Schlick T. Molecular Modeling and Simulation: An Interdisciplinary Guide
Подождите немного. Документ загружается.

210 7. Topics in Nucleic Acids Structure: Noncanonical Helices and RNA Structure
is shifted in the bp plane relative to its partner due to steric misalignment of the
bases. Experimental evidence for bp wobbling has come from tRNA structural
analysis.
Thus, various non-WC base-pairing schemes can occur when bases are chemi-
cally modified, found in the rare enol and imino tautomeric forms, or mismatched,
for example. The alternative arrangements for mispaired bases, in particular, can
in turn lead to mutations upon DNA replication if not corrected by the elaborate
repair machinery of the cell.
Other Patterns
For further details regarding these and many other base-pairing schemes in nu-
cleic acids, see [1191]and[737] for an early encyclopedic compendium of
the “bewildering” variety of observed canonical and non-canonical nucleic acid
arrangements, as compiled from RNA base-pairing interactions. This variety
led to many further works and a proposal for new nomenclature for RNAs
[738] which stimulated a large international initiative termed the RNA Ontol-
ogy Consortium (ROC) [735]. ROC aims to establish a standard vocabulary for
studying RNA by creating a common system to describe RNA sequences, 2D, and
3D structures and by developingsoftware tools that merge sequence and structural
databases for RNA for use by the general scientific community. See ROC website
(http://roc.bgsu.edu/) for details.
In general, non WC bps are generally lower in stability because they typically
incur a greater distortion in the sugar/phosphate backbone (assuming that the rest
of the DNA is in a canonical A, B, Z-type conformation). Though they are more
difficult to accommodate in general, many local alterations in sequence compo-
sition, environmental factors, and conformational patterns favor these alternative
base-pairing arrangements.
7.2.2 Hybrid Helical/Nonhelical Forms
Alternative Helical Geometries
Numerous variations are noted in the helical structures of polynucleotides (see
[139, 893, 1080], for example). For example, C-DNA, D-DNA, and T-DNA
helices have been defined with 9.3, 8.5, and 8.0 bps/turn, respectively. The
C-DNA motif forms in fibers of low humidity (57–66%), D-DNA is observed
in poly(dA)·poly(dT) strands, and T-DNA has been noted in bacteriophages that
contain modified C residues and glucose derivatives for the sugar attachments.
Within polynucleotide structures, local distortions can also produce hairpins
(DNA and RNA strands that fold back on themselves), cruciforms (intrastrand
hydrogen bonds between complementary bases, often leaving a few unpaired
bases at the hairpin tip), inlcuding bulges and loops (unpaired extrusions)
(Figure 7.4). Such motifs are especially important in stabilizing the folding of
single-stranded RNA.
7.2. Variations on a Theme 211
DNA Triplexes and Quadruplexes
Though Linus Pauling was ridiculed for his early suggestion of a triple helix
structure for DNA [975], we now recognize the existence or the possibility for for-
mation of various DNA triplexes (involving WC and Hoogsteen bps) [421,1008].
Other existing or possible forms are quadruplexes (stabilized by hydrogen bond-
ing and cations as in guanine-quartets; see Figure 7.1)[961,968,998,1160,1183,
1319,1320,1337]; parallel DNA (requiring reverse WC base pairing); DNA ana-
logues; and hybrids of RNA, DNA, and other polymers such as oligonucleotide
mimics (see below) [712].
Triplex forming oligonucleotides are promising pharmaceutical targets since
they modulate gene activity in vivo through recognition of duplex DNA [905].
Triplexes and other unusual oligonucleotides are good subjects for simulation
techniques since the factors that govern sequence-dependent properties can be
explored [784].
Triple-helical DNA gained wide attention in the mid-1980s when various
groups demonstrated that homopyrimidine (polynucleotide chains containing
pyrimidines, such as poly(dT) or poly(dCT)) and some purine-rich oligomers
(such as poly(dA) or poly(dAG)) can form stable and sequence-specific triple-
stranded complexes with corresponding sites on duplex DNA; the third strand
associates with the duplex via Hoogsteen pairing (see Figure 7.1)[421].
Homopurine/homopyrimidine stretches in supercoiled DNA were also found to
adopt an unusual structure which includes a triplex called H-DNA.
Four-stranded DNA structures called G-quadruplexes are believed to play im-
portant functional roles in genome maintenance through their stabilization of
chromosome ends or telomeres. Likened to plastic caps firming shoe-lace ends
(by Stu Borman, C&EN writer), telomeres are protein/DNA assemblies that con-
fine chromosome ends. Such protection from the possible fusing of chromosomes
into one another helps maintain the integrity of the genome in living cells.
Telomere structure is of great biomedical importance since aging and cancer
can be associated with malfunctioning telomeres. Thus, drugs targeted to
telomeres can affect telomere function in DNA synthesis and transcription.
Telomeric DNA contains short guanine-rich repeats. At the very end of telom-
eres, G-quadruplexes are found, elaborately folded stacks of G-quartets (also
known as GGGG tetrads) — the planar arrays of hydrogen-bonded guanines as
shown in Figure 7.1.
Direct evidence that quadruplexes exist in human cells was provided by Hurley
and coworkers [1183], who reported a chair-shaped quadruplex in a purine-rich
strand of DNA in a promoter region of the oncogene c-Myc activated in cancer
cells. Their work also highlights the potential role of quadruplex-targeted com-
pounds as agents to combat disease, for example through control of oncogene
expression to induce tumor shrinkage [601].
The architecture of G-quartets in G-quadruplexes remains an area of intense
study. The planar G-quartets, as shown in Figure 7.1, can stack vertically with
monovalent ions (Na
+
or K
+
) sandwiched in between the layers. Wang and
212 7. Topics in Nucleic Acids Structure: Noncanonical Helices and RNA Structure
O
O
POO
O
O
Base
O
O
POS
O
O
Base
O
POO
O
O
Base
BaseBase
DNA
Phosphate changed
(phosphorothioate)
Sugar changed
(homo-DNA)
Linkage changed
(3'-thioformacetal)
Backbone changed
(PNA)
O
S
C
O
C
O
Base
Base
O Base
NH
N
NH
N
Base
Base
O
O
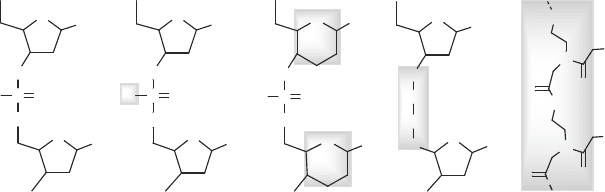
OO
Figure 7.3. Various oligonucleotide analogues that involve modifications of the DNA
phosphate unit, sugar, entire phosphodiester link, or the entire sugar/phosphate backbone.
Patel [1337] revealed in 1993 an NMR structure of a G-quadruplex in sodium
solution. The sequence studied, with three repeats of TTAGGG, has a topol-
ogy of G-quartets connected by loops (the TTA sequences that join each GGG
series to the next) at the top and bottom of the quadruplex rather than the
sides (see original article for figures). An exciting 2002 crystal structure by the
Neidle group [961] of the same single-stranded sequence motif (specifically,
TTAGGG
2
and TTAGGG
4
) in the presence of potassium ions reveals a radi-
cally different arrangement, with propeller-like connections through the sides of
the stack (double-chain reversal topology [968]) rather than top and bottom (or
one diagonal and two edgewise loops [968]).
Since mammalian cells are likely to have quadruplexes coordinated by potas-
sium ions (since K
+
concentration exceeds that of Na
+
by an order of magnitude),
this propeller-form quadruplex structure is of great importance and has clinical
implications as well [961]. The topologies of quadruplexes and their interactions
with other DNA, RNA, and proteins continue to define an active research fron-
tier with significant potential for biomedical applications, for example in cancer
therapy. See [1319,1320] for recent computational studies.
DNA Mimics
A growing number of DNA analogues (or ‘DNA mimics’) is also appearing (see
Figure 7.3)[905]. Analogues can be sought in several ways (see Figure 7.3 and
Box 7.1):
• by modifying the phosphate backbone segment (e.g., one P–O link changed
to P–S or P–CH
3
);
• by modifying the sugar (e.g., 6-membered instead of 5-membered ring);
• by modifying the entire phosphodiester linkage of 4 atoms, O3
through
C5
(to increase DNA hybrid stability through charge neutrality); or
• by replacing the entire sugar/phosphate backbone, retaining just the bases
as in PNA, peptide (or polyamide) nucleic acids.

7.2. Variations on a Theme 213
Such close analogues of oligonucleotides are designed to bind selectively to DNA
bps through triple strands or duplexes. Various duplex DNA/RNA hybrids are
key elements in transcription and replication and thus are potential therapeutic
agents in anti-sense techniques. Triplex structures are also potential agents for
sequence-specific cutting of DNA and drugs (by attacking various disease targets
at the genetic level). Non-complementary or ‘anti-sense’ binding to a sequence of
complementary messenger RNA or single-stranded DNA can also be exploited in
antisense technology to suppress gene expression.
To be suitable, DNA analogues must also be biologically stable (i.e., resistant
to nuclease digestion) and have favorable cellular absorption properties. All issues
of geometry, flexibility, hydrophobicity, and triplex stability must be weighed in
designing DNA mimics with practical utility.
Box 7.1: PNA (Peptide Nucleic Acid)
PNA is an especially interesting oligonucleotide analogue that forms very stable com-
plexes with double-stranded DNA [228, 350, 351, 905, 907, 1161]. In studies of evolution,
PNA has been proposed as a candidate for the backbone of the first genetic material
that predates the “RNA world”. While RNA is unstable and difficult to synthesize, the
polymeric constituent of PNA, N-(2-aminoethyl)glycine (or AEG), may spontaneously
polymerize and is accessible in prebiotic syntheses. No firm evidence, however, exists
regarding this hypothesis [662].
The sugar/phosphate backbone of oligonucleotides is replaced in PNA by a pseudopep-
tide chain of AEG units with the N-acetic acids of the bases (N9 for purines and N1 for
pyrimidines) linked via amide bonds (Figure 7.3). This substitution of the DNA backbone
makes the PNA backbone electrically neutral rather than negatively charged; though the
same number of chemical bonds as found in DNA and RNA exist in PNA (albeit different
types), the replacement of the sugar ring by a linear bond sequence introduces additional
torsional flexibility.
Stable hybrids between PNA and complementary DNA or RNA oligonucleotides form
in a sequence-selective manner. Binding of PNA to double-stranded DNA leads not only
to triple helices but also to P-loops strand-displacement complexes [905]. Model building,
molecular mechanics and dynamics calculations, as well as experimental studies have
shown how the replacement of DNA by PNA disrupts the canonical DNA triple helix, by
displacing the hydrogen-bonded bases away from the global helical axis with respect to
their positions in B-DNA helices, and changes the groove structure substantially [1216].
This makes PNA/DNA hybrids (involving both single and double helical DNA) A-like
rather than an intermediate between A and B-DNA forms, as is typical for canonical DNA
triplexes. Helix stabilization in PNA/DNA hybrids stems from favorable base pairing,
stacking, and (reduced) electrostatic interactions.
PNA molecules have already found practical applications as probes — an alternative
to conventional DNA probes — for the detection of bacteria like Salmonella enterica or
Staphylococcus aureus; chemical advantages stem from PNA’s strong resistance to degra-
dation by proteases and nucleases in the cell and its salt-independent hybridization kinetics.

214 7. Topics in Nucleic Acids Structure: Noncanonical Helices and RNA Structure
Triplexes and DNA analogues like PNA also have potential as drugs in gene therapy [905].
In particular, PNA is quite attractive for drug development, given its high triplex stability.
However, given the A-like form of PNA/DNA hybrids, the design of stable B-like hybrids
requires a different chemical approach than that used to construct PNA.
7.2.3 Unusual Forms: Overstretched and Understretched DNA
Single-Molecule Manipulations
Recently, a new experimental achievement of single-molecule observation and
manipulation experiments using laser tweezers or single glass fibers [187, 1196]
has been applied to DNA [682,1049], re-invigorating interest in DNA’s structural
versatility [120,182,188,250,990,1203,1233]. Such experiments can also be used
to determine DNA force constants, such as torsional rigidity [182]. It has been
found that DNA subjected to previously unattainable forces of 10–150 picoNew-
tons (pN) displays a highly cooperative, sharp, and reversible transition to a new
DNA structure at around 70 pN. This new overstretched helical structure, termed
the S-DNA ladder, has been suggested to have a rise per residue of around 5.8
˚
A
(compare to values in Table 5.3 of Chapter 5) and a notable inclination (possibly
as large as 70
o
)[671], but structural details remain controversial. The transition
under a force of extension is also affected by changes in the ionic strength of the
medium, sequence, and addition of intercalators. Interestingly, DNA can sustain
extensions to roughly twice its original length without severe distortions to the
base pairing.
Modeling studies soon followed these experiments to investigate global con-
formational features of overstretched DNA duplexes [671, 711] and the local
morphological features associated with the deformation process [678](see
Box 7.2).
Single-force spectroscopy has also been applied to study the mechanical prop-
erties of nucleosome arrays of the chromatin fiber (see Section 6.5), to suggest
chromatin organization and to examine the effect of linker histones on fiber
compaction [682].
Similar experiments have also been applied to RNA [759, 771, 1452]and
proteins [1453].
Biological Relevance and Other Applications
The extreme deformations of DNA are of great interest because flexible DNA
molecules undergo a variety of distortions in biological environments. Notable
examples occur during DNA recombination and cell division. It has been sug-
gested, for example, that a helix to ladder transition in DNA near the chromosomal
centromere region may occur during cell division and thus play a regulatory
role [671]. Intriguingly, the overstretched DNA in recombination filaments of
5.1
˚
A per bp is close to that associated with the phase transition observed in
single-molecular micro-manipulation experiments.

7.2. Variations on a Theme 215
It has also been suggested that longitudinal deformations of DNA might be
associated with many DNA/protein binding events, such as DNA binding to the
TATA-box binding protein TBP (where DNA is locally compressed, strongly bent,
and unwound) and DNA binding to nucleosomes [1050], where variable lengths
of DNA associated with nucleosome wrapping might accommodate sequence and
ionic variations [678].
More generally, single-molecule biochemistry experiments help investigate the
energetics and dynamics of folding and unfolding, the stability of various 2D and
3D intermediates, conformational landscapes, and folding pathways.
Box 7.2: Modeling Overstretched DNA
Results of modeling studies of overstretched DNA — by energy minimization or molec-
ular dynamics — are protocol dependent. For example, results and interpretations depend
on which end of the DNA is pulled, what minimization method is used, how minimization
is implemented (e.g., how fine the helical rise increments are), what force field is em-
ployed, and how solvent is treated.
The minimization work of Lavery and coworkers [711] suggested that the stretched con-
formation may be a flat, unwound duplex or a narrow fiber with substantial bp inclination
[711]. The molecular dynamics simulations of Konrad and Bolonick [671] reproduced the
helix to ladder transition and analyzed the geometric and energetic properties stabilizing
the S-ladder. The constrained minimization studies of Olson, Zhurkin and coworkers [678]
produced a wealth of structural analyses for both compressed and stretched DNA duplexes
(from 2 to 7
˚
A per bp) of poly(dA)·poly(dT) and poly(dG)·poly(dC) homopolymers under
high and low salt conditions. It was found that DNA can stretch to about double, and
compress to half, its length before the internal energy rises sharply. Energy profiles span-
ning four families of right-handed structures revealed that DNA extension/compression
deformations can be related to concerted changes in rise, twist, roll, and slide parameters.
The lowest energy configurations correspond to canonical A and B-DNA. These models
may be relevant to nucleoprotein filaments between bacterial Rec-A-like proteins and
overstretched, undertwisted DNA [349].
Such fascinating single-molecule biochemistry manipulations have also shown that un-
der much smaller forces (<3 pN), a new DNA phase is achieved, with about 2.6 bps/turn
and thus about 75% more extended than B-DNA [21]. This new DNA conformation,
termed P-DNA (for Pauling, see below), occurs at moderate positive supercoiling for
molecules that cannot relieve the torsional stress via writhing (nonplanar bending). (For
negative supercoiling, DNA denatures in this force range). Intriguingly, some modeling
suggests that such a structure is ‘inside-out’, with bases on the helical exterior and the
sugar/phosphate backbone at the center, as Linus Pauling once suggested for the structure
of the double helix based on a model with un-ionized bases [975]. Such a conformational
transition arrives from major rotation of torsion angles (notably α and γ,towardthetrans
conformation) and is compatible with both C2
-endo and C3
-endo sugar puckers. The
intrastrand P–P distances in P-DNA (around 7.5
˚
A) are larger than the corresponding
values in canonical A and B-DNA (5.8 and 6.6
˚
A, respectively). Evidence shows that this
P-DNA conformation is found in the packed DNA inside some virus complexes, where

216 7. Topics in Nucleic Acids Structure: Noncanonical Helices and RNA Structure
the DNA is constrained by the helical coat protein [289, 775]. Still, the interpretation of
positively-supercoiled, overstretched DNA as an ‘inside-out’ model remains controversial.
Polymer statistical-mechanics calculations suggest that the experimental data involved
in these force-versus-extension measurements of DNA can be fit to the elastic theory of
the entropic force required to extend a worm-like chain [190]. However, such studies
of entropic force are only relevant to small fluctuations about the B-DNA equilibrium
conformation.
7.3 RNA Structure and Function
7.3.1 DNA’s Cousin Shines
While proteins are household words and DNA is an icon, in science as well as
art (for the latter, see [1112] for an overview), their biomolecular cousin, RNA,
was largely left behind until recently. Indeed, RNA’s starring role in the cell has
emerged with new discoveries concerning RNA’s vital regulatory roles, as in-
troduced in Chapter 1. Namely, our appreciation for RNA has heightened with
discoveries that RNA molecules are integral components of the cellular machin-
ery for protein synthesis and transport, RNA editing, chromosome replication and
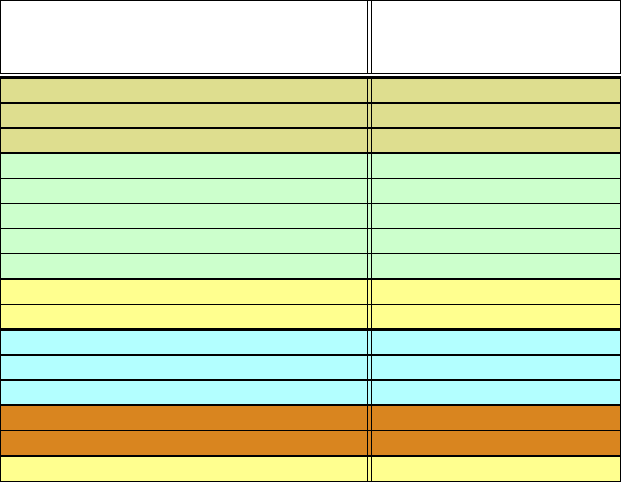
regulation, catalysis, and many other functions (see Table 7.1 for some of RNA’s
diverse roles).
At a 2006 symposium organized by NCI at Cold Spring Harbor, Susan
Gottesman suggested on a presentation slide: “Anything that DNA can do, RNA
candobetter”. At the same symposium, Gary Ruvkun announced 23 challenges
facing scientists concerning regulatory RNAs. James Watson simply summed the
atmosphere: “This is a revolution”. Indeed, we are now discovering that bio-
logical and synthetic RNAs prepared in the laboratory perform a broad range
of functions and have numerous applications. This comes with the discovery not
only of regulatory RNAs but also of many synthetic RNAs developed from in vitro
selection experiments that have significantly expanded our knowledge of RNA’s
repertoire.
7.3.2 RNA Chains Fold Upon Themselves
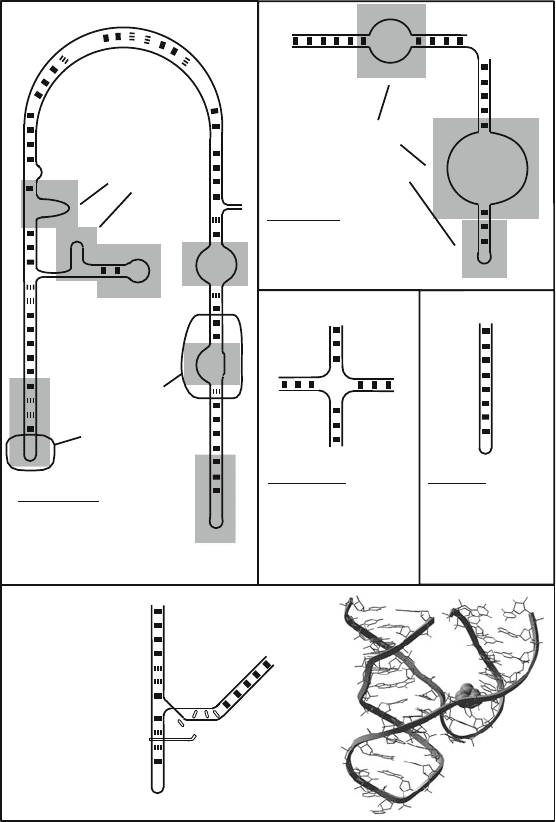
Many of the base pairing variations described for DNA in the prior sections
are common for RNA, the single-stranded polynucleotide chain, which can fold
upon itself to form double-stranded segments interspersed with loops. Such pat-
terns are governed by the primary sequence and can accommodate a variety of
hydrogen bonding patterns. The double stranded regions (stems) can be imper-
fect with bulges, mismatched pairs, and unusual hydrogen bond schemes, as
shown in Figure 7.4. The stem/loop structures themselves can fold further, seeking
7.3. RNA Structure and Function 217
Table 7.1. Some classes of non-coding RNA (ncRNA).
RNA Function
transfer RNA (tRNA) protein synthesis
ribosomal RNA (rRNA) protein synthesis
Signal recognition particle (SRP) protein recognition
small nucleolar RNA (snoRNA) rRNA modification
micro RNA (miRNA) translation regulation
transfer-messenger RNA (tmRNA) protein stability in ribosome
telomerase RNA replication
guide RNA (gRNA) mRNA editing
spliced leader RNA (SL RNA) mRNA trans-splicing
small nuclear RNA (snRNA) RNA splicing
hairpin, hammerhead, and HDV ribozymes self-cleavage
Group I intron self-splicing
Group II intron self-splicing
RNase P pre-tRNA processing
23S rRNA peptide bond formation
G, A, glmS, TPP and other riboswitches gene regulation
favorable stacking, hydrogen bonding, and other interactions between distant part-
ners. See [1113] for an introduction to RNA structure and associated biological
problems.
The multitude of RNA secondary structures and tertiary interactions are sta-
bilized by various double-stranded regions, interior and terminal loops, bulges,
K-turns, U-turns, S-turns, A-platforms, tetraloops, and other motifs (e.g., [102,
545,654,736,739,740,875]).
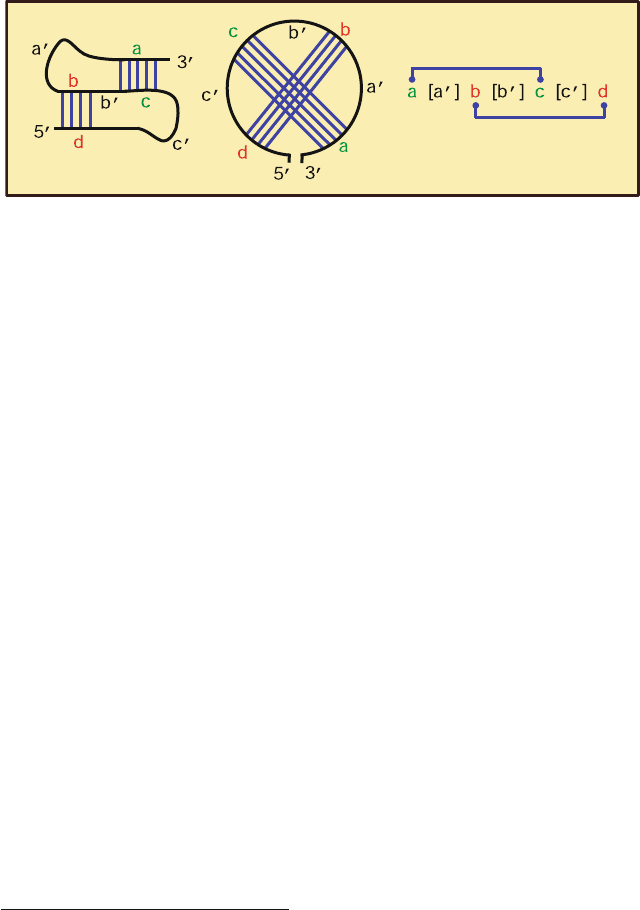
RNA pseudoknot motifs, which can be considered supersecondary structural
elements, are produced from a special intertwining of secondary structural ele-
ments by Watson-Crick base pairing (see Figure 7.5) Namely, pseudoknots form
when a consecutive single-stranded domain with segments a,a
, b,b
, c,c
and
d (a
,b
,andc
are connectors) folds to form two hydrogen bonds: a with c,and
b with d. This is a pseudoknot rather than a knot since the strands do not actu-
ally pass through one another [1367]. See Figure 7.6 for examples of RNAs with
pseudoknots.
7.3.3 RNA’s Diversity
The wonderful capacity of RNA to form complex, stable tertiary structures has
been exploited by evolution. RNA molecules are integral components of the cel-
lular machinery for protein synthesis and transport, RNA editing, chromosome
218 7. Topics in Nucleic Acids Structure: Noncanonical Helices and RNA Structure
Stem I
Stem II
Stem III
C
17
Hammerhead
ribozyme
UA
CG
GA
AU
CG
AG
AU
CG
GA
CG
GC
GC
UA
AUG
U
U
G
G
G
C
C
C
C
C
A
A
Stem I
Stem II
Stem III
5
’
3
’
5
’
3
’
17
Hairpin:
SL3 element of
Ψ site of
human immuno−
deficiency virus
type−1
GC
GC
5
’
3
’
GC
AU
UA
AU
CG
CG
G
G
G
A
Cruciform:
4−way branch
junction
C
U
U
C
G
G
G
C
C
U
UC
AG
A
AGC
UG
C
G
A
A
3
’
5
’
3
’
5
’
3
’
5
’
3
’
5
’
A
A
B
Bulge loops
& Hairpin:
hairpin ribozyme
of tobacco ringspot
satellite virus
Bulge
loops
Hairpin
3
’
U
G
UUU UC
GAA
A
A
C
C
GU
A
A
A
A
G
G
CA
U
GU
AC
CG
U
CG
GC
A
U
U
G
CG
AU
CG
A
A
A
A
A
A
A
A
G
G
C
C
U
U
U
U
5
’
3
’
5
’
U
C
U
AU
GC
AU
AU
CG
CG
AU
GU
UA
G
U
GC
GA
AU
GU
UA
CG
UA
CG
GU
AG
AG
AA
CG
AU
CG
U
A
U
G
GG
C
C
A
A
A
U
U
G
C
A
A
A
U
AU
A
G
U
C
G
C
U
A
G
U
A
U
A
A
CG
CG
UA
GU
A
G
C
C
A
A
G
U
CG
AU
GC
CG
CG
C
G
G
A
C
A
U
G
G
U
C
C
U
A
A
C
C
A
U
U
A
A
A
A
A
A
G
G
G
G
G
G
G
G
C
C
C
C
5
’
3
’
Loops, Bulge Loops
& Hairpins:
P4−P6 domain of
the group I intron of
Tetrahymena
thermophila
Loops
Tetraloop
Receptor
Tetraloop
P6b
P6a
P6
P4
P5
P5a
P5c
P5b
Figure 7.4. Various nucleotide-chain folding motifs from solved RNA systems: loops,
bulge loops, hairpins, hairpin loops, cruciforms (four-way junctions, found in recombi-
nation intermediates and synthetic designs), and the complex tertiary contacts found in
RNA (illustrated on the hammerhead ribozyme), which compact the secondary-structure
elements. Broken connections are used for non-WC bps, including mismatches. The PDB
files for the structures (clockwise from left) are as follows: tetrahymena intron, 1GID; to-
bacco ringspot ribozyme, 1B36; HIV-1 Ψ-stem loop 3, 1A1T; and hammerhead ribozyme,
1MME. The tertiary interactions shown for the hammerhead drawing (ellipses or connect-
ing line segments) are based on [1154]. The nucleotide C17 shown in space-filling form is
the strand cleavage site of the ribozyme.
7.3. RNA Structure and Function 219
Figure 7.5. RNA pseudoknots have an intertwined form of base pairing, which can be
evident from a circular representation of base pairing.
replication and regulation, catalysis and many other functions
2
. Indeed, some
scientists hypothesize that life was based on RNA (‘RNA World’) before the
evolution of the modern nucleoprotein universe [454, 906]. RNA/DNA hybrid
double helices also occur in transcription of RNA onto DNA templates and in
the initiation of DNA replication by short RNA segments.
As mentioned in Chapter 1, a highlight of RNA’s functional diversity is its
catalytic capability, as discovered in the 1980s and recognized in the 1989 No-
bel Prize in Chemistry to Thomas Cech and Sidney Altman. Such catalysis is
performed by RNA molecules termed ribozymes; hundreds of ribozyme types
are now known in a diverse range of organisms (e.g., [1142, 1208, 1251, 1275],
and some have been designed in the laboratory by in vitro selection (e.g., [1228,
1245]), as spare in composition as two base building-block units (rather than four)
[1042].
3
Many ribozymes make or break phosphodiester bonds in nucleic acid
backbones, but other biological and chemical functions are continuously being
discovered.
RNA’s conformational flexibility, modularity, and versatility [493] are impor-
tant for recognition by, and interactions with, ligands (e.g., as in the RNA of the
human immunodeficiency virus, HIV) and other molecules. These features are
of practical importance for the design of therapeutic agents that exploit RNA’s
functional sites as potential drug targets (e.g., [76,211,539,546,725,981,1268]).
The construction of ligands that bind RNA and interfere with protein synthesis,
transcription, or viral replication have potential as new therapeutic agents, such
as antibiotic/antiviral drugs [984]. These pharmaceuticals are urgent given the
resurgence of previously known viral and bacterial diseases and the emergence of
resistant mutants of common antibiotic and antiviral drugs.
2
For example, tRNA molecules carry amino acids and deposit them in correct order, mRNAs
translate hereditary information from DNA into protein, rRNA are involved in protein biosynthesis
(within a complex of ribosomal RNA and numerous proteins), gRNAs edit RNA messages (by ‘guide’
sequences), cRNAs play roles in catalysis and autocatalysis, and snRNAs are splicing agents (by
‘small nuclear’ components of the mRNA splicing machinery known as the splicosome).
3
The 83-nucleotide ribozyme composed only of two different building blocks — uracil and 2,6-
diaminopurine — was shown to catalyze the ligation of two RNA molecules with a rate 36,000 times
faster than the uncatalyzed reaction [1042]. The fact that RNA’s genetic code may be simpler than
today’s four bases lends further support to the “RNA world” hypothesis.
