Schlick T. Molecular Modeling and Simulation: An Interdisciplinary Guide
Подождите немного. Документ загружается.


200 6. Topics in Nucleic Acids Structure: DNA Interactions and Folding
Box 6.5: Elastic Constants and Persistence Length
As discussed in the text (eq. (6.5)), the elastic bending constant of DNA, A, can be
linearly related to the bending persistence length of DNA, p
b
,as:A = k
B
Tp
b
,where
k
B
is Boltzmann’s constant and T is the temperature. In addition, A can be related to the
root-mean-square (RMS) bending angle θ
2
b
1/2
of a semiflexible chain with a preferred
axis of bending (perpendicular to the helix axis), for which the angular fluctuations are
independent from one another [694]:
A =(2k
B
Th)/θ
2
b
. (6.7)
Here h is the bp-separation distance in DNA (around 3.4
˚
A) [491, 1095]. (Box 6.6 de-
scribes how θ
b
is related to the roll and tilt variables introduced earlier).
Similarly, the torsional rigidity constant can be related to the persistence length of
twisting p
tw
and the RMS twist angle Ω as [830]:
C =(p
tw
k
B
T)/2 (6.8)
and
C =(k
B
Th)/Ω
2
. (6.9)
For the bending to torsional rigidity constant ratio r = A/C, the relationship
Ω
2
1/2
= θ
2
b
1/2
r
2
(6.10)
follows.
The typical values for DNA (A =2.0 × 10
−19
erg cm and C =3.0 × 10
−19
erg cm)
correspond to persistence lengths for bending and twisting of p
b
= 500
˚
A[491], p
tw
=
750
˚
A, and r =2/3. These values correspond to RMS bend and twist-angle values of
θ
2
b
1/2
=6.7
o
and Ω
2
1/2
=3.9
o
at room temperature, in the context of isotropic
bending. The bending constant A can also be related to measured local angular variations in
solved DNA structures, as detailed in Box 6.6. The torsional rigidity constant has recently
been estimated from pulling experiments on single molecules of DNA to be 40% higher
than generally accepted values [182].
6.7.3 Simulations of DNA Supercoiling
Many groups are studying the geometric, thermodynamic, statistical, and dy-
namical properties associated with supercoiling using a variety of models both
for representing the DNA and for simulating conformations. See, for example,
[103,107,108,145,231–233,512,577,656,657,697–699,830,939,947,1036,1036,
1052,1105,1119,1124,1127–1129,1144,1175,1211,1249,1250,1256,1274,1306–
1308,1308,1310,1311, 1339,1366,1410].

6.7. Computational Treatments of DNA Supercoiling 201
Box 6.6: Relationship between the DNA Bending Constant and Local Angular
Measurements in Solved Nucleic Acid Structures
To relate the elastic bending constant A and measured standard deviations of DNA
bending angles, the two components of bending — namely roll (ρ) and tilt (τ ), each with
associated stiffness constants A
ρ
and A
τ
[943] — can be related to A via:
A =
2k
B
Th
ρ
2
+ τ
2
. (6.11)
As above, h is the bp-separation distance in DNA (around 3.4
˚
A). By comparing the above
to eq. (6.7), we see that the bending persistence length of p
b
= 500
˚
A corresponds to an
isotropic model where θ
2
b
1/2
=6.7
o
, as well as to an anisotropic bending model where
ρ
2
1/2
=5.7
o
and τ
2
1/2
=3.6
o
. Values from analysis of B-form crystal structures
yield somewhat smaller values for these roll and tilt fluctuations (e.g., ρ
2
1/2
=5
o
and
τ
2
1/2
=3
o
) and thus a larger effective bending persistence length and rigidity constant
(by about 1.3,orA =2.6 × 10
−19
erg cm and p
b
= 500
˚
A). However, the static fluc-
tuations in the crystal structures do not directly correspond to the dynamic range of DNA
flexibility.
For sufficiently small angular deflections, we can partition the total bending magni-
tude as
θ
2
b
= ρ
2
+ τ
2
=[k
B
Th(A
ρ
+ A
τ
)]/(A
ρ
A
τ
) . (6.12)
This first-order model does not account for the well-recognized preferential direc-
tions of bending of DNA into the major and minor grooves with respect to other
directions [473, 940, 1096, 1285, 1454]. A non-uniform bending chain view is re-
quired for better representations, especially for sequences with intrinsic curvature, like
A-tracts.
A few of the many topics that have been studied using such models are the
behavior of supercoiled DNA as a function of salt [1125, 1309], solvent [1036],
superhelical density [1129],andlength[830, 1214, 1250]; extended theoretical
treatments based on elastic rod mechanics and dynamics [305, 651, 1175]; the
response of supercoiled DNA to constraints imposed by proteins [945]; theory
of elastic rods with applications to DNA [255, 256]; the geometry of DNA in
small minichromosome systems [834]; and the site juxtaposition time (the bring-
ing together in space of linearly-distant DNA segments due to supercoiling)
[607, 1211, 1310]. See Boxes 6.4 and 6.7 for examples of nucleosome folding
and site juxtaposition studies, respectively, as well as corresponding Figures 6.12
and 6.14–6.15 and related experimental studies of EM and nucleosome interaction
measurements [483].
Many models for studying nucleosomes and oligonucleosome arrays have also
been developed (e.g., [64, 108, 353, 883, 1219, 1240, 1312, 1441]) and should of-
fer further insights into the nature of the complex folding and dynamics of the
chromatin fiber.
202 6. Topics in Nucleic Acids Structure: DNA Interactions and Folding
0 1 2 453 6 7 8 9 10
0
1
2
3
1.6 ms
3.4 ms
4.2 ms4.0 ms
2.4 ms
3.0 ms
1.8 ms 2.0 ms 2.2 ms 2.6 ms
3.8 ms
2.8 ms
3.2 ms
time (ms)
i
2
, kb
bp 1
bp 900
3.6 ms
c
s
= 0.2 M
s
= −0.06
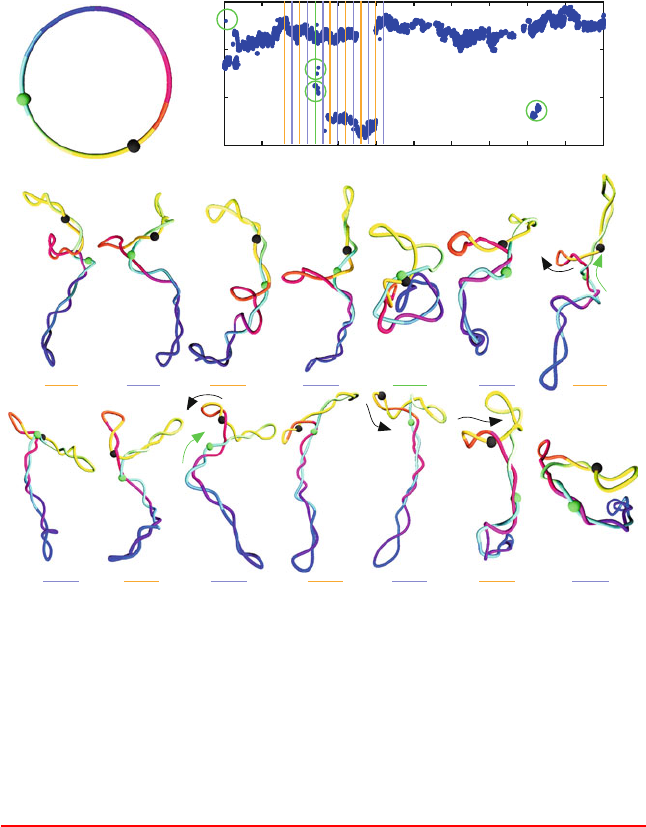
Figure 6.14. Juxtaposition kinetics, as analyzed for two beads separated by 900 bps in
circular DNA of length 3000 bps and σ = −0.06 by Brownian dynamics simulations at
high salt (c
s
=0.2 M sodium ions) [577]. (Top) Juxtaposition event plot, showing bps
i
2
that juxtapose with bp 1; the circled values correspond to more random juxtaposition
events, as illustrated for the snapshot at 2.4 ms. (Bottom) BD snapshots, showing ordered
motions along the DNA contour and rarer random collision events.
Box 6.7: Simulations of DNA Site Juxtaposition
Illustrative results from computer simulations of supercoiled DNA are shown in
Figure 6.14. These snapshots are taken from a Brownian dynamics simulation of a 3000-bp
DNA system at the superhelical density of σ = −0.06 at high salt using a homogeneous
bead/wormlike model [577]. Simulations have tabulated the times for distant sites to juxta-
pose due to the ambient floppiness of DNA as a function of the superhelical density, DNA
length, site separation, and the salt concentration [607].
6.7. Computational Treatments of DNA Supercoiling 203
0 0.02 0.04 0.06
0
10
20
30
40
〈T
c
〉 (ms)
0 0.01 0.02
0
5
10
C
S
= 0.01 M
C
S
= 0.2 M
0 0.02 0.04 0.06
0
20
40
60
80
100
Reaction, normalized %
synapsis
SIMULATION DATA
EXPERIMENTAL DATA
L = 3000 bp
d
L
= 600 bp
|s |
|s |
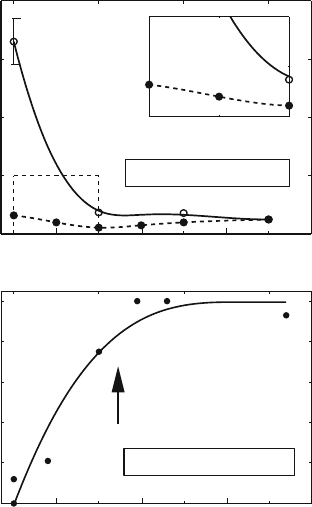
Figure 6.15. (Top): Site juxtaposition time measurements, T
c
, defined as the mean time
for two sites d
L
bps apart to come within 100
˚
A, as averaged over all such separated pairs,
as a function of |σ| for d
L
= 600 bps for a 3000 bp system and two salt concentrations
[577]. The inset focuses on the large salt effect at low |σ|. Note also the plateauing effect
of T
c
around |σ| =0.03 for both salt concentrations. This is also the experimental
observation (bottom)[119] for the synapsis-time dependence on the superhelical density.
In the study of juxtaposition, two linearly-distant sites are considered juxtaposed when
the distance between them is 100
˚
A or less. Interestingly, we find (Figure 6.15)thatatlow
salt [608] juxtaposition times are accelerated by a factor of 10 or more due to supercoiling
(i.e., juxtaposition times for relaxed DNA are much slower), but that supercoiling beyond
|σ| =0.03 does not accelerate such site synapsis times [608]. Such critical behavior at
moderate |σ|agrees with experimental findings regarding supercoiling-dependent synapsis
rates for the resolvase recombination system [119], as also displayed in Figure 6.15.The
explanation for the fact that further supercoiling does not enhance juxtaposition times
significantly emerges from our simulations: the balance between flexibility and tight su-
percoiling is optimized at this mean value of superhelicity.
At high salt, the effect of superhelicity is far less dramatic. Further analysis of the mech-
anism for juxtaposition shows that large conformational rearrangements due to Brownian
motion operate at low superhelical density and low salt, whereas systematic slithering mo-
tions (reptation, or bidirectional conveyer-like motion) along the polymer contour [118]
are more prominent at high salt, as shown in Figure 6.14. At high salt, site juxtaposition
204 6. Topics in Nucleic Acids Structure: DNA Interactions and Folding
can be described by a combination of slithering, branch rearrangements, and interbranch
collisions, as also shown in the figure. The figure, prepared for studying site juxtaposition
kinetics [577], shows at top a plot in time of the bp i
2
that juxtaposes with the first bp (i.e.,
is within 100
˚
A of it). The continuous pattern in i
2
in time indicates ordered slithering; at
low salt, the patterns are much more random.

7
Topics in Nucleic Acids Structure:
Noncanonical Helices and RNA
Structure
Chapter 7 Notation
S
YMBOL DEFINITION
α polynucleotide backbone torsion angle
γ
polynucleotide backbone torsion angle
χ
glycosyl (sugar/base) torsion angle
Anything that DNA can do, RNA can do better.
Susan Gottesman, Cold Spring Harbor symposium, 2006.
7.1 Introduction
This chapter builds upon nucleic acid concepts introduced in the prior two
chapters to include a description of alternative hydrogen bonding schemes in
nucleic acids, non-canonical helical and hybrid structures, DNA mimics, over-
stretched and understretched DNA, and RNA structure and folding, including
secondary and tertiary-structure RNA modeling.
Besides the excellent texts [139, 195, 1080, 1191], the reader is invited to ex-
plore the wealth of related structural data on the nucleic acid database NDB
T. Schlick, Molecular Modeling and Simulation: An Interdisciplinary Guide, 205
Interdisciplinary Applied Mathematics 21, DOI 10.1007/978-1-4419-6351-2
7,
c
Springer Science+Business Media, LLC 2010

206 7. Topics in Nucleic Acids Structure: Noncanonical Helices and RNA Structure
(ndbserver.rutgers.edu/) and the protein data bank/research collaboratory for
structural bioinformatics (PDB/RCS) resource [128,1365], as well as information
concerning RNA on the RNA Ontology Consortium (ROC) [735].
As in the previous chapter, we abbreviate base pair and base pairs as bp and
bps, respectively.
7.2 Variations on a Theme
As evident from the previous chapter, nucleic acids are extremely versatile. The
classic right-handed double helical structure described by Watson and Crick is
an excellent textbook model. In Nature, however, polynucleotide helices have
developed an enormous repertoire of sequence-dependent structures, helical
arrangements, and folding motifs on both the bp and higher level of folding.
7.2.1 Hydrogen Bonding Patterns in Polynucleotides
Hydrogen-bonding variations are important for DNA’s adaptability to base mod-
ifications (e.g., methylation, drug or carcinogen binding), bp mutations and
mismatches, and various interactions between DNA and proteins and among
polynucleotide helices.
Classic Watson-Crick (WC)
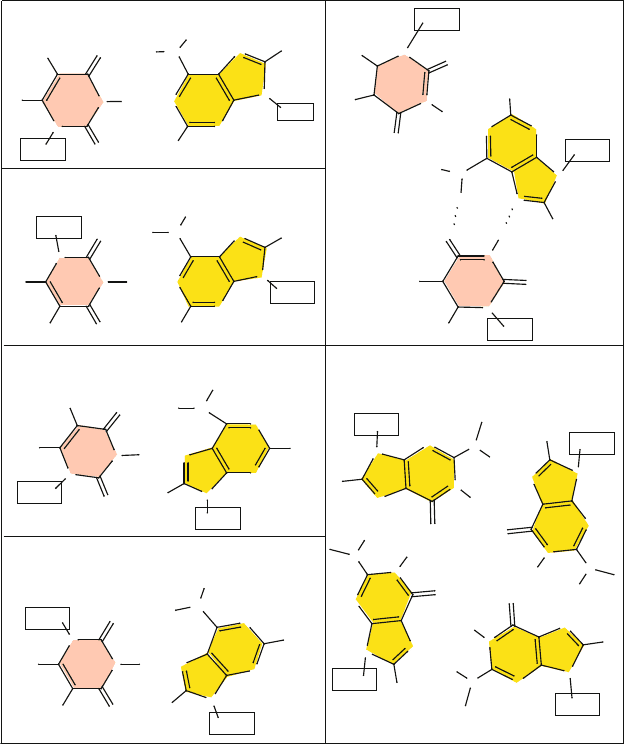
The classic WC hydrogen-bonding arrangement as shown in Figure 5.3 of
Chapter 5 is a particularly beautiful arrangement. It can be appreciated by not-
ing that the C1
(pyrimidine) to C1
(purine) distance is around 10.6
˚
A in both the
AT and GC pairs, in good preparation for helix stacking. The intrinsic energy of
hydrogen bonds — that is, relative to vacuum — is generally in the 3–7 kcal/mol
range but only up to 3 kcal/mol in nucleic acids due to geometric constraints.
1
Though much weaker than covalent bonds (80–100 kcal/mol) [1191, p. 12–14],
the cumulative effect in the double helix is substantial due to the collective im-
pact of hydrogen-bonding interactions resulting from cooperative effects [1018].
Based on theoretical studies, the strength of the hydrogen-bond energy depends
on base composition but is very similar for the two WC bps. The energy of bp
stacking, 4–15 kcal/mol, substantially contributes to overall helix stability and
depends on the nucleotide sequence [169].
Yet, hydrogen-bonding patterns in polynucleotides are extremely versatile.
There are several nitrogens and oxygens on the bases, allowing various donor and
acceptor combinations involving different interface portions of the aromatic rings.
1
Estimates of base-pairing energetics in solvated nucleic acids come mainly from theoretical cal-
culations [1225]. This is because the resolution of macromolecular thermodynamic measurements into
subcontributions (hydrogen bonding, stacking, and electrostatic forces) remains a challenge, despite
numerous thermodynamic studies on nucleic acid systems [169, 449, 1007].
7.2. Variations on a Theme 207
To appreciate this versatility, we first examine the classic WC pattern in detail.
For an AT pair, two hydrogen bonds form between the C4–N3 face of T
and the C6=N1 face of A (see Figure 5.3 of Chapter 5 and Figure 7.1 here).
One hydrogen bond involves the sequence O4 (T)···H–N6 (A), i.e., between
the thymine carbonyl oxygen O4 attached to ring atom C4 and the adenine N6
atom, which is attached to the ring C6 atom. The other hydrogen bond is N3–H
(T)···N1 (A), between the thymine ring nitrogen N3 and the adenine N1.
For the GC pair, three hydrogen bonds form between the C4=N3–C2 face of
C and the C6–N1–C2 face of G: N4–H (C)···O6 (G), N3 (C)···H–N1 (G), and
O2 (C)···H–N2 (G). All hydrogen-bond lengths in both the AT and GC bps are
very similar, with individual lengths ranging from about 2.85 to 2.95
˚
A between
the heavy atoms.
Reverse WC
Reverse WC base-pairing schemes can result when one nucleotide rotates 180
o
with respect to its partner nucleotide, as shown in Figure 7.1. Thus, one hydrogen
bond from the original WC scheme might be retained, but another can involve
a different atom combination. This flip of one bp changes the position of the
glycosyl linkage. Hence, an AT pair can more easily accommodate this flip due to
near symmetry about the N3–C6 axis of T. That is, since the ring face C4–N3–C2
of T has carbonyl attachments at both C4 and C2, the hydrogen bonding face can
be changed from C4–N3 to C2–N3 (see Figure 7.1). This type of reverse WC base
pairing exists in parallel DNA [1080].
Hoogsteen
Hoogsteen bps utilize a different part of the aromatic ring for hydrogen bonding.
Specifically, the N7–C5–C6 face of A and G, where N7 is in the 5-membered
ring and C6 is in the 6-membered ring of the purine, is used in the Hoogsteen
arrangement (see Figure 7.1). This is instead of the C6=N1 face of A and G —
both atoms of which are in the 6-membered ring of the fused-ring compound —
in combination with the C4–N3 face of T and C — as in WC interactions.
The Hoogsteen arrangement also implies a change in the glycosyl torsion angle
χ, from anti to syn, and a shortening of the C1
–C1
distance, from about 10.5
˚
A
in WC to about 8.5
˚
A in Hoogsteen base pairs (e.g., [12]).
Hoogsteen base pairing helps stabilize structures of highly bent DNA (e.g.,
TATA elements [971]), tRNAs, and DNA triplexes where two strands are held
by WC hydrogen bonds and where two strands are stabilized by Hoogsteen-type
pairing (see Figure 7.1). They also appear occasionally in complexes of DNA with
anti-cancer drugs. Recently, an unusual Hoogsteen A–T bp was discovered in a
high-resolution complex of unbent DNA bound to 4 MATα2 homeodomains (see
Figure 7.2)[12, 13].
208 7. Topics in Nucleic Acids Structure: Noncanonical Helices and RNA Structure
N
N
O4
N
N
N
N
H
N6
H
T
A
N
N
O2
N
N
N
N
N6
H
H
N
N
O2
N
N
N
N
N6
H
H
N
N
N
N
N6
N
N
O4
H
3
4
5
6
2
Sugar
H
1
A
T
1
2
3
45
6
T
Watson-Crick
65
4
3
2
1
Reverse Watson-Crick
7
8
9
A
5
6
1
2
Hoogsteen
Reverse Hoogsteen
4
3
8
7
9
5
6
4 3
2
1
A
T
54
3
2
1
6
7
9
8
4
5
1
6
2
3
H
3
C
H
O2
H
H
Sugar
Sugar
H
H
3
C O4
H
Sugar
H
H
H
3
C
H
O2
Sugar
Sugar
H
H
H
Sugar
H
H
3
C O4
H
H
Sugar
H
H
. . .
. . .
. . .
. . .
. . .
. . .
Triplex
N
N
N
N
N6
N
N
H
8
7
9
5
4
3
1
6
2
H
3
C
A
T
H
H
H
H
H
O2
2
4
6
5
3
1
N
N
O2
H
3
C
T
H
H
4
2
6
1
3
5
Sugar
O4
O4
Sugar
N
N
N
N
O6
G
H
N
H
H
N
N
N
N
O6
G
H
N
H
H
N
N
N
N
O6
G
H
N
H
H
N
N
N
N
O6
G
H
N
H
H
Guanine Quartet
8
7
9
5
4
3
1
6
2
. . .
. . .
. . .
. . .
Sugar
Sugar
Sugar
Sugar
1
6
3
5
2
4
7
8
9
1
6
3
5
2
4
7
8
9
1
6
3
5
2
4
7
8
9
1
6
3
5
2
4
7
8
9
. . .
. . .
. . .
. . .
. . .
. . .
. . .
.
. .
Sugar
WC
Hoog
Figure 7.1. Various hydrogen-bonding schemes: (left) classic Watson-Crick, reverse
Watson-Crick, Hoogsteen, and reverse Hoogsteen in base pairs; (right) WC/Hoogsteen
in a triplex (from an H-DNA structure, which forms in sequences that have stretches of
purine followed by stretches of pyrimidines, PDB code 1B4Y) [1290], and patterns in a
guanine-quartet quadruplex (or guanine tetraplex, PDB code 1JB7) [569].
Reverse Hoogsteen
Similarly, a reverse Hoogsteen arrangement involves the flipping of one nucleo-
tide by 180
o
with respect to its partner, by analogy to reverse WC, as shown in
Figure 7.1.
7.2. Variations on a Theme 209
F
D
E
C
G
42
A
1
C
21
T
22
T
22
C G
G C
C G
A T
C G
A T
T A
T A
T A
A T
C G
T A
T A
A T
A T
T A
G C
T A
A T
C G
A
Hoogsteen
base pair
1
21
42
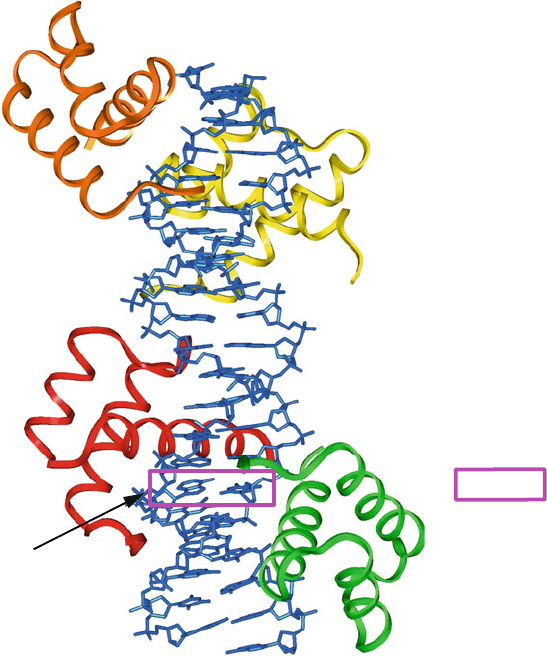
Figure 7.2. Unusual DNA/protein complex in which a Hoogsteen A–T base pair is found
in unbent DNA. The complex is a high-resolution crystal of four MATα2 homeodomains
bound to DNA [12,13].
Mismatches and Wobbles
Other bp arrangements involve anti/syn bps between two purines (a mismatch),
in which one base adopts an atypical syn glycosyl configuration. This type of
pairing can occur due to protonation of one base (favored at acidic conditions) or
keto-enol tautomerizations. (See introduction to the term in Chapter 1, subsection
on the discovery of DNA structure, and Chapter 5, subsection on nitrogenous
bases).
Such ionized bases reverse the polarity of the hydrogen-bondingsites and hence
lead to various hydrogen-bonding schemes, such as between G
+
·C (Hoogsteen)
or the ionized mispair A
+
·C(wobble). Wobble pairs are formed when one base
