
syndrome.
2 Synthesis of Pyrimidine Ribonucleotides Cells also
synthesize pyrimidines de novo but, in this six-step process, a
free base is formed before it is converted to a nucleotide,
UMP. UTP is then formed by phosphorylation of UMP, and
CTP is synthesized by the amination of UTP. Pyrimidine
biosynthesis is regulated by feedback inhibition as well as by
the concentrations of purine nucleotides.
3 Formation of Deoxyribonucleotides Deoxyribonu-
cleotides are formed by reduction of the corresponding ri-
bonucleotides. Three classes of ribonucleotide reductase
(RNR) have been characterized: Class I RNR, which occurs in
nearly all eukaryotes and many prokaryotes, contains an
Fe(III)¬O
2–
¬Fe(III) group and a tyrosyl free radical; Class II
and III RNRs, which occur only in prokaryotes, contain,
respectively, a coenzyme B
12
cofactor, and a [4Fe–4S] cluster
together with a glycyl radical.All of them catalyze free radical–
based reductions. The substrates for Class I and II RNRs are
NDPs, whereas those for Class III RNRs are NTPs. Class I
RNR has three independent regulatory sites that control its
substrate specificity and its catalytic activity in part via its
oligomerization state, thereby generating deoxynucleotides in
the amounts required for DNA synthesis. The E. coli Class I
RNR is reduced to its original state by electron-transport
chains involving either thioredoxin, thioredoxin reductase,
and NADPH; or glutaredoxin, glutathione, glutathione reduc-
tase, and NADPH. Thymine is synthesized by the methylation
of dUMP by thymidylate synthase to form dTMP. The reac-
tion’s methyl source, N
5
,N
10
-methylene-THF, is oxidized in the
reaction to yield dihydrofolate. N
5
,N
10
-Methylene-THF is sub-
sequently regenerated through the sequential actions of dihy-
drofolate reductase and serine hydroxymethyltransferase.
Since this sequence of reactions is required for DNA biosyn-
thesis, it presents an excellent target for chemotherapy.
FdUMP, a mechanism-based inhibitor of thymidylate syn-
thase, and methotrexate, an antifolate that essentially irre-
versibly inhibits dihydrofolate reductase, are both highly ef-
fective anticancer agents.
4 Nucleotide Degradation Purine nucleotides are catab-
olized to yield uric acid. Depending on the species, the uric
acid is either directly excreted or first degraded to simpler
nitrogen-containing substances. Overproduction or underex-
cretion of uric acid in humans causes gout. Pyrimidines are
catabolized in animal cells to amino acids.
5 Biosynthesis of Nucleotide Coenzymes The nucleotide
coenzymes NAD
and NADP
, FMN and FAD, and coenzyme
A are synthesized in animals from vitamin precursors.
CHAPTER SUMMARY
General
Nyhan, W.L., Disorders of purine and pyrimidine metabolism,
Mol. Genet. Metab. 86, 25–33 (2005).
Valle,D. (Ed.), The Online Metabolic & Molecular Bases of Inher-
ited Disease, http://www.ommbid.com/. [Part 11 contains chap-
ters on defects in purine and pyrimidine metabolism.]
Purine Nucleotide Biosynthesis
Almassey, R.J., Janson, C.A., Kan, C.-C., and Hostomska, Z.,
Structures of the apo and complexed Escherichia coli glycin-
amide ribonucleotide transformylase, Proc. Natl. Acad. Sci.
89, 6114–6118 (1992).
Eriksen, T.A., Kadziola, A., Bentsen, A.-K., Harlow, K.W., and
Larsen, S., Structural basis for the function of Bacillus subtilis
phosphoribosylpyrophosphate synthetase, Nature Struct. Biol.
7, 303–308 (2000).
Greasley, S.E., Horton, P., Ramcharan, J., Beardsley, G.P.,
Benkovic, S.J., and Wilson, I.A., Crystal structure of a bifunc-
tional transformylase and cyclohydrolase enzyme in purine
biosynthesis, Nature Struct. Biol. 8, 402–406 (2001).
Kappock, T.J., Ealick, S.E., and Stubbe, J., Modular evolution of
the purine biosynthetic pathway, Curr. Opin. Chem. Biol. 4,
567–572 (2000).
Levdikov, V.M., Barynin, V.V., Grebenko, A.I., Melik-Adamyan,
W.R., Lamzin,V.S., and Wilson, K.S.,The structure of SAICAR
synthase:An enzyme in the de novo pathway of purine biosyn-
thesis, Structure 6, 363–376 (1998).
Li, C., Kappock, T.J., Stubbe, J., Weaver, T.M., and Ealick, S.E.,
X-ray crystal structure of aminoimidazole ribonucleotide syn-
thetase (PurM) from the Escherichia coli purine biosynthetic
pathway at 2.5 Å resolution, Structure 7, 1155–1166 (1999).
Löffler, M., Fairbanks, L.D., Zameitat, E., Marinaki, A.M., and
Simmonds, H.A., Pyrimidine pathways in health and disease,
Trends Mol. Med. 11, 430–437 (2005).
Mathews, I.I., Kappock, T.J., Stubbe, J., and Ealick, S.E., Crystal
structure of Escherichia coli PurE, an unusual mutase in the
purine biosynthetic pathway, Structure 7, 1395–1406 (1999).
Poland, B.W., Fromm, H.J., and Honzatko, R.B., Crystal structures
of adenylosuccinate synthetase from Escherichia coli com-
plexed with GDP, IMP, hadacidin, NO
3
,and Mg
2
, J. Mol. Biol.
264, 1013–1027 (1996).
Sintchak, M.D., Fleming, M.A., Futer, O., Raybuck, S.A., Cham-
bers, S.P., Caron, P.R., Murcko, M.A., and Wilson, K.P., Struc-
ture and mechanism of inosine monophosphate dehydroge-
nase in complex with the immunosuppressant mycophenolic
acid, Cell 85, 921– 930 (1996).
Smith, J.L., Glutamine PRPP amidotransferase: Snapshots of an
enzyme in action, Curr. Opin. Struct. Biol. 8, 686–694 (1998).
Tesmer, J.J., Klem,T.J., Deras, M.L., Davisson, V.J., and Smith, J.L.,
The crystal structure of GMP synthetase reveals a novel
REFERENCES
JWCL281_c28_1107-1142.qxd 6/8/10 10:40 AM Page 1140
