Schlick T. Molecular Modeling and Simulation: An Interdisciplinary Guide
Подождите немного. Документ загружается.

220 7. Topics in Nucleic Acids Structure: Noncanonical Helices and RNA Structure
P
D
o
P9
P7
P3
P8
P1
P2
P3
P1.1
P
U1ARBD
J1/2
L3
J1.1/4
J4/2
P1
P2
P3
P1.1
P
U1ARBD
J1/2
L3
J1.1/4
J4/2
C
A
A
C
C
C
G
U
UU
AG
CG
AU
CG
CG
CGAA
U
G
G
G
A
C
A
C
C
C
U
G
G
U
A
G
C
G
G
G
C
C
G
G
G
C
C
C
U
A
C
G
G
G
C
C
G
C
C
U
U
U
C
C
C
G
G
G
5
’
3
’
P3P8 P7
P9.0
P9
A
5
’
3
’
GG
AAACC CU
P4 P6
Domain
GG G
CCUUUA
GGGGG
AAA
CUCUUCU
U
U
G
U
A
AA ACU
G
AC
AUA
G
CU
G
A
U
A
C
U
AAG
UUA
G
A
C
C
U
C
U
C
C
G
AA
A
G
G
C
G
G
A
C
U
C
A
U
U
U
G
G
G
A
J8/7
J3/4
J6/7
P1
P1.1
P2
P2
P1
P1.1
P1
P1
U1ARBD
U1ARBD
P
D
o
P8
P8
P3
P3
P7
P7
P9
P9
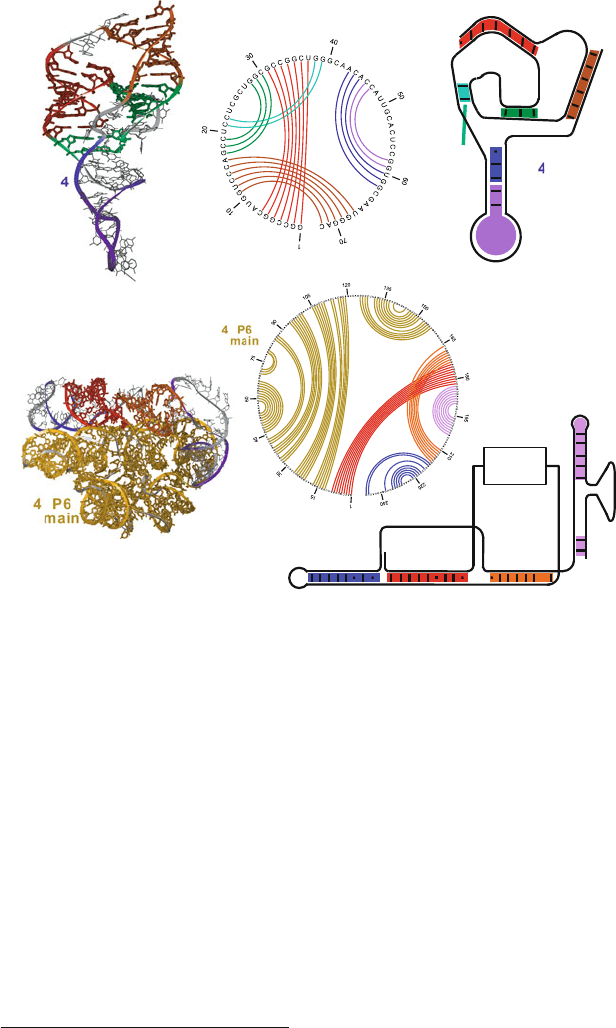
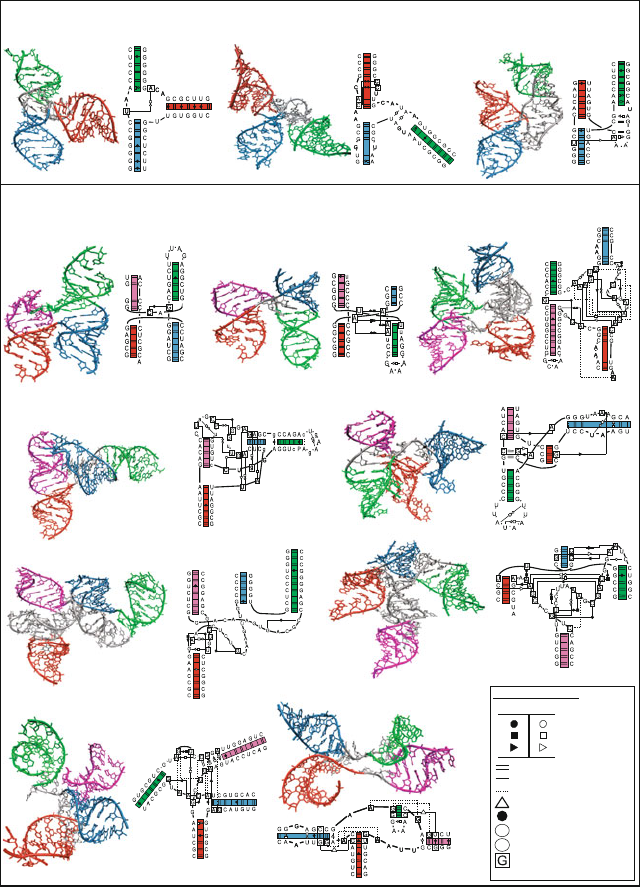
Figure 7.6. Ribozymes with pseudoknots. Top: Hepatitis Delta virus (HDV) ribozyme
(PDB code 1DRZ) shows two pseudoknots, with pseudoknot basepairing pattern drawn
in the circular diagram with basepair number along the perimeter: a main one formed by
regions P1 (red) and P2 (orange-red), and a minor one formed by regions P1.1 (cyan) and
P3 (green). A hairpin formed by the P4 helix (blue) and the U1A ribonucleoprotein binding
domain (purple) is also present. Bottom: P3-P9 domain of the group I intron Tetrahymena
thermophila ribozyme (PDB code 1GRZ) shows one pseudoknot formed by regions P3
(red) and P7 (orange-red). The P4-P6 domain (yellow) folds independently. See Box 7.3
for a discussion of some of those structures. The intertwined or pseudoknotted regions are
clearly seen by the crossings in the circular diagrams.
These applications — together with design of novel RNAs and of RNA se-
quences called aptamers that bind specific molecular targets (e.g., [166, 199,
724]) — have important potential as regulators of gene expression, therapeutic
agents, molecular switches, and molecular sensors
4
.See[546, 889, 1207, 1208,
1275,1381], for some examples.
4
RNA sensors are RNA molecules that are turned on or off when they contact a target, like small
organic molecules, and can trigger a chemical reaction.
7.3. RNA Structure and Function 221
Designing new RNAs based on this idea of aptamers has led to the exciting
experimental field of in vitro selection. This experimental technology involves
generating and screening large random-sequence libraries of nucleic acid mole-
cules for a specific function, such as binding or catalysis. Numerous nucleic acid
molecules binding targets (aptamers) have been developed, as diverse as organic
molecules, antibiotics, proteins, and whole viruses. In addition, new classes of
RNA enzymes (ribozymes) have been discovered by this approach, with applica-
tions in biomolecular engineering, such as in the design of allosteric ribozymes
and aptamer-based biosensors, aptamers capable of inhibiting protein function,
and therapeutic aptamers (e.g., inhibiting the TAR RNA element of HIV-1). Other
emerging applications of engineered RNAs include RNA nanotechnology, where
RNAs are assembled into functional arrays, and RNA synthetic biology, where
designed RNAs are used to control cellular functions (e.g., regulate gene ex-
pression). See Subsection 7.5.2 on analysis of random nucleotide pools to better
understand the relationship between sequence and structure/function and devel-
opment of a computational procedure for mimicking aspects of this selection
approach in silico.
7.3.4 Non-Coding and Micro-RNAs
Such newly found roles for RNAs, especially concerning tiny RNAs that do not
encode proteins (ncRNA for non-coding RNAs) but can influence gene action won
DNA’s cousin the venerable trophy of “Breakthrough of The Year” by Science
editors in 2002 (see the 20 December 2002 issue of Science, volume 298). Non-
protein coding stretches of mRNAs range in size from only 20 nucleotides to over
10000 nucleotides [1228]; they are required to control the translation from the
mRNA transcript into protein.
The 2002 award recognized a large group of papers that unraveled various fas-
cinating features of small RNAs (affectionately termed nanoRNAs). Micro-RNAs
(miRNAs), generally 21 to 25 nucleotides long, form a regulatory class of
ncRNAs [200]. These RNAs control gene expression by repressing translation
of target genes through, for example, binding to 3
untranslated regions of the
messenger-RNA targets.
Such small RNAs in animals, plants, and fungi collectively became associ-
ated with the title RNAi (for RNA interference). The agents that initiate RNAi
in a sequence-specific manner are double-stranded RNA segments that silence
gene expression (siRNAs, for small interfering RNAs). For example, they may
seek out the messenger RNA and destroy it, or they may bind to chromatin
and/or modify chromatin structure [383, 1004, 1432]. Though initially regarded
as anomalies, work is revealing that such siRNAs regulate gene expression in a
variety of organisms.
Such interference mechanisms by RNA silencing can provide an organism a
natural defense against invading viruses and transposons (DNA segments that mi-
grate within and across organisms and are associated with bacterial pathogenicity)
[11]. Consequently, this natural protection is being exploited by scientists using
222 7. Topics in Nucleic Acids Structure: Noncanonical Helices and RNA Structure
siRNAs to target viral genes that can inhibit the replication of HIV-1, polio, or
other viruses (e.g., [757]). Moreover, RNA interference mechanisms are being in-
vestigated by companies who apply them to discover the functions of genes by
turning them off to determine the effect on the plant or the animal. For exam-
ple, a landmark study on obesity employed RNA interference [626] to inactivate
about 85% of the roundworm’s predicted 19,757 genes that code for proteins in
a single experiment [66]. More recent studies have used RNA interference to un-
cover genes involved in regulating the immune response to pathogenic bacterial
infections [273] and genes regulating programmed cell death (or apoptosis)[229].
These fascinating discoveries regarding RNA’s interference with gene activity
are associated with many epigenetic phenomena — changes in gene expression
(inheritance of features) that do not involve alterations in the genome and persist
across at least one generation [31]. A different kind of epigenetic control was also
discovered by Breaker, Nudler, and coworkers [863,864,889,926,1275,1381]in
some bacterial messenger RNAs containing sequences that sense small molecules
directly to control translation of mRNA into protein. Namely, specific control
regions of mRNA can bind directly to metabolites associated with vitamin B syn-
thesis and import, and induce a conformational change in RNA’s folding state; this
metabolite-triggered conformational change acts as part of the signal transduction
pathway that senses vitamin level and controls enzyme production. Such RNAs
that control gene expression upon binding metabolites or other ligands have been
termed riboswitches.
A riboswitch can undergo local and global conformational changes upon bind-
ing to its substrate. The secondary and tertiary structures of these natural aptamers
can be very diverse, and several structures have been solved [873]. Examples in-
clude the type-I glmS ribozyme and purine and SAM-II riboswitches in which one
binding region induces local conformational changes upon binding. TPP, SAM-I,
and the M-box magnesium are examples of type II riboswitches because of their
two regions which undergo conformational changes upon binding their metabo-
lite, making possible global as well as local conformational rearrangements (see
Figure 7.7).
Such a switch of RNA conformation between two states in a ligand-dependent
manner (see Figure 7.7) also opens new avenues for thinking about RNA design
in a variety of contexts (e.g., [166,199,242,599,1028,1245]). Like the Paracelsus
challenge for proteins (see Chapter 2), one can formulate a similar challenge for
RNA design — describe minimal changes in the nucleotide sequence to trigger
a conformational rearrangement in the folding of a given RNA molecule — that
can be approached by a combination of computational and experimental wizardry.
Indeed, Science editor Jennifer Couzin [267] writes: “Having exposed RNAs’
hidden talents, scientists now hope to put them to work”.
7.3.5 RNA at Atomic Resolution
Until fairly recently, less was known at atomic resolution on RNA structure
in comparison to DNA, but this has changed as the ‘RNA era’ has begun.

7.3. RNA Structure and Function 223
Progress in RNA structure elucidation can be attributed to vast improvements in
crystallization procedures (e.g., RNA structure determination through crystalliza-
tion with a protein that would not interfere with the enzyme’s activity [391]), as
well as alternative approaches for studying RNAs such as high-resolution NMR,
spectroscopy, crosslinking reactions, and phylogenetic data analysis. Our knowl-
edge of RNA structures has also increased dramatically with recent solutions of
ribosomes [85, 201, 206, 210, 910, 922, 1135, 1374, 1379, 1427], since ribosomes
contain numerous tertiary motifs of RNAs and therefore provide a rich resource
of information on RNA structural elements and organization.
The clover-leaf structure of the tRNA molecule has been known for decades,
and for a long time was the only well-characterized major structure of an RNA
molecule [1173]. Its structure whet our appetite for RNA appreciation by reveal-
ing the long-distance tertiary interactions. By now, RNA folds characterized by
X-ray crystallography include the tRNA, hammerhead ribozyme, Tetrahymena
group I intron, hepatitis delta virus ribozyme, Group I intron from Azoarcus and
Twort, Group II intron, various ribozymes (e.g., hairpin, GlmS, Diels-Alder), var-
ious riboswitches (e.g., purine, M-box, and TPP), RNase P types A and B, signal
recognition particle, and various fragments such as kissing hairpin and sarcin/ricin
motifs. See some examples in Figure 7.7 and details of some solved catalytic
RNAs in Box 7.3.
Box 7.3: RNAs at Atomic Resolution
The hammerhead [1005, 1282]andHepatitis Delta Helper virus (HDV) [391]ri-
bozymes, in the family of self-cleaving catalytic RNA, were solved in 1994 and 1998,
respectively. The hammerhead’s Y, or wishbone-shaped, structure has three base-paired
stems resembling the head and handle of a carpenter’s hammer (see Figure 7.4). The RNA
is unpaired in its U-turn core and stabilized by non-WC, non-wobble bps in the stems.
Visualizing its structure led to further analysis of the mechanism of RNA self cleavage
through trapping of intermediates in the ribozyme reaction pathway [331, 886]. While the
hammerhead’s active site is open, that of HDV is hidden (see Figure 7.6), resembling the
catalytic sites of globular proteins, and contains two pseudoknots.
The crystal structure of one self-folding Tetrahymena thermophila Group I intron [488],
was solved in 2004. Its secondary structure consists of nine regions (P1-P9), which fold
into two major domains: P3-P9 and P4-P6. Both domains are stabilized through extensive
inter-domain tertiary interactions. The P3-P9 domain is formed by a coaxial helix between
P3 and P8, with a slight bend with respect to P7, while helix P9 is oriented perpendicular
to P7 (see Figure 7.6). The P4-P6 domain has a helical tetraloop region connected to
another helical segment, the tetraloop receptor,byalargebend(≈150
o
) (see Figure 7.4).
Thus, its active site is hidden and exemplifies the complexity of RNA structure, involving
complex intertwining of secondary structure elements, including pseudoknots. Indeed,
both WC and non-canonical base-paired regions are interspersed with internal loops, and
an adenosine-rich bulge region mediates the long-range tertiary contacts in the RNA,
improving base stacking interactions. The adenosine platform motif emerges from this
structure as an important architectural component of RNA that might have arisen early
224 7. Topics in Nucleic Acids Structure: Noncanonical Helices and RNA Structure
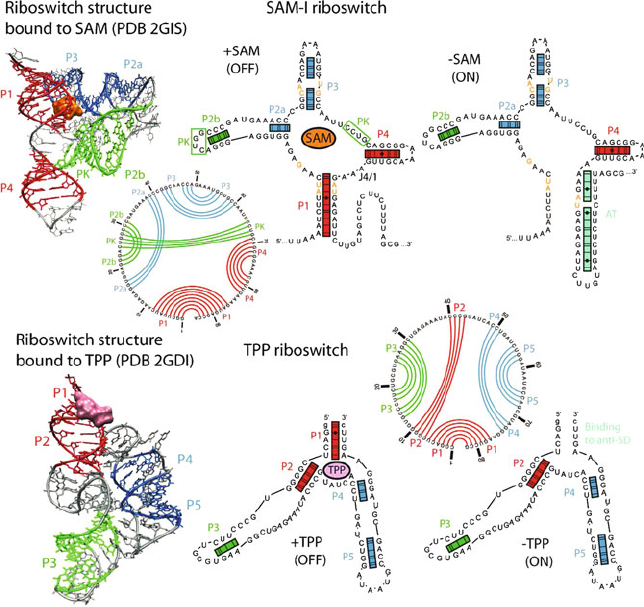
Figure 7.7. Examples of two riboswitches: SAM-I and TPP riboswitch. In the presence
of its substrate, S-adenosynlmethionine (SAM), the SAM riboswitch is composed of a
four-way junction forming a pseudoknot and a pair of coaxial helices P1–P4 and P2a–P3.
The junction is stable due to hydrogen bonding between nucleotides (orange) in helices P1
and P3 which bind to SAM. The bound state of the riboswitch was solved by crystallogra-
phy [872]. In the absence of SAM, the 3
-side of helix P1 forms an anti-terminator element
(AT), allowing the mRNA to be fully transcribed. The TPP riboswitch The thiamine py-
rophosphate (TPP) riboswitch from Escherichia coli thiM mRNA also has two forms
depending on the presence of TPP. The bound-state form has been solved by X-ray crys-
tallography [1166]. Without TPP, the 3
-side of helix P1 binds to an anti-ShineDalgarno
(anti-SD) region, allowing the ribosome to access the SD element so that translation can
start as proposed by the Breaker group [1381].
in evolution. As found in other RNA structures, metal-phosphate coordination is also
important in stabilizing tertiary contacts in this intron RNA domain. In addition to the
Group I intron of the Tetrahymena thermophila, the Group I intron of the Azoarcus [3]and
Twort [466] species are also available.

7.4. Current Challenges in RNA Modeling 225
The complete Group II intron solved in 2008 [1266] is a self-splicing ribozyme that
catalyze their own excision from precursor mRNAs and joins together flanking exons with-
out the help of proteins. The secondary structure is characterized by six domains (I to VI)
composed of step-loop structures connected by a common central core and stabilized by
long-range tertiary interactions. Domain I is the largest element and contains recognition
sequences responsible for the correct assemble of the intron in its active form. Domains
II and III enhance the catalytic efficiency of the splicing, while Domain IV contains the
open reading frame (ORF) for expression of a reverse-transcriptase enzyme. Domain V is
the most phylogenetically conserved element and contains a hairpin loop involved in catal-
ysis. Domain VI contains an adenosine that interacts with the splice site during splicing.
The Group II intron and the spliceosome share common structural and functional features,
supporting the hypothesis of a common ancestor [1266].
7.4 Current Challenges in RNA Modeling
7.4.1 RNA Folding
Deducing the functional structure of RNA molecules from the primary sequence
has been called the RNA folding problem [172, 241, 771, 1261]. The challenge
is to understand how the strong electrostatic repulsions between closely packed
phosphates in RNA are alleviated. Indeed, the stability of compact RNA forms
is strongly maintained through interactions with both monovalent and divalent
cations and by pseudoknotting.
As mentioned in connection to the multitude of hydrogen bonding patterns for
polynucleotides in Section 7.2.1, the rich variety of possibilities in RNA, as re-
viewed in [736, 739, 740], has led to proposal for new nomenclature for RNAs
and the annotation of RNA structure through the international RNA Ontology
Consortium,ROC(http://roc.bgsu.edu/)[735].
7.4.2 RNA Motifs
We now recognize at least seven major tertiary interaction motifs for RNA, as
shown in Figure 7.8 — subdivided into three classes. These classes separate in-
teractions between two double stranded helices, between a single strand and a
helix, and between two single stranded regions. A recent annotation study of 54
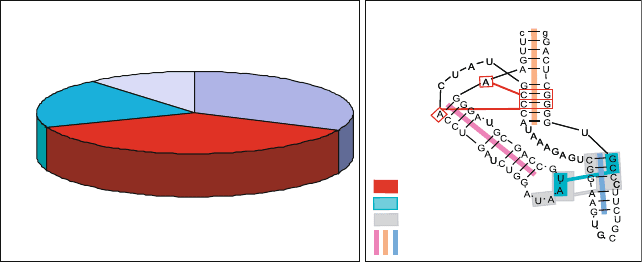
representative high-resolution solved RNA structures showed the dominance of
A-minor motifs (37%), coaxial helices (32%), and ribose zippers (20%), which
together account for 89% of the total motifs (which number 613) [1403](see
Figure 7.9). Correlations among motifs, such as a pseudoknot or coaxial helix
with A-minor, reveal patterns of higher organization and underscore RNA’s hier-
archical structure (Figure 7.9). Analyses of RNA junctions of orders 4 through 10
further suggested global patterns and subnetworks that organize RNA in complex
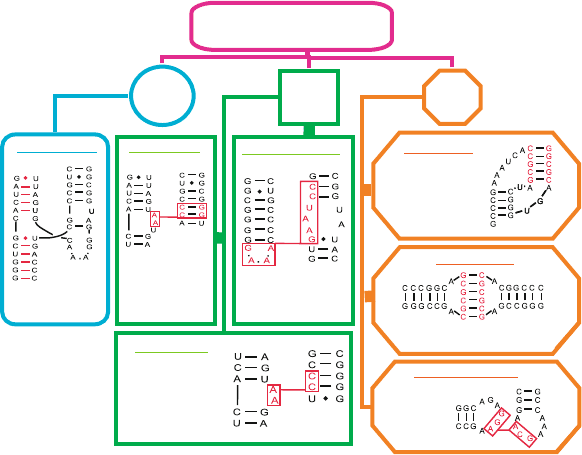
226 7. Topics in Nucleic Acids Structure: Noncanonical Helices and RNA Structure
ways as well as a classification into distinct families (Figure 7.10)[690, 691].
Such studies can help in the goal of RNA structure prediction by defining major
motifs in RNA and pinpointing specific sequence/structure relations.
7.4.3 RNA Structure Prediction
Predicting the secondary and tertiary folding of RNA is a difficult and ongoing
enterprise [172, 225, 241, 277, 689, 1022, 1145, 1465]. Secondary structure ele-
ments are easier to identify through free energy minimization combined with
comparative analysis (sequence alignment) using evolutionary and database rela-
tionships [838,839,1460,1461]. The minimized objective function is empirically
derived based on basic physiochemical laws. However, caution is warranted in the
S/S
tRNA D-loop:T-loop
Kissing hairpin
Pseudoknots
Self-complementary nucleotides
Two intertwining
regions forming
Complementary
basepairs
D/T loop
interaction
Ribose zipper
Antiparallel
stem/loop
interaction
5'-CC-3' (Stem)
3'-AA-5' (Loop)
A-minor motif
Clustering
of adenines
G-C preferred
Tetraloop receptor
Tetraloop/internal loop
5'- GAAA -3'
5'-CC-UAAG-3'
S/H
H/H
Coaxial helices
Junction,
“pseudo-stem”
5'
3' 5'
3'5'
3'
5'
3' 5'
3'5'
3'
3'
5'
5'
3' 5'
3'5'
3'
3'
5'
5'
3'5'
3' 5'
3'
3'
5'
3'
5'
3'
5'
Two hairpins
3' 5'
3'
5'
RNA Tertiary Motifs
Figure 7.8. Seven major motifs of tertiary interactions in RNA, involving two double
stranded helices (H/H), single strand and a helix (S/H), and two single stranded regions
(S/S). Coaxial helices form when nucleotide bases from two separate helices stack and
align axes to form a pseudo-continuous coaxial helix. A-minor motifs originate from clus-
tering of adenosine interactions, often present within other motifs such as coaxial helices.
Tetraloop receptor is an interaction between a tetraloop (GNRA, where N = A,G,C,U and
R = A,G) and an internal loop plus two GC bps. Ribose zippers form when two consecutive
residues from one chain segment interact in an antiparallel fashion with two consecu-
tive residues from another chain segment distant in sequence but close in space. Kissing
hairpins form by base pairing between single-stranded residues of two hairpins with com-
plementary sequences, and tRNA D-loop:T-loop are interactions between two conserved
hairpins in tRNA. See glossary of [1403] for more details and references to original motif
definitions.
7.4. Current Challenges in RNA Modeling 227
Coaxial helix
32%
A-minor motif
37%
Ribose zipper
20%
Other
11%
TPP riboswitch
(2GDI)
5’
3’
A-minor motif
Ribose Zipper
Loop-loop receptor
Coaxial helix
P1
P2
P3
P4
P5
Figure 7.9. The distribution of RNA tertiary motifs in a non-redundant set of 54 high-
-resolution crystal structures and annotated diagram of the TPP riboswitch (PDB 2GDI)
showing correlated motifs [1403].
interpretations of such 2D structures since predictions are imperfect, especially
for more than 200 nucleotides. The new structures, however, provide opportuni-
ties for learning what works, as well as what fails, in structure prediction. Thus,
discriminating among the possible tertiary interactions to obtain the final folded
state remains a challenge [277, 1115,1261]. Still, findings concerning the folding
kinetics of Tetrahymena ribozyme [1149] have suggested that, as thermodynamic
data on tertiary structure interactions become available [1261], the RNA folding
problem might be easier to solve than protein folding [101,1261]. Therefore, with
advances in RNA synthesis and structure determination [558] and the availability
of thermodynamic data on tertiary interactions [1261], it is likely that our under-
standing of RNA structure, RNA folding, and RNA’s role in enzyme evolution
will dramatically increase in the coming decade. These developments are pro-
pelling studies in RNA informatics (or ribonomics)[332] and RNA design (e.g.,
[166,199,241,242,359,599,600,689,1115,1142,1245, 1245, 1251, 1281]).
Emerging themes in RNA structure include the importance of metal ions and
loops for structural stability, hidden active sites in some ribozymes like cat-
alytic sites of proteins, various groove binding motifs (e.g., adenines interacting
with minor-groove helices to stabilize tertiary contacts, as deduced from crys-
tal structures [327, 911] and statistical analyses [1383]), architectural motifs
tailored for intermolecular interactions [546], hierarchical folding, fast estab-
lishment of 2D elements, and extreme flexibility of the molecule as a whole
[172,277,390,493,1261]. See recent review [1465].
Some key challenges concerning RNA include finding novel RNA genes,
identifying the biological roles of these RNA genes, determining the structural
repertoire of RNA, determining RNA tertiary folds from sequence, and designing
novel RNAs [1112]. Bioinformatics tools hold great promise in addressing these
challenges.
228 7. Topics in Nucleic Acids Structure: Noncanonical Helices and RNA Structure
P64
P66
P65
P67
P61
P63
P62P64
Three-Way Junctions
P57
P59
P58
Family A
23S rRNA (2J01)
P75
P79
P76
P28
P43
P29
P75
P79
P76
5'
3'
5'
3'
5'
3'
P28
P43
P29
5'
3'
5'
3'5'
3'
Family B
16S rRNA (2J00)
P57
P59
P58
5'
3' 5'
3'5'
3'
Family C
23S rRNA (1S72)
Four-Way Junctions
Family H
(RNase P type B 1NBS)
P3
P3b
P3aP3c
P7
P9
P8
P10
P1
P3P2
P4
P51
P53
P52
P54
P11
P13
P12
P14
5'
3'
5'3'
5'
3'
P3
P3b
P3a
P3c
5'3'
P1
P3P2
P4
Family cH
(HCV IRES domain 1KH6)
Family cL
(tRNA-Phe 1EHZ)
P29
P41
P30
P42
P27
P29
P28
P31
P75
P79
P76
P79
3'
5'
3'
5'
5'
3'
Family cK
(23S rRNA 1S72)
Family π
(Ribonuclease P 1U9S)
5'
3'
5' 3'
P11
P13
P12
P14
5' 3'
5'
3'
5'3'
5'3'
P61
P63
P62
P64
5'3'
Family cW
(23S rRNA 2J01)
Family
ψ
(23S rRNA 2J01)
5'
3'
5'
3'
P64
P66
P65
P67
3' 5'
3' 5'
Family cX
(16S rRNA 2J00)
5'3'
5'
3'
5'
3'
P29
P41
P30
P42
5'
3'
5' 3'
P
P
P27
P29
P28
P31
5'
3'
5'
3'
Family X
(16S rRNA 2J00)
P7
P9
P8
5'
3'
3'
5'
3' 5'
P10
5' 3'
Base pair interactions
Watson-Crick edge
Hoogsteen edge
Sugar edge
Base involved in
tertiary interaction
Cis Trans
Hydrogen bond interaction
AU cis Watson-Crick base pair
GC cis Watson-Crick base pair
P
Helix-packing interaction
RI
RII
Bifurcated base pair
Ribo-base interaction type I
Ribo-base interaction type II
B
Figure 7.10. Classification of RNA junctions into families according to coaxial stacking
properties, perpendicular helical configurations, and flexible helical arms. Lines inside the
helices represent the canonical WC bps GC, AU, and the GU wobble bp. The network
symbology follows the Leontis Westhof notation [738].
7.5. Application of Graph Theory to Studies of RNA Structureand Function 229
P1
P2
P2 6
P4 7
P2 5
P7 2
P7 3
P9 4
P9 8
P9 9
P3 6
P3 7
P3 9
P4 0
P3 8
P4 1
P4 5
P4
P4 A
P11
P12
P13
P14
P8 0
P7 4
P7 5
P8 1
P8 2
P8 8
P2
P3
P15
P15.1
P5
P15.2
P3
P2
P4
P5
P1
P2
P8
P3
P7.1
P10
P14
P16
P15
P21
P22
Five-Way Junctions
Group I Azoarcus
intron (1U6B)
23S rRNA
(
1S72
)
tRNA
(2BTE)
5'3'
P3
P5
P1
P2
P4
P5
P7
P6
P13
P14
P7
P9
P8
P10
P8
P27
P29
P28
P30
P31
5'3'
5' 3'
5'
3'
5'
3'
P2
P3
P8
P7 . 1
P10
5' 3'
5' 3'
5' 3'
5'
3'
3'
5'
5'3'
P14
P15
P16
P2 1
P2 2
5'
3'
16S rRNA
(2AVY)
5'
3'
3'
5'
5'
3'
B
P5
P6P7
P14
P13
Group II
Intron (3EOH)
5' 3'
3'
5'
Pi i
PA
PB
PC
PD
23S rRNA
(1S72)
3'
5'
3'
5'
P
RI
P
RI
P3 0
P2 7
P2 8
P3 1
P2 9
3' 5'
Ribonuclease P
(2A64)
23S rRNA
(2AW4)
23S rRNA
(2J01)
5'
3'
C
C
A
C
C
C
G
C
G
G
G
U
G
C
A
G
C
U
C
C
G
G
A
A
C
G
G
A
C
U
G
G
G
C
C
C
A
G
G
A
U
C
U
C
C
C
U
G
G
G
G
A
G
C
C
G
G
C
A
C
C
G
G
A
A
G
A
C
U
A
A
G
U
A
A
G
A
A
5'
5'
5'
5'
3'
3'
3'
3'
G
A
P4
P4 A
P11
P12
P13
P14
G
G
U
A
A
U
C
G
A
U
U
A
C
C
C
U
G
C
G
G
C
C
G
C
G
G
C
C
G
U
A
G
G
A
C
A
A
A
G
C
U
C
U
C
G
C
G
C
G
A
G
A
C
C
C
G
G
G
G
C
A
C
C
U
U
C
C
C
G
G
G
A
A
G
G
A
G
A
U
A
G
A
U
A
C
A
G
G
C
G
A
G
C
C
U
G
U
5'
3'
3'
5'
5'
3'
5'
3'
A
A
G
A
G
C
C
G
C
G
U
A
A
U
U
G
AA
A
A
G
P2
P3
P15
P15.1
P5
P15.2
5'
3'
5'
3'
5'
3'
C
A
C
C
U
A
G
G
U
A
C
U
A
U
G
U
A
G
U
G
A
C
G
U
C
A
G
U
G
A
U
C
A
U
G
C
A
U
A
U
G
A
U
U
C
A
A
C
G
G
A
C
G
G
U
G
G
U
G
C
C
A
U
C
G
U
U
5'
3'
5'
5'
5'
5'
3'
3'
3'
3'
U
U
C
G
U
A
G
C
P
U
C
C
U
C
C
G
G
A
G
G
A
A
A
A
U
P8 0
P7 4
P7 5
P8 1
P8 2
P8 8
5'3'
5'
3'
23S rRNA
(2AW4)
5'
C
U
C
C
C
C
G
A
U
A
G
G
G
G
A
G
G
C
U
A
U
G
U
A
G
C
U
C
G
U
G
A
U
C
C
G
G
G
C
A
C
G
G
C
G
G
G
U
U
C
C
G
U
C
G
U
G
G
G
G
C
C
C
C
C
G
C
G
C
G
G
C
A
G
C
G
A
C
G
C
G
C
G
U
U
U
U
G
G
C
A
C
G
A
A
A
G
G
A
A
A
A
A
G
G
C
5'
5'
3'
A
G
P
RII
P3 6
P3 7
P3 9
P4 0
P3 8
P4 1
P4 5
5'
5'
3'
5'
5'
3'
23S rRNA
(2AW4)
G
G
U
U
A
A
G
C
G
C
U
U
A
A
C
C
G
C
G
U
A
C
A
C
G
G
U
G
G
A
U
G
C
U
G
A
A
A
C
C
G
U
G
U
A
C
G
U
G
U
G
G
C
U
G
C
U
U
U
G
U
A
U
A
A
C
C
U
A
U
G
G
G
U
C
A
G
C
G
C
U
G
A
C
A
A
G
U
A
A
A
A
U
U
U
A
A
G
U
A
A
A
A
A
C
G
U
G
A
A
U
G
C
G
C
U
G
C
A
G
U
G
C
C
C
G
C
G
G
C
G
C
C
G
U
G
G
G
G
G
A
G
A
A
C
G
U
U
C
U
C
C
G
A
C
C
C
U
A
G
G
G
U
C
G
A
C
U
U
G
G
A
A
G
G
C
C
G
G
C
U
U
A
C
C
G
U
C
U
G
A
A
A
U
C
A
G
A
C
A
A
G
C
A
5'
5'
5'
5'
5'
5'
5'
3'
3'
3'
3'
3'
3'
3'
3'
5'
GU
P1
P2
P2 6
P4 7
P2 5
P7 2
P7 3
P9 4
P9 8
5'
3'
5'
3'
P9 9
Six-Way Junctions
Seven-Way
Junctions
Ten-Way Junctions
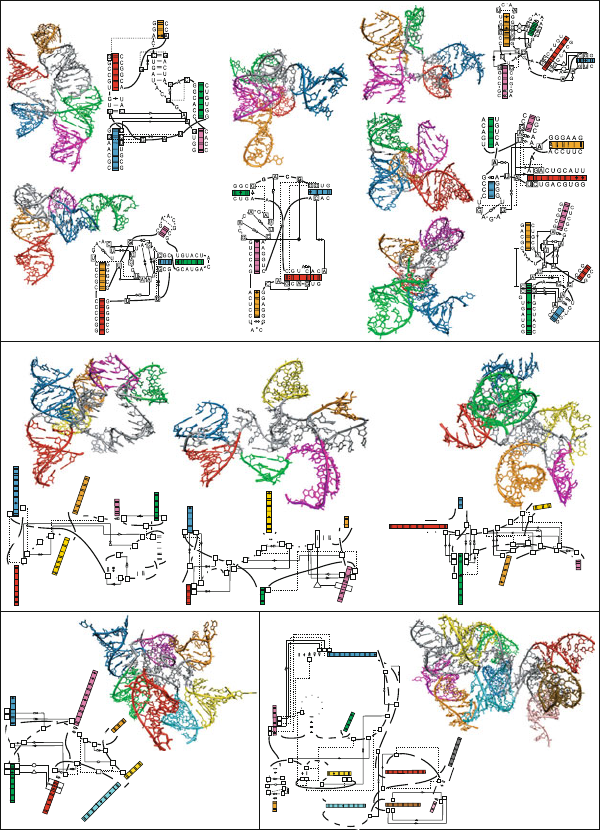
Classification of RNA junctions (continued).
7.5 Application of Graph Theory to Studies of RNA
Structure and Function
7.5.1 Graph Theory
The application of graph theory to describe secondary structure motifs of RNA,
as pioneered by Waterman in 1978 and extended by others [116, 438, 708, 1350],
