Schlick T. Molecular Modeling and Simulation: An Interdisciplinary Guide
Подождите немного. Документ загружается.


170 6. Topics in Nucleic Acids Structure: DNA Interactions and Folding
in the sense of causing large global distortions. However, this would not be the
case if the sequence composition was very regular, as in poly(AT), or contains a
motif (such as A
n
) that recurs every about 10 bps (i.e., within the repeating unit
of the DNA helix); such ‘phased’ sequences may lead to substantial global effects
as a sum of local variations.
A dramatic sequence-dependent pattern of this type from the sum of small net
curvature was noted in the early 1980s in Paul Englund’s laboratory [829]in
segments of kinetoplast DNA, which displayed anomalously slow gel migration
rates.
1
Besides kinetoplast DNA, A-tracts are associated with other structures and
functions in vivo, such as core regulatory elements (found in DNA origins of repli-
cation and recombination), intergenic regions in prokaryotes, tips of supercoiled
DNA, DNA loops, and nucleosomal DNA in chromatin.
Studies of this intriguing phenomena continued in Don Crothers’ laboratory
[276, 741]. Upon close analysis, this behavior was explained by the AT-bp rich
content of these DNA sequences. Specifically, kinetoplast DNA contains stretches
of consecutive groups of adenines residues (4 to 6), separated by other sequences,
phased with the helical repeat. This composition of blocks of adenine repeated
every ≈10 bps is known as phased A-tracts. For example, a representative part of
the sequence is:
CCCAAAAATG|TCAAAAAATA|GGCAAAAAAT|GCCAAAAATC|,
where the vertical bars mark each 10 residues. Thus, the 5 or 6-membered A-tracts
in this sequence occupy positions 3–8, 4–8, or 4–9 within each 10 residues. The
significant curvature observedin association with each A-tract (roughly 15
o
)[276,
514] thus translates into a highly-curved helix when viewed over hundreds of
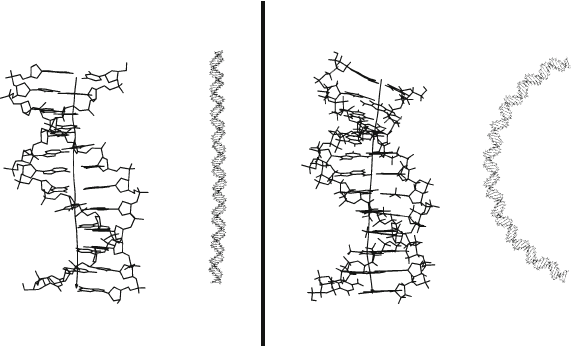
residues, as shown in Figure 6.1 (right).
The models in Figure 6.1 were built from dodecamer curvature as deduced
from MD simulations of two solvated systems [1230]: the A-tract system
CGCGAAAAAACG solved by crystallography (1D89) [312] and a control se-
quence CGCGAATTCGCG (known as the Dickerson/Drew dodecamer) [334,
1380]. The control sequence — though containing two adenines and thus ex-
hibiting limited aspects of intrinsic bending — has a small overall helix bend.
The A-tract oligomer, in contrast, exhibits an overall helix bend of about 14
o
per dodecamer. This curvature has profound implications on the plasmid level,
as seen in the figure, and explains the anomalous migration rates observed in
kinetoplast DNA.
Though we now recognize the bent helical axes in DNAs containing phased
A-tracts, a clear understanding of the origin and details of intrinsic bending has
not yet been achieved despite numerous experimental and theoretical studies [310,
492,514, 551, 800, 946, for example]. Part of the dilemma in explaining bending
involves reconciling experimental data obtained by crystallography [310]versus
1
The anomaly is a deviation from the usual linear relationship between DNA length and the
logarithm of the distance migrated on the gel.
6.2. DNA Sequence Effects 171
5’−CGCGAAAAAACG−3’
14.1
o
/12 bp
140.1
o
/
104 bp
{
10 bp
repeat
5’−CGCGAATTCGCG−3’
27.8
o
/
104 bp
6.9
o
/12 bp
{
10 bp
repeat
Figure 6.1. Bending on the dodecamer and longer-DNA levels: models of 110 bp DNA
were constructed from dodecamer systems with differing bending trends: a gentle degree of
helix bending (7
o
per dodecamer) with no preferred direction of bending, and more sub-
stantial bending (14
o
per dodecamer) with preferred bending realized as minor groove
compression, for an A-tract system (1D89 in Figure 6.2). Data are based on molecular
dynamics simulations [1230]. Scales are different for the long and short DNAs.
solution studies since environmental effects can be critical; another component is
the consideration of static versus dynamic structures; finally, there is a technical
difficulty in quantifying large helical bending in DNA; see Box 6.2.
Indeed, various bending models have been proposed for adenine-rich sequences
[1191]. At the two extremes, the junction-type model (developed from models of
junctions between two types of helices [1159]) suggests that a localized bend-
ing between two bps at regions separating A-tract regions from others largely
explains the large degree of bending; the wedge-type model proposes that bend-
ing is smooth, a cumulative effect from the small bending associated with each
AA dinucleotide step. Clearly, the realistic model involves a combination of sharp
and smooth bending models and depends on the residues that flank both the 5
and
3
sides of the A-tract [1230]. A growing body of simulations and experiments are
suggesting that the A-tract moieties themselves are relatively straight and that the
overall curvature results from substantial rolls at the A-tract junctions, between
A-tracts and external sequences, the magnitude of which is sequence modulated
[800,1230, for example].
Moreover, the factors that induce and stabilize bending include the tendency
of AT bps to be propeller-twisted and exhibit systematic differences in sugar
conformations between the adenine and thymine sugars (∼15
o
pucker angle)
[1171,1212,1230,1417,1422]. These factors combine to compress the DNA mi-
nor groove and stabilize it by bifurcated hydrogen bonds (between thymines on
one strand and adenines on the opposite strand at successive steps), as illustrated
in Figure 6.3. This geometry also leads to a stabilizing, ordered spine of hydration
172 6. Topics in Nucleic Acids Structure: DNA Interactions and Folding
−30 −20 −10 0 10 20 30
−100
−80
−60
−40
−20
0
20
40
60
80
100
θ
R
θ
T
1CGP:
CAP Binding
Protein
1CDW:
TATA−Box
Binding
Protein
93.1
o
: 2.4
o
A
1BNA: DNA −
Dickerson/Drew
18.0
o
: 3.1
o
A
1LMB:
cI repressor
1D98
1D89
11.4
o
: 3.0
o
A
27.0
o
: 3.2
o
A
90.1
o
: 2.8
o
A
14.8
o
: 3.1
o
A
Bend mag.
(
q
T
2
+
q
R
2
)
1/2
End−to−end dist per bp
CGCGAAAAAACG
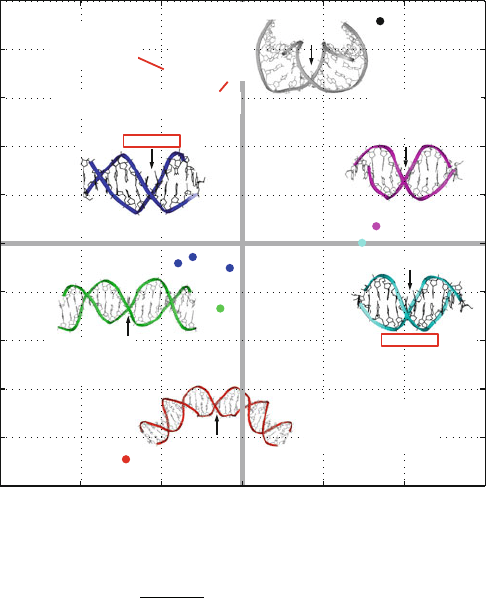
CGCAAAAAAGCG
Figure 6.2. Macromolecular examples of net tilt (θ
T
)andnet roll (θ
R
) combinations, as cal-
culated by eqs. (6.1)and(6.2), for each system in the reference plane indicated by arrows.
The bending angle magnitude (
+
θ
2
T
+ θ
2
R
) and the end-to-end distance (normalized per
bp) are also shown. Regions of positive and negative θ
R
are often termed “major-groove
compression” or “minor-groove compression”, respectively. Co-crystals of dimeric ma-
jor-groove binding proteins (like CAP and cI repressor) tend to wrap the DNA around the
protein with the minor groove at the center of curvature (θ
R
< 0). Structures with bends
toward the major groove, such as the Dickerson/Drew dodecamer or co-crystals of minor–
groove binding proteins like the TATA-box binding protein (TBP), have θ
R
> 0. The net
bend angles for the two A-tract crystal structures (1D89 and 1D98) correspond to near-zero
θ
R
(and opposite magnitudes of θ
T
) rather than bending toward the minor groove (θ
R
< 0),
as suggested by simulations and solution models. For 1D89, three crystal forms have been
solved (three filled blue circles).
in the minor groove of the A-tracts, with counterion coordination playing a role
[1182,1417]. In addition, many factors such as temperature, organic solvent, and
monovalent and divalent ions influence A-tract structure.
In their recent extensive review of the entire field of A-tract structures and DNA
bending, Haran and Mohanty postulate based on the accumulating evidence that
A-tracts appear to serve as a multi-tasking DNA element involved in various in
vivo functions and that it is the unique structure of A-tracts — rather than their
intrinsic bending per se — that explains their features and effects [515]. They
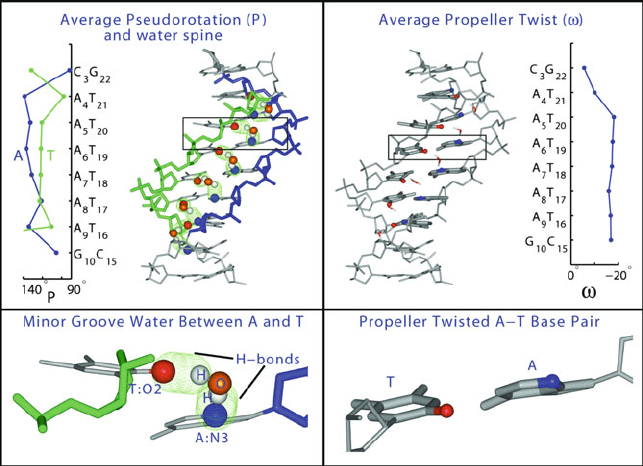
6.2. DNA Sequence Effects 173
Figure 6.3. Bend-stabilizing interactions in A-tract DNA as revealed by MD simulations
[1230] of two A-tract DNA dodecamers, 1D89 and 1D98, also shown in Figure 6.2: sugar
puckering differences between adenines and thymines on opposite strands (left plot), or-
dered minor-groove water spine (left dodecamer), and propeller twisting in the A-tract
region (right dodecamer and right plot).
rationalize their conjecture by the fact that since sequence mutations can easily
modify curvature and thus affect function drastically, this would not be advanta-
geous if A-tract’s role was primarily functional. In contrast, protein binding is a
much more effective vehicle for affecting global DNA structure and hence func-
tion, especially in eukaryotes where the DNA is packaged in the chromatin fiber
around nucleosomes.
Research continues on this important front.
6.2.5 Sequence Deformability Analysis Continues
While Crothers’ sentiment [274] is now well-accepted: “the days are over when
one dinucleotide step was as good as another in determining the structure and
mechanical properties of DNA”, our understanding of sequence-dependent de-
formability in DNA and DNA complexes is far from complete. While clearly
much effort has focused on finding a ‘set of rules’ relating nucleic-acid se-
quence to preferred structure, it is probably fair to say that we have not yet been
illuminated by a useful set of predictive rules.
Consideration of longer bp-step regions, such as trinucleotide and tetranu-
cleotide steps, as described above, is one obvious route for establishing clearer

174 6. Topics in Nucleic Acids Structure: DNA Interactions and Folding
predictive rules (and exceptions to the rules); the increasing resolution of DNA
crystal structures will undoubtedly facilitate such analyses. Still, even if a set of
rules cannot be identified, the local tendencies of DNA are important, as they are
known to work along with the deformations induced by proteins.
6.3 DNA Hydration and Ion Interactions
The binding of water molecules and ions to DNA atoms in the cellular environ-
ment is not only important for explaining DNA’s conformational stability and
energetic properties; it can also help interpret the dependence of DNA con-
formation on the environment. For example, differences in hydration patterns
(specifically with regard to accessible surface area in the grooves) are believed to
be a factor in DNA’s preference for the canonical A-form at low humidity, higher
salt concentrations, and larger amounts of organic solvents, and that of Z-DNA at
increased ionic strengths for certain sequences. Both bases and phosphate groups
have organized hydration shells [1141].
Protein/DNA and drug/DNA interactions are also dependent on water contacts.
Proteins can displace water molecules that surround certain DNA regions and
replace them with interactions involving amino acid side chains. Thus, hydra-
tion sites represent preferential, energetically favored binding sites for hydrophilic
DNA binders, like proteins and drugs [1382].
Positive counterions are fundamentally important in shielding the negatively
charged DNA backbone and allowing DNA segments to come closer together.
Dramatic effects are manifested in long supercoiled DNA molecules, as observed
experimentally and studied by simulation (e.g., [1125, 1309]).
Box 6.2: Net DNA Bending and A-Tract Analysis
A quantitative description of helical DNA bending is far from trivial. Indeed, depending
on the choice of the analysis program used for computing DNA parameters, differences
in measurements frequently result when the DNA is significantly curved. Part of these
differences can be explained by the various protocols used to define reference frames for
each base and base pair [74]. This can be resolved by using standard definitions [74]. Still,
helical bending is often measured as the angle between two extremal vectors connecting
two separated bp steps [292,405, 406,1284,1285, 1455].
As an alternative, introduced in [1230] are two net-bend angles termed net tilt and net
roll (θ
T
, θ
R
). These angles are calculated by considering the cumulative projection of tilt
and roll at each dinucleotide step, adjusted for twist, into the plane of a reference ‘average’
bp (typically at the oligomer center). From the tilt, roll, and twist at each bp-step j, namely
τ
j
, ρ
j
,andΩ
j
, we compute the net tilt and roll angles as:
θ
T
=
j
τ
j
cos
j
i=N
c
Ω
i
+ ρ
j
sin
j
i=N
c
Ω
i
, (6.1)

6.3. DNA Hydration and Ion Interactions 175
θ
R
=
j
−τ
j
sin
j
i=N
c
Ω
i
+ ρ
j
cos
j
i=N
c
Ω
i
, (6.2)
where
j
i=N
c
Ω
i
is the cumulative twist over i bp-steps from the plane of the reference
bp, N
c
, to bp-step j [1230]. Bends in the helical axis defined by θ
R
< 0 are equivalent to
‘minor groove compression’, a property associated with A-tracts [276] or with the center
oligonucleotide of the CAP/DNA complex [163, 1143]; bends defined by θ
R
> 0 are
equivalent to ‘major groove compression’, as noted for the TATA-box binding protein TBP
[163, 971] (see Figure 6.2).
The crystal versus solution data of A-tract systems differ in their characterization of
the degree of bending. Crystallographic studies have indicated various bend directions for
solved A-tract systems: negative [312, for example] versus positive [896, for example]
net tilt, as shown in Figure 6.2. Solution models of A-tract oligomers, in contrast, have
suggested a unique bend direction along negative net roll. Such data come from gel elec-
trophoresis (mobility data), NMR (minor-groove water life times), and hydroxyl radical
footprinting (compressed minor groove, as inferred from the reduced accessible surface
area in the A-tract minor groove to hydroxyl radical [OH
−
] cleavage of the DNA back-
bone). See [186, 276, 310–312, 324, 439, 520, 1017, 1213] and other citations collected in
[1230]. Simulations have shown, however, that small changes from those initially dissim-
ilar bends (from crystallographic models) can yield similar bend directions, equivalent to
minor groove compression, in accord with solution data [1230]. The crystal models may
reflect the inherent conformational disorder (multiple conformations) in A-tract systems, as
well as effects by the organic solvent needed for crystallization (2-methyl-2,4-pentanediol
or MPD). See also discussion in [946] emphasizing the importance of considering the
role of thermal fluctuations and the local chemical environment on DNA curvature and its
interpretation.
6.3.1 Resolution Difficulties
Though fundamentally important, it has been a challenge in the past to estab-
lish precise hydration and ion patterns for biomolecules (see [352], for example,
concerning the latter).
First, the number of water molecules, captured in crystal structures of DNA
and DNA complexes, is highly dependent on structure resolution, though the
high-resolution (e.g., better than 2
˚
A) structures now appearing should alleviate
this problem (e.g., [641]). Counterions, which in the past could not be located
in the electron density maps, can also be detected in ultra-high resolution DNA
structures [234, 641, 1182]. See [352], for example, for a perspective, and recent
overviews [216,675,1076].
Second, the crystal environment of the lattice — periodicity or organic sol-
vent — can also introduce artifacts, so interpretations must be cautious (see also
[946]).
176 6. Topics in Nucleic Acids Structure: DNA Interactions and Folding
Third, NMR spectroscopy measurements for locating protons and other tech-
niques are complicated by the relatively fast, reorganization component of water
dynamics (0.1 ps and longer) that can lead to overlap of spectra signals; such
experiments can yield more detailed information on the less-transient, and more
structured, water molecules, such as the minor groove hydration patterns in DNA
A-tracts.
Fourth, though computer simulations are vulnerable to the usual approxima-
tions (force fields, protocols, etc.), they are nonetheless becoming important in
deducing biomolecular hydration and ion patterns [216,380,675,1412], especially
in identifying counterion distribution patterns for DNA [239, 1076, 1419, 1421]
and RNA [69, 70, 72, 73, 172, 225, 240, 557]. Continuum solvent models are
also beginning to provide information on the relative stabilities of different se-
quences and different helical forms (e.g., [203, 1273]), as are innovative methods
such as integral methods using a 3D reference interaction-site model [1412],
modified Poisson-Boltzmann implementation [239], and grand canonical Monte
Carlo [675].
6.3.2 Basic Patterns
From all available techniques, the following facts have now been established
regarding DNA hydration and ion patterns:
1. Multiple Layers. Hydration patterns are inhomogeneous and multi-
layered, from the first layer closest to the DNA (including the nucleotide-
specific hydration patterns and minor-groove‘spine of hydration’ [125,126,
1140]) to the outermost layer of the highly transient bulk water molecules.
(Intermediate layers are characterized by fast exchange of water molecules
and ions with bulk solvent). Water interactions around nucleic acids are
important for stabilizing secondary and tertiary structure.
2. Local Patterns. Hydration patterns are mostly local, i.e., short range and
largely dependent on the neighboring atoms [1138]. For example, the water
patterns around guanines and adenines are very similar and there are clear
differences in the distributions of hydration sites around guanines and cy-
tosines that canonical A, B, and Z-DNA helices share [1140]. (Conclusions
for adenine and thymine bps are not available due to their strong prefer-
ence for B-form DNA). The strong local patterns generated near individual
nucleotides permit canonical helices to be reconstructed, complete with
preferential hydration sites [1140].
To analyze hydration patterns and ions around nucleic acids, thermody-
namic, spectroscopic, and theoretical calculations have been used. A use-
ful concept for quantifying hydration patterns is the solvent-accessible
surface — introduced to describe the proportion of buried to accessible
atomic groups [723]. The three-dimensional quantification of hydration is
typically computed as a volume-dependent probability [653,1138,1140].

6.3. DNA Hydration and Ion Interactions 177
3. Structured First Layer. The first hydration shell is generally more struc-
tured. It contains roughly 15–25 water molecules per nucleotide, with
varying associated life times. The accessible electronegative atoms of DNA
are water bound in this model, including the backbone phosphate region
(around phosphate oxygens), sugar oxygen atoms (especially O4
), and
most suitable base atoms. In particular, hydrogen bonds form in roughly
one quarter of these molecules, bridging base and backbone atoms or two
base sites. Interestingly, the accessible, hydrophilic O3
and guanine N2
groups are statistically un-hydrated [1140, 1141].
4. Cation Interactions and Counterion Condensation Theory. Debye-
H¨uckel theory, along with the counterion condensation theory (see
Box 6.3), are invaluable frameworks for understanding how counterions
distribute in electrolyte solutions.
Counterions tend to cluster around a central ion of opposite charge. The
Debye length κ
−1
(or Debye radius) is the salt-dependent distance at
which the probability of finding a counterion from a central ion is great-
est. Debye-H¨uckel theory predicts the screening factor exp(−κr) (see
Chapter 10, Subsection 10.6.4 on continuum electrostatics) by which the
ordinary Coulomb electrostatic potential is reduced (see Figure 10.10 in
Chapter 10). For DNA, the negatively charged (and thus mutually repul-
sive) phosphate residues on different strands — separated from each other
by about 10
˚
A — are effectively screened by added salts, thereby stabi-
lizing the helix and allowing the phosphates to come closer together. At
physiological ionic concentrations, the Debye length for DNA is about 8
˚
A.
Along with Debye-H¨uckel theory, counterion condensation theory can be
used to analyze the ionic environment around polyelectrolyte DNA and its
effect on thermodynamic processes [386, 822,824,1040].
As a result of screened Coulomb effects, the association of ions with DNA
spreads over a broad range, from very loose associations to tight binding,
and the timescale ranges are also broad. Experimental methods tend to
capture only average effects, but some insights are emerging from simu-
lation studies. For example, the counterion distribution around DNA that
emerged from Monte Carlo simulations [1419] revealed two distinct lay-
ers of sodium ions, the innermost of which corresponds to the condensed
counterion layer, and the outer layer more diffusive [1419, Figure 1].
2
It is well appreciated that cations interact in a non-uniform and sequence-
specific manner with the DNA grooves and the phosphate backbone
[234, 1417]. Major groove cation binding appears to be a more general
2
In nucleic acid simulations, typically one sodium ion per base is incorporated into the first hydra-
tion shell for charge neutralization; additional ions (positive and negative) are placed, with positions
adjusted by minimization or Monte Carlo procedures so as to produce ions in the most probable
regions compatible with the salt concentration of the solution (e.g., [1421]).
178 6. Topics in Nucleic Acids Structure: DNA Interactions and Folding
motif than the more sequence-specific minor-groove cation binding, as ob-
served in A-tracts [578, 850, 1255]. Furthermore, divalent cations bind to
duplex DNA in a more sequence-specific manner than univalent ions since
the former tend to be fully hydrated and thus can donate or accept hydrogen
bonds to base atoms through their water ligands [234].
Such details of cation binding to DNA have come from nucleic acid sim-
ulations and are beginning to emerge from ultra-high resolution crystal
structures of DNA [234, 641, 1182]. The facilitation of DNA bending mo-
tions by proper cation shielding was demonstrated by classic studies of
Mirzabekov, Rich, and Manning on the role of asymmetrically neutralized
phosphates in promoting bending around nucleosomes [823, 865]. These
findings were highlighted by Maher and co-workers, who examined bend-
ing promoted by neutral phosphate analogs [815], and by Williams and
co-workers, who examined ion distribution and binding in crystallographic
nucleic acid structures [850, 1182]. See [352] for a perspective.
5. Second Hydration Layer. The second water shell mainly stabilizes the
first shell, allowing it to have a favorable tetrahedral structure (see box
on water structure in Chapter 3). The outermost, bulk solvent hydration
shell is generally more disordered, with its network of interactions rapidly
changing.
6. Hydrated Grooves. The DNA major and minor grooves are well hydrated.
Though the wider major groove can accommodate more water molecules,
the minor groove hydration patterns tend to be more structured. The dif-
ferent groove shapes in canonical A, B, and Z-DNA lead to systematic
differences in hydration patterns [1139, 1140, 1182]. For example, hydra-
tion sites are better defined and form an extensive network in the very deep
major groove surface of A-DNA than those in B-DNA (less deep major
groove) and Z-DNA (very flat major groove surface); hydration sites have
higher densities in the narrower minor groove of B and Z-DNA than in
corresponding A-DNA minor-groove sites.
7. Sequence-Dependent Hydration. Hydration pattern details depend on the
sequence and helical context [1139, 1140]. The compressed minor groove
waters of adenine-rich sequences (A-tracts) are particularly associated with
long lifetimes. This results from well-ordered water interactions (‘spine of
hydration’) stabilized by cross-strand, bifurcated hydrogen bonds (e.g., N3
of A at step n +1with O2 of T on the opposite strand at step n), where the
adenine and thymines are on opposite strands and offset by one step), as
showninFigure6.3. Such patterns have been extensively studied in asso-
ciation with the central AT-rich region of the Dickerson-Drew dodecamer
(CGCGAATTCGCG) [334,763,1380]. An example of sequence-dependent
local water is shown in Figure 6.4 for three DNA sequences that differ by
one bp from one another.
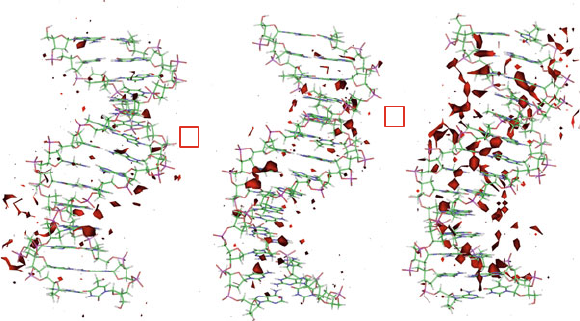
6.3. DNA Hydration and Ion Interactions 179
WB − 100% TE
ρ
max
= 0.133
T28 − 14% TE
ρ
max
= 0.180
A29 − 0% TE
ρ
max
= 0.287
G
C
T
A
T
A
A
A
A
G
G
G
C
A
G
C
T
A
T
T
A
A
A
G
G
G
C
A
G
C
T
A
A
A
A
A
A
G
G
G
C
A
−31
−31 −31
−28
−28
−28
−24 −24
−24
Figure 6.4. Local hydration patterns in three DNA systems (TATA elements differing
by one bp) as deduced from molecular dynamics simulations [1026] based on crys-
tallographic studies [971]. The sequences are variants of the promoter octamers called
TATA elements (for their rich A and T content) [163]. The experimental studies [971]
examined the structures of resulting DNA/TBP complexes (TBP is the eukaryotic tran-
scription ‘TATA-box Binding Protein’) and their relative transcriptional activities (TE
values given in figure). The simulations [1026] discerned sequence-dependent structural
and flexibility patterns, like the maximal water density around the DNA, as shown here
for three TATA sequences. The transcriptionally inactive variant shown at right, an A-tract
(GCTA
6
GGGCA), has a much denser local water environment than the wildtype sequence
(WB, GCTATA
4
GGGCA, left). The maximal water densities (number of oxygens) were
computed on cubic grids of 1
˚
A.
8. Varying Lifetimes. Water lifetimes vary, from residence times in the
several-hundred picosecond range for major groove waters associated with
thymine methyl groups to life times of order nanosecond in the minor
groove of A-tracts [763,995,1230].
9. Phosphate Localization. Hydration patterns around the phosphate groups
exhibit characteristic hydration ‘cones’ with three tetrahedrally-arranged
water molecules in the first hydration shell of each phosphate-charged
oxygen [1141]. Furthermore, the arrangements depend on the DNA helix
type. For example, the shorter distances between successive phosphate
groups in A-DNA (with respect to B-DNA) encourage water molecules
to form bridges between them. In B-DNA, these waters favor forming
hydrogen bonds with the anionic phosphate oxygens O1P and O2P; O5
is
inaccessible and O3
is surprisingly under-hydrated [1382].
