Schlick T. Molecular Modeling and Simulation: An Interdisciplinary Guide
Подождите немного. Документ загружается.

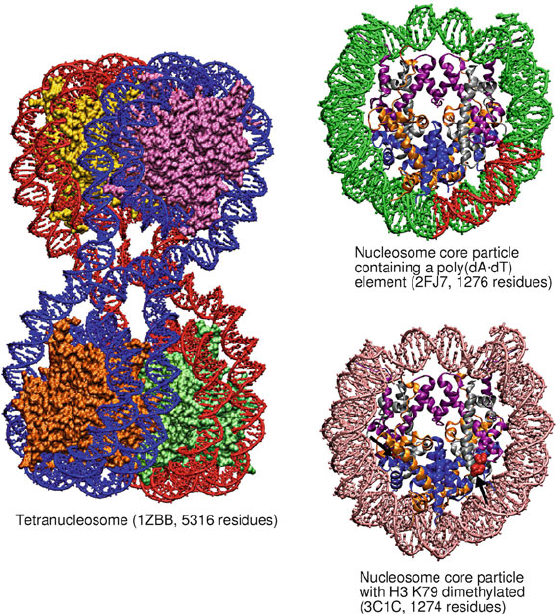
190 6. Topics in Nucleic Acids Structure: DNA Interactions and Folding
Figure 6.9. Solved crystal structures for the tetranucleosome and several nucleosomes:
tetranucleosome complex [1094]; nucleosome core particle containing a poly(dA•dT)
tract [88] (red); nucleosome core particle with H3 K79 dimethylated residues [786]. In
single nucleosome particles, the histone proteins are colored by type (i.e., H3, purple; H4,
silver; H2A, orange; H2B, blue). Arrows point to dimethylated H3 K79 residues (red).
fibers with long linker DNAs and with linker histone and divalent ions produced
strong evidence for solenoid model [1055]. Studies also suggest that, depending
on the linker DNA length and presence of linker histone, different fiber dimen-
sions are produced; in particular, short linker DNAs cannot produce compact
fibers [1055].
Modeling by Wong et al. [1388] also showed the dependence of fiber width on
the linker DNA length and the orientation of linker histones. Modeling of simpli-
fied coarse-grained nucleosome models by the Rippe group reinforced the large
effect of the linker length and nucleosome twist angles on the extent of fiber com-
paction [1219]. Our mesoscale modeling (Fig. 6.11) combined with experimental
studies of EM and nucleosome interaction measurements [483](seeFig.6.12 and
Box 6.4) suggest a compact zigzag organization for the chromatin fiber at typical
linker DNA lengths with linker histones and a more heteromorphic architecture
6.5. Cellular Organization of DNA 191
0.2
0 2 4 6 8 10 12 0 2 4 6 8 10 12
024681012
1
2
3
4
5
6
7
8
9
1
2
7
1
2
3
4
5
6
1
2
3
6
1
2
3
4
5
6
7
8
9
10
11
3
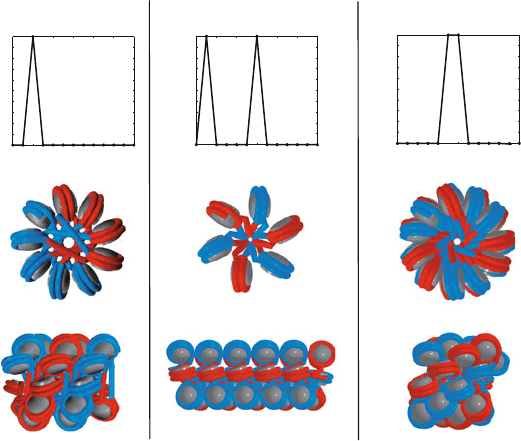
Two-start zigzag One-start solenoid Interdigitated solenoid
5
0.2
0.4
0.6
0.8
1
0
0.4
0.6
0.8
1
0
0.2
0.4
0.6
0.8
1
0
I(k)
I(k)
I(k)
kk k
6
2
1
3
5
9
8
10
9
10
11
4
5
4
9
8
7
10
7
Figure 6.10. Various hypothetical models for the structure of the chromatin fiber. Three
hypothesized chromatin arrays are shown from the top and side with black nucleosome
cores and alternating colors for the wound DNA in consecutive nucleosomes. In the classic
two-start zigzag model [1094], nucleosomes i interact with i ± 2. In the classic one-start
solenoid model [851], nucleosomes i interact with i ± 1 and i ± 6. In the interdigitated
solenoid model [1055], nucleosomes i interact on the flat sides with i ± 5, i ± 6, and on
the narrow edges at i ± 11.
of zigzag forms with straight linker DNA interspersed with bent DNA linkers at
divalent ion environments. Single-molecule force microscope studies [682] sub-
jecting 25-nucleosome arrays with two linker DNA lengths (167 and 197 bp for
the wrapped plus linker DNA) to forces up to 4 pN suggested a fundamental
solenoid organization, stiffer fibers with short linkers, and only a stabilizing but
not structure-determining effect of the linker histone. While studies are ongoing
and no consensus has been reached regarding the structure of the 30-nm fiber,
both the zigzag and solenoid models appear to be viable and relevant at various
external and internal conditions (e.g., presence of linker histones, length of linker
DNA, monovalent and divalent salt concentrations). Indeed, chromatin structure
is likely heteromorphic [483,1388].
Still, the condensation at this polynucleosomal level does not achieve the 5
orders of spatial compaction realized by the chromatin fiber near the end of the
cell cycle (metaphase chromosomes).Various looping, scaffolding, wrapping, and
specific contacts with other proteins and possibly RNA have been suggested for
this higher folding to occur. Beyond the 30-nm fiber, a higher degree of coiling is
thought to lead to fibers of dimension about 130 nm in diameter, and these forms
in turn are condensed to chromatids of dimension 200–400 nm ([844] and refs.
quoted therein).
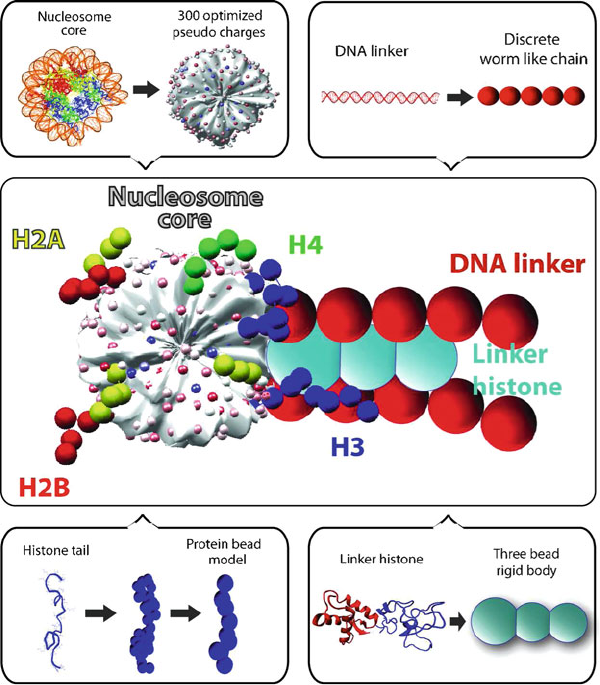
192 6. Topics in Nucleic Acids Structure: DNA Interactions and Folding
Figure 6.11. The mesoscale oligonucleosome model in which the nucleosome core with
wrapped DNA (excluding histone tails) is treated as an electrostatically charged object
with Debye-H¨uckel charges optimized to approximate the electric field using the Dis-
crete Surface Charge Optimization (DiSCO) algorithm [108,109,1240,1441]; linker DNA
are treated as beads using the wormlike chain model for supercoiled DNA; and the histone
tails and linker histones are coarse grained to approximate atomistic models [64,65, 483].
Undoubtedly, in the next decade, we will begin to understand better how this
nucleoprotein chromosomalmaterial is stabilized and packed in the cells, and how
packaging works with biological processes, such as replication and transcription,
that require full access to the DNA. Much work is in progress on understanding
the biochemical mechanisms by which the electrostatic charge density of polynu-
cleosomes is modulated (e.g., by acetylation and phosphorylation mechanisms) to
regulate transcription, and the sequence-dependent “code” in the DNA that binds
to nucleosomes.

6.5. Cellular Organization of DNA 193
Box 6.4: Simulations of Polynucleosome Folding
As mentioned in the text, little is known about the detailed organization of the chromatin
fiber — the DNA/protein complex in eukaryotes made of DNA wound around histone pro-
teins (see Figure 6.7) — both in terms of the linker DNA geometry and the nucleosome
packing arrangement in the fiber. However, the crystallographic triumphs that produced
an atomic-level view of the basic building block of the chromatin — the nucleosome core
[55, 287, 790, 1094] — provide firm foundations for complementary modeling work.
Over the past several years, our group has developed a mesoscale model of chromatin
(see recent overview [1114]) in which the nucleosome, excluding the histone tails, with
wrapped DNA is treated as an electrostatically charged object with Debye-H¨uckel charges
to approximate the atomistic electric field computed by the Poisson-Boltzmann equation
using the DiSCO (Discrete Surface Charge Optimization) algorithm [108,109,1240,1441].
In this model, each nucleosome unit is represented by several hundred charges, as shown
in the Figure 6.11, optimized so that the effective Debye-H¨uckel electrostatic field matches
that predicted by the nonlinear Poisson-Boltzmann equation [108]. This electrostatic rep-
resentation is important, since properties of chromatin are sensitively dependent on the
internal charges as well as on the ionic concentration of the medium.
Connecting these charged bodies of nucleosomes is the linker DNA, which we treat
as beads in the wormlike chain model used for supercoiled DNA. The histone tails
[62, 65] and linker histones [64] are coarse grained by beads from united-atom protein
model (Figure 6.11). Following detailed model validation against available experimental
data (e.g., translational-diffusion constants, radii of gyration, sedimentation coefficients,
etc.), as recently summarized [64], such a model simulated using Monte Carlo [63]and
Brownian dynamics permits detailed analysis of many structural, energetic, and dynamic
questions, such as the folding and unfolding of dinucleosomes and trinucleosomes as a
function of salt [109], the dynamics and energetics of nucleosome arrays as a function of
salt [1240], the roles of histone tails in stabilizing fiber architecture [64, 65], the influence
of linker histones and divalent ions in compacting the fiber structure [483], and the influ-
ence of linker DNA length on fiber organization [989].
Recent studies show that without linker histones, the fiber structure has a loose zigzag
conformation [483]. Linker histones further compact the fiber by forming stems in the
linker DNA where they closely interact with the linker histone, thereby promoting an or-
dered zigzag fiber organization. When divalent ions are introduced, bending in some linker
DNAs results to minimize steric clashes along the fiber axis; this produces a mostly zigzag
fiber accented with some bent linker DNA, resembling the solenoid form. These studies
thus lend support for the both solenoid and zigzag models; moreover, they underscore the
heterogeneous and polymorphic nature of the chromatin fiber. These dynamic fiber struc-
tures and their dependence on the ionic strength and linker histones can be seen from
Figure 6.12, which shows snapshots at low and high salt conditions (including divalent
ions), with and without linker histones. A condensation trend is apparent as the ionic con-
centration increases and as linker histones provide additional screening of the negatively
charged linker DNA.
194 6. Topics in Nucleic Acids Structure: DNA Interactions and Folding

6.6. Mathematical Characterization of DNA Supercoiling 195
6.6 Mathematical Characterization of DNA
Supercoiling
Now that we have covered many local features of nucleic acids as well as basic
global characteristics, I will introduce quantitative tools to describe how DNA is
condensed in the cell.
6.6.1 DNA Topology and Geometry
Topological methods are important for analyzing some reactions of supercoiled
DNA. For example, successful collaborations between biologists and mathemati-
cians have led to techniques that establish mechanisms for various reactions
that produce knots and catenanes (linked DNA rings) from supercoiled DNA
substrates by recombination [1239].
Basic Topological Identity
The topological method, as well as the many computational methods used to study
DNA supercoiling, rely on the fundamental identity attributed to Grigore Calu-
gareanu, Jim White, and F. Brock Fuller [279, 431, 1369]. This equation relates
the topological invariant Lk,orlinking number, to the geometric quantities twist,
Tw,andwrithe, Wr as:
Lk = Tw+ Wr. (6.3)
Linking Number
Essentially, Lk characterizes the order of linkage of two closed-oriented curves in
space (Figure 6.13). It is unchanged by continuous deformations but altered when
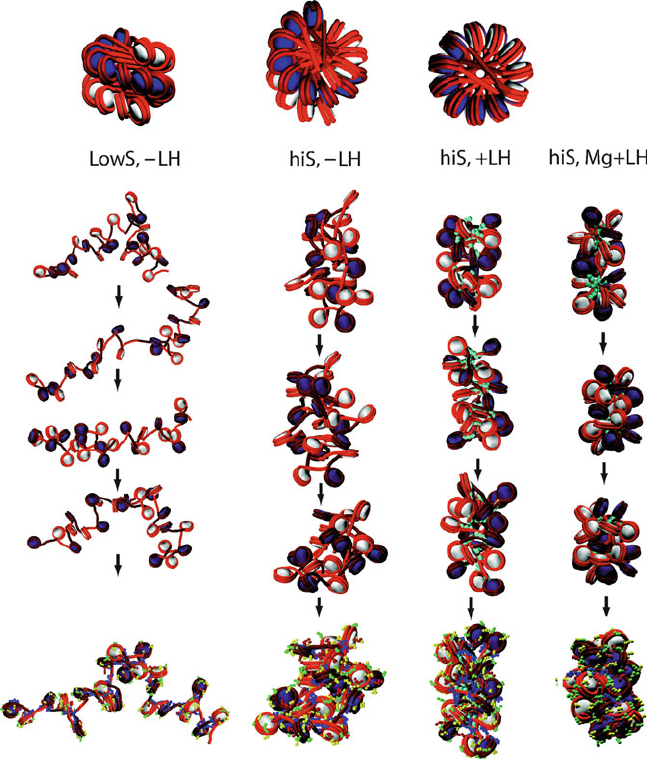
Figure 6.12. Configurational snapshots of 24-core oligonucleosomes simulated at various
conditions [989]. The top shows three different orientations of the zigzag starting config-
uration used for all series. The four simulated conditions from left to right involve: low
(0.01 M) monovalent salt without linker histone, physiological monovalent salt (0.15 M)
without linker histone, physiological monovalent salt (0.15 M) with linker histone, and
physiological monovalent salt (0.15 M) with moderate levels of divalent ions (as modeled
to a first order approximation [483]) and linker histones. The configurations are simu-
lated for 10 million steps using Monte Carlo simulations [63]. The nucleosomes are colored
white and navy in alternating order, and linker DNA segments are rendered red. The linker
histones (each represented by 3 beads) are colored in turquoise. The final configurations at
bottom also show the other tail beads colored as: H2A—yellow, H2B—red, H3—blue, and
H4—green (see mesoscale model in Fig. 6.11). Overall, we see the increased compaction
trend from left to right. At low salt, the negatively charged linker DNAs (red segments)
repel one another and expand the array, producing the beads-on-a-string form shown also
in Figure 6.7. At higher salt concentrations, the chromatin fiber condenses. Note the com-
paction as enhanced by linker histone as well as divalent ions. With divalent ions, the
mostly irregular zigzag conformation is accented by some linker DNA bending [483].
196 6. Topics in Nucleic Acids Structure: DNA Interactions and Folding
(+)
(+)
(+)
(+)
(+)
(+)
(+)
(+)
CCW (+)
CW (−)
global
helix
axis
shown
Relaxed Circle
Lk = +4 = Lk
0
Tw = +4 = Tw
0
Wr = 0
Underwound Circle
Left-handed supercoil
Lk = +3 = Lk
0
-
1
Tw = +3 = Tw
0
-
1
Wr = 0
Lk = +3
Tw = +4
Wr =−1
Figure 6.13. Descriptors of supercoiling topology and geometry. For the relaxed and un-
derwound circles of DNA, each line represents one polynucleotide strand of the double
helix. For the supercoil, the curve represents the double helical axis of the DNA.
strand breaks occur. By convention, for right-handed DNA, the linking number of
the relaxed state, Lk
0
, is the number of primary turns: Lk
0
= n/n
b
where n is
the number of bps and n
b
is the number of bps/turn (e.g., 10.5).
Lk can be rigorously computed from any planar projection of these curves onto
the plane by summing all signed crossings (ignoring self interaction) and dividing
the result by two, as shown in Figure 6.13. By convention, a crossover index is
considered positive if the tangent vector of the top vector is rotated counterclock-
wise to coincide with the tangent of the bottom curve; the sign is negative if this
rotation is clockwise. For precise mathematical definitions, refer to [431, 1370].
Twist
The variable Tw reflects the familiar concept of the helical twist angle intro-
duced earlier, namely the winding angle per residue. For the case of a helix
spinning about a global helical axis, Tw measures the number of times the he-
6.7. Computational Treatments of DNA Supercoiling 197
lix revolves about that axis. As conventional for Lk, Tw is positive if the helix is
right-handed and negative if it is left-handed (see Figure 6.13).
When the global helix axis is a closed circle and the DNA winds n times around
this planar axis, Tw = Lk ≡ Lk
0
= n/n
b
.Fromeq.(6.3), this implies that
Wr =0. This case is shown for the relaxed circle in Figure 6.13.
A nonzero writhing number is introduced by the freeing of the two ends of
the closed ring and overwinding or underwinding the duplex before resealing the
ends. Topoisomerase enzymes perform this magical act in the cell. The unwrithed
(planar) state is unfavored because there is substantial local disruption to the natu-
ral twist of the DNA (Figure 6.13). The induced torsional stress can be alleviated
through nonplanar writhing, or supercoiling, namely the twisting of the global
helix axis itself. This is illustrated in Figure 6.13 where a figure-8 interwound
forms the path of the double helical axis.
Writhe
The writhing number describes the geometry of this global helix axis in space.
Essentially, it is the average number of crossings in space of this curve. Like
the linking number, Wr can be obtained for each planar projection and averaged
over all these projections. The same sign convention used for Lk is used for Wr
(Figure 6.13).
Rigorously, the writhe can be computed by the Gauss double integral [1370]
(mathematically speaking, a map that associates each pair of curve points with
the unit sphere). For example, the simplest interwound — a figure-8 form — has
Wr = ± 1; the interwound with two self-crossings, separating the chain into
three regions, has Wr = ±2. The figure-8 interwound in Figure 6.13 (bottom)
has Wr = −1, and the interwound supercoil in Figure 6.6 (left) has Wr ≈−13.
See [1127, for example] for computational determination of Wr.
Typically, DNA is subjected to a linking number deficit,orunderwinding, with
respect to its relaxed state: ΔLk = Lk − Lk
0
< 0. This state is often described
by the normalized quantity σ =ΔLk/Lk
0
.Thevalue−0.06 is typical of DNA
in vivo. In the absence of loop-constraining agents like histone proteins, DNA
tends to absorb most of this stress into writhing, by forming left-handed supercoils
(negative Wr).
6.7 Computational Treatments of DNA Supercoiling
While the mathematical concepts of Lk, Tw,andWr have been invaluable for
interpreting supercoiling, the energetic and geometric details of the process re-
main unknown and require further computational treatments. The processes of
interest involve fundamental reactions such as replication, recombination, and
transcription, that often require, or are facilitated by, a supercoiled DNA substrate
[631,1307]. Experimental techniques such as gel electrophoresis and electron mi-
croscopy (atomic, scanning, and cryo) have been crucial for studying properties
198 6. Topics in Nucleic Acids Structure: DNA Interactions and Folding
associated with large supercoiled DNA molecules. The former allows separation
of DNA molecules into topological isomers, and the latter offers views of over-
all shapes of supercoiled DNA. However, the experimental resolution is limited
due to the large size (thousands of bps) of the DNA subjects and their extreme
floppiness in solution. Hence, theoretical tools based on numerical analysis and
computational modeling have been useful.
Computational treatments of the energetics and dynamics of supercoiled DNA
have largely relied on the theories of polymer statistical mechanics and elas-
ticity in the framework of elastic rod mechanics and dynamics. For realistic
results, non-elastic contributions — electrostatics and hydrodynamics — must
be included. These can be easily incorporated into computational models and in-
vestigated using various configurational sampling techniques (Metropolis/Monte
Carlo) [1308] and dynamic techniques (by molecular, Langevin, and Brownian
dynamics) [1105].
6.7.1 DNA as a Flexible Polymer
Very long DNA can be described by concepts from polymer statistical mechan-
ics [410]. The DNA polymer is characterized by two key quantities: a contour
length L, and a bending rigidity A. This view of a random coil slithering
through space is reasonable only for naturally occurring DNA of mixed sequences
that are not intrinsically bent at lengths much greater than DNA’s persistence
length, p
b
.
The persistence length is essentially the length scale on which the polymer
directionality is maintained. It can be computed as the average projection angle
between the end-to-end distance vector on the first bond vector of a polymer, in
the infinite-length limit [410]. The main result from polymer statistical mechanics
is that the length of this vector is proportional to the square root of the contour
length; that is, the mean square displacement, R
2
, satisfies:
R
2
=2p
b
L (6.4)
with 2p
b
as the proportionality constant.
Hence, for lengths p
b
, the DNA can be considered straight, but for lengths
p
b
, a better description is a bent random coil undergoing Brownian motion.
This is evident from the views of DNA on different length scales, as shown in
Figure 6.7. For DNA, the persistence length in vivo is around 500
˚
A or about
150 bps at physiological monovalent salt concentrations [491]. A salt dependence
of the persistence length is recognized for DNA [104], but the magnitude of this
effect is not currently understood.
The persistence length is also related to the bending force constant of a long
flexible polymer of length L and bending rigidity A via
A = p
b
k
B
T (6.5)

6.7. Computational Treatments of DNA Supercoiling 199
(k
B
is Boltzmann’s constant and T is the temperature). In this description, the
floppy polymer writhes through space as a worm-like chain. The bending rigid-
ity — which tries to keep the DNA straight — is balanced by thermal forces —
whichtendtobenditinalldirections.
A decomposition of the persistence length into static and dynamic components
has been developed [1097,1272].
6.7.2 Elasticity Theory Framework
Since the pioneering applications of Fuller [431, 432], the elastic energy ap-
proximation has proven valuable for studying global (i.e., long range and time
flexibility) features of superhelical DNA. In this approximation, the DNA poly-
mer is idealized as a long, thin and naturally straight (i.e., no intrinsic curvature)
elastic isotropic rod with a circular cross section. Homogeneous bending (i.e.,
equal flexibility in all directions) is often assumed as a first approximation,
though current computational models follow twist locally, for example by using
body-centered coordinate frames along the helical axis with associated Euler
transformations.
The elastic deformation energy can then be written as a sum of bending
and twisting potentials, with bending and torsional-rigidity constants (A and C,
respectively) deduced from experimental measurements of DNA bending and
twisting [491]. The bending term is proportional to the square of the curvature
κ, and the twisting energy is proportional to the twist deformation:
E = E
B
+ E
T
=
A
2
κ
2
(s) ds +
C
2
(ω − ω
0
)
2
ds. (6.6)
In these equations, s denotes arclength, and the integrals are computed over the
entire closed DNA curve of length L
0
. The intrinsic twist rate of the DNA is
ω
0
(e.g., 2π/10.5 radians between successive bps). The parameters A and C,
denoting the bending rigidity and the torsional rigidity constants, respectively,
can be estimated from various experimental measurements of DNA, such as the
persistence length. These force constants are key characteristics of an elastic ma-
terial.
6
See Box 6.5 for the relationship between the force constants A and C
and the bending and twisting persistence lengths, and Box 6.6 for the relation-
ship between A and measured variations in roll and tilt angles in solved DNA
structures.
6
For example, a rubber band, for which bending is facile, has a small ratio r = A/C, while
a stiff material with strong bending resistance has large r. This bending to torsional-rigidity ratio is
a key descriptor of an elastic material. It is related to both Poisson’s ratio (σ
e
) — a characteristic
of a homogeneous isotropic material — and the geometry of the rod cross section. Specifically, for
a homogeneous elastic rod of circular cross section, r =1+σ
e
. Note: the ratio A/C is usually
designated by the symbol ρ, but here we reserve ρ for the roll angle.
