Schlick T. Molecular Modeling and Simulation: An Interdisciplinary Guide
Подождите немного. Документ загружается.


180 6. Topics in Nucleic Acids Structure: DNA Interactions and Folding
Given this multilayered organization of hydration shells in DNA and the large
costs associated with fully-solvated simulations of nucleic acids, practitioners
have attempted to economize on system size by modeling a limited number of
water molecules to represent only the thin, ordered first solvation shell around the
DNA [230,843].
Box 6.3: Counterion Condensation Theory
Counterion condensation theory essentially predicts formation of a condensed layer of
mobile and hydrated counterions at a critical polyionic charge density in the vicinity
of the DNA surface (∼7
˚
A). For monovalent cations, the concentration of this cloud is
relatively independent of the bulk cation concentration. This concentrated cloud of ions
effectively neutralizes ∼76% of the DNA charge, thereby reducing the negative charge of
each phosphate to about one quarter its magnitude (hence the charge scaling done in early
nucleic-acid simulations [523, 1013, 1260]). Divalent and trivalent counterions reduce the
residual charge further according to this model (to about one tenth of its magnitude). The
resulting phosphate screening reduces the electrostatic stiffness of DNA and also explains
the favorable entropy of binding by cation ligands to DNA, since they lead to release of
counterions from the condensed ion layer to the bulk solvent.
Various experimental and theoretical models of DNA electrostatics support counterion con-
densation of polyion charge above a critical threshold of polyion charge density. Theories
differ mainly in the structure of the condensed layer [1419]. Work on assessment of models
in light of experimental observations continues; see [850,1373], for example, for reviews.
6.4 DNA/Protein Interactions
Fundamental biological processes involve the interaction of nucleic acids and
proteins. These complexes may involve various types of proteins: regulatory,
packaging, replication, repair, recombination, and more. Many questions are now
being addressed in this large area of research involving protein/DNA complexes,
including:
• How do these proteins recognize DNA? That is, how do the protein and the
DNA accommodate each other? What geometric, energetic mechanisms are
involved?
• How are the mutual interactions stabilized? What are the specific and
nonspecific interactions involved?
• How are nucleotide and/or amino acid substitutions tolerated in the target
DNA (e.g., [971]) and bound protein regions (e.g., [734,1100])? Numerous
mutation studies (experimental and theoretical) are shedding insights into
this question and highlighting the residues that are essential to structure
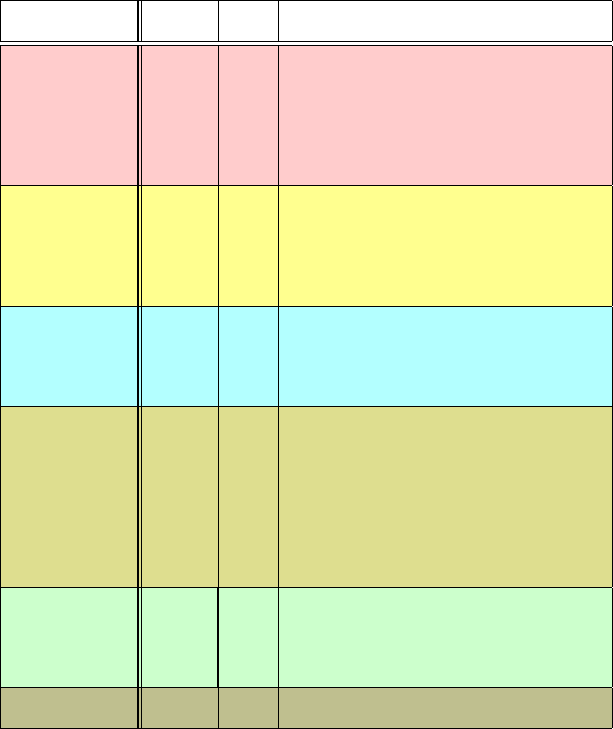
6.4. DNA/Protein Interactions 181
Table 6.2. Representative protein/DNA and drug/DNA complexes.
Complex Binding Binding Details of Complex
Motif
a
Groove
b
λ repressor HTH Mgr Canonical HTH; homodimers; 2 helices of Cro
dimer cradle Mgr, stabilized by direct H-bond
and vdW contacts; little DNA distortion.
CAP repressor HTH Mgr About 90
◦
bend.
trp repressor HTH Mgr Indirect, water-mediated base contacts.
Purine rep. HTH Mm α-helices inserted in mgr.
Yeast MATα2 HTH Mgr Homeobox domains bind as monomers.
Zif268 Zn Mgr Zinc finger subfamily; each Zn finger recognizes
3 bps.
GATA-1 Zn Mm Transcription factors subfamily; single domain
coordinated by 4 cysteines.
GAL4 Zn Mgr Metal binding subfamily; each of two Zn ions,
coordinated by 6 cysteines, recognizes 3 bps.
GCN4 Leu/Zip Mgr Canonical; basic region/leucine zipper (α he-
lices) motif; slight DNA bending.
fos/jun Leu/Zip Mgr α-helices resemble GCN4; unstructured basic
region folds upon DNA binding.
fos/jun/NFAT Leu/Zip Mgr α-helices bend to interact with NFAT.
MetJ β-ribbon Mgr Two anti-parallel β-strands in Mgr; bends each
DNA end by 25
o
.
papillomavirus
E2 DNA target
β-barrel Mgr Domed β-sheets form an 8-strand β-barrel dimer
interface with 2 α-helices in Mgr; strong tailored
fit for every base of the recognition element; bent
DNA; compressed mgr; DNA target crystallized
without protein.
TBP β-saddle mgr Ten-β-strand saddle binds in Mgr; significant
distortion, ≈90
◦
bend.
p53 tumor supp. Loop/other Mm Binds to DNA via protruding loop and helix
anchored to anti-parallel β-barrel.
SRY Loop/other mgr Isoleucine intercalated into mgr.
NFAT Loop/other Mm Flexible binding loop stabilized by DNA.
histones Loop/other Mm Nonspecific PO
4
interactions.
distamycin (drug) mgr Selective to AT bps; binds in mgr without
distortion.
a
HTH: helix/turn/helix; Zn: Zinc-binding; β: β-strand motif; Leu/Zip: Leucine zipper/bZIP;
Loop/other: motifs with few representative members.
b
Mgr: binding mainly to major groove; mgr: binding mainly to minor groove; Mm: binding to
both grooves.
and function and those that are more variable, as evidenced by evolutionary
trends. Enhancing interactions by design is also an exciting application of
such studies.
• What is the relation between structural stability of the complex and biolog-
ical function? For example, Burley and co-workers [971] showed that TBP
can successfully bind to several single-bp mutation variants of the wild type
182 6. Topics in Nucleic Acids Structure: DNA Interactions and Folding
TATA-box octamer element of adenovirus major late promoter (AdMLP),
5
TATAAAAG 3
, but that the biological transcriptional activity can be
sensitively compromised by these mutations.
These are complex questions, but we are addressing them steadily as our
collection of complexes between proteins and DNA and between DNA and
other molecules (drugs, various polymers) is rapidly growing. See [808],
for example, for a review of protein/DNA and protein/RNA molecular dy-
namics simulations and [1313] for a comprehensive review of DNA/protein
interactions.
There are now hundreds of solved protein/DNA complexes known, a repre-
sentation of which is collected in Table 6.2 and illustrated in Figure 6.5.What
has certainly been established is that the observed protein/DNA interactions span
the whole gamut of possibilities — regarding binding specificity for sequence
or grooves, stabilizing motifs, protein topology, overall deformation, tolerance to
mutations, and more.
6.5 Cellular Organization of DNA
Thus far we have discussed the structure of DNA at the atomic and molecular
levels. Understanding the organization of DNA in the cell is important for appre-
ciating DNA’s role as the hereditary material and its versatility in structure and
function.
6.5.1 Compaction of Genomic DNA
DNA’s cellular organization is critical because of the enormous content of ge-
nomic DNA. The genome size — in terms of nucleotide bps per chromosome
haploid (eukaryotic chromosomes are each made of two haploids) — varies from
organism to organism. Though the number of bps that specifies our makeup basi-
cally increases with the number of different cell types present in each organism,
it ranges greatly within an organism. Some organisms also have more genomic
content than mammals. For example, bacterial genomes have about 10
6
–10
7
bps
per haploid; for algae, the range is 10
8
to 10
11
; for mammals it is around 10
9
;yet
salamanders reach the large number of 10
10
–10
11
residues.
Table 6.3 shows representative examples of the genomic content of different or-
ganisms, along with the corresponding total length of DNA (assuming the DNA
is fully stretched). We see that this total DNA length isolated for bacterial chro-
mosomes is only of order 1 mm, but for human DNA the comparative length
is 3 orders of magnitude greater. In fact, if the genomic content in each human
chromosome would be stretched, 4 cm of DNA would result; stretching out all
6.5. Cellular Organization of DNA 183
Helix-turn-helix: λ cI repressor
Leucine zipper: fos/jun heterodimer
barrel: papilloma virus E2 factorβ
SRY (mgr intercalator)
Zinc finger: zif268
Zinc-stabilized with leucine zipper: GAL4
saddle: TATA-box binding proteinβ
Nucleosome (histones/DNA)
Figure 6.5. DNA/protein binding motifs for various complexes, with secondary structural
elements such as α-helices (red), β-strands (blue), metal ions (green), and DNA (grey)
shown.
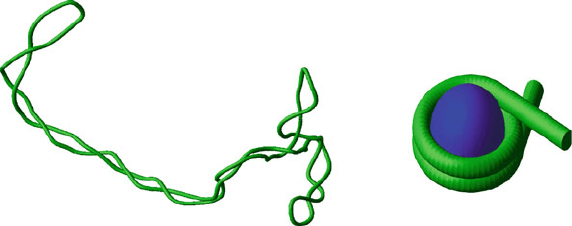
184 6. Topics in Nucleic Acids Structure: DNA Interactions and Folding
Interwound Supercoil Solenoid Supercoil
Figure 6.6. Interwound and toroidal supercoiling. Interwound supercoiling with writhing
number Wr ≈−13 (left), and a solenoidal supercoil, drawn wrapped around a protein
core to mimic the nucleosome (right).
the DNA (two haploids for each of the 23 diploid human chromosomes) would
produce 2 meters of DNA! Yet, the eukaryotic nucleus size (or the cell size in
prokaryotes) is much smaller, around 5 μm, thus more than 5 orders of magnitude
smaller. This necessitates extreme condensation of the DNA.
This condensation is largely achieved by supercoiling, the coiling imposed on
top of the double-helical coiling, involving the twisting and bending of the DNA
about the global double helix axis itself. The right-handed B-DNA form is most
suitable for this coiling, as first shown via modeling by Levitt [745]: B-DNA
can be bent smoothly about itself to form a (left-handed) superhelical structure
with minimal changes in the local structure. This property facilitates the cellular
packaging of DNA, not only by reducing the overall volume DNA occupies but
also by promoting protein wrapping in the chromatin fiber complex.
Different types of DNA compaction can be distinguished for prokaryotes and
eukaryotes. The former have supercoiled closed circular genomes (leading to the
solenoids introduced below), while the latter have linear DNA duplexes and nu-
cleosomes (leading to the toroids discussed below). Multivalent cations, such as
polyamines, are known to be important in the spontaneous condensation of DNA
to form compact, orderly toroids [138] and are likely to have a significant role in
chromosomal condensation.
6.5.2 Coiling of the DNA Helix Itself
Supercoiling is a property of both free and protein-bound DNA. When this
wrapping of the global DNA helix itself involves self-interaction, like a braid,
a plectoneme or interwound configuration results. If instead, the winding oc-
curs around the imaginary axis of a torus, a toroidal or solenoidal supercoil (or
superhelix) is formed (see Figure 6.6).
Interwound supercoiling is common for circular DNA, such as found in the
genomes of many viruses and bacteria, as well as for topologically constrained
DNA in higher organisms. In addition, eukaryotic DNA is circular in certain
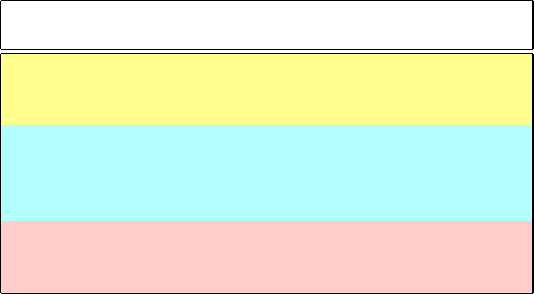
6.5. Cellular Organization of DNA 185
Table 6.3. DNA content of representative genomes.
Organism
a
kb
b
Total DNA
(# haploids)
c
Bacteriophage λ virus 49 17 μm
Haemophilus influenzae bacterium 1800 0.6 mm
E. coli bacterium 5000 1.6 mm
Yeast (S. cerevisiae) 13,000 4.6 mm (16)
Roundworm (C. elegans) 100,000 3.4 cm (6)
Mustard plant (Arabidopsis thaliana) 135,000 4.6 cm (5)
Fruitfly (Drosophila) 137,000 4.7 cm (4)
Mouse (M. musculus) 3,100,000 1.1 m (21)
Human (H. sapiens) 3,300,000 1.1 m (23)
Salamander (A. tigrinum, axolotl) 42,000,000 14.3 m (14)
a
All listed are eukaryotes except the virus and bacteria; a bacteriophage is a virus that invades
bacteria.
b
One kb = 1000 bps.
c
Contour lengths of stretched DNA are calculated based on 3.4
˚
A per bp. The number of haploids
reflected in this total DNA length is given in parentheses.
energy-producing organelles, the mitochondria. Toroidal-type supercoiling is
characteristic of the packaged form of chromosomal DNA in the chromatin fiber
(see below).
The orderly packaging of the DNA in the cell has two major roles. It con-
tributes to the flexibility of the DNA fiber and to the accessibility of DNA for
performing vital biological processes — replication, transcription, and transla-
tion — all of which require the DNA to unwind. The packaging also compacts the
chromosomal material by orders of magnitude, as required.
6.5.3 Chromosomal Packaging of Coiled DNA
Cellular DNA is organized in the chromosomes. Each chromosome in eukary-
otes (two haploids) is made up of a fiber called the chromatin that contains DNA
wound around proteins. Specifically, in this packaged form of the DNA, DNA
wraps around many large histone protein aggregates like a long piece of yarn
around many spools (see Figure 6.7). The diameter of this DNA/protein fiber in
its compact form is around 300
˚
A, an order of magnitude greater than the diameter
of the double helix [1384].
Several levels of folding are recognized for the chromatin fiber, but only the
basic units of the chromatin fiber and the associated low-level packaging are
well characterized; see [383, 567] for an overview of the basic components of
chromatin and levels of structure. This view has been deduced from various exper-
imental studies of the individual globular histone proteins (class types H1, H2A,
H2B, H3, H4) as well as of the chromatin fiber. Techniques used for the fiber

186 6. Topics in Nucleic Acids Structure: DNA Interactions and Folding
analysis include electron microscopy, X-ray diffraction, neutron diffraction, nu-
clease digestion combined with gel electrophoresis,
3
and chromatin reconstitution
in vitro.
4
The Nucleosome: DNA + Histones
A key fact established in 1974 by Roger Kornberg and Jean Thomas was that
the repeating unit of the chromatin is the nucleosome [674]. This unit consists
of about 200 bps of DNA, most of which is wound around the outside core of
histones; the remainder, linker DNA, joins adjacent nucleosomes (Figures 6.6 and
6.7).
5
The histone proteins have a large proportion of the positively-charged residues
Arg and Lys. Both residues make up between 20 and 30% of all residues: the
percentages of Lys/Arg residues for H1, H2A, H2B, H3, and H4 are 29/1, 11/9,
16/6, 10/13, and 11/14, respectively. (These proteins have 215, 129, 125, 135, and
102 amino acids, respectively). Therefore, electrostatic interactions between the
negatively charged DNA backbone and the positively-charged histone side chains
are thought to stabilize this protein/DNA complex.
In addition, as mentioned in connection to our heightening appreciation for the
subtle, sequence-dependent effects in nucleic acids, the DNA wrapped around
nucleosomes has specific regions that are more favorable to bind to nucleosomes.
Such recent discoveries concerning a “second genetic code”, the nucleosome
positioning code [1157, 1288,1425], are actively being pursued.
Nucleosome Structure
The earlier works, combined with recent chemical, enzymatic, and structural
studies (e.g., [55, 790, 1020]) suggest detailed organization of the nucleosome
units and reveal some aspects of stabilizing electrostatic interactions. For ex-
ample, based on a nucleosome crystal structure without the wound DNA [55],
Moudrianakis and collaborators have shown that the nucleosome has a tripartite
organization — assembly of two dimers of H2A–H2B, one on each side of a
centrally located H3–H4 tetramer [55]. The nucleosome was later shown to be
surrounded by a positive ion cloud with an average local density exceeding the
bulk ion concentration significantly.
In 1997, the 11-nanometer nucleosome core particle, including the wrapped
DNA, was solved by X-ray crystallography at 1.9
˚
A resolution [790](see
Figure 6.8 for a rendering of the nucleosome refined later [287] with the histone
tails), revealing further details. Namely, 146 bps of core DNA are wound on
3
In this procedure, the phosphodiester bond of DNA in solution is cleaved. This leaves chromatin
protected and therefore reveals overall chromatin organization when analyzed by gel electrophoresis.
4
Reconstitution can involve the construction of a chromatin-like fiber by adding histones to
specific sequences of DNA.
5
The lengths associated with the total nucleosomal DNA and with the linker component vary
from organism to organism and tissue to tissue. Specifically, the total length ranges from around 160
to 260 bps; the length of linker DNA ranges broadly from about 10 to 110 bps, though it is usually
around 55 bps.
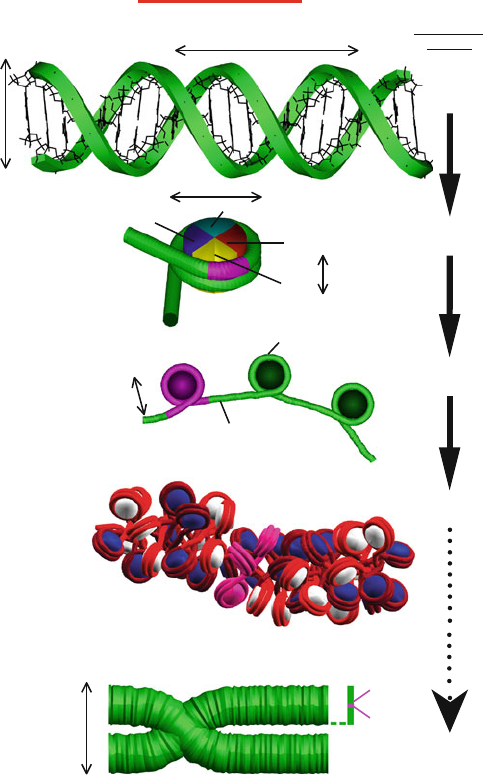
6.5. Cellular Organization of DNA 187
H2A
H2B
H4
H3
6 nm
Linker DNA ~60 bp
Wrapped DNA
~146 bp
12 nm
10 bp 3.4 nm
~
~
Duplex DNA
Nucleosome Core
146 bp wrapped DNA/
histone core
Compact Chromatin
(hypothetical structure)
Chromatin
Extended ‘Beadsonastring’
12 nm
Total DNA
Length
~0.1 p
b
(7 nm)
~1 p
b
(60 nm)
~4 p
b
(200 nm)
~10
6
p
b
(2 cm)
2.4 nm
DNA in the Cell
1500 nm
Metaphase Chromosome
30 nm
Figure 6.7. Schematic view of DNA’s many levels of folding. On length scales much
smaller than the persistence length, p
b
, DNA can be considered straight. In eukaryotic
cells, DNA wraps around a core of histone proteins (see also Figure 6.8) to form the chro-
matin fiber. The fiber is shown in both the extended view and a hypothetical compact zigzag
view (the ‘30-nm fiber’) deduced from a modeling study [483]; the compact structure of the
chromatin fiber is unknown. Chromosomes are made up of a dense chromatin fiber, shown
here in the metaphase stage. For reference, we highlight with pink in all the DNA/protein
views the hierarchical organizational unit preceding it. The length scale at right indicates
the level of compaction involved.
188 6. Topics in Nucleic Acids Structure: DNA Interactions and Folding
the outside of an octamer core of the histone proteins (two dimers each of proteins
H2A, H2B, H3, and H4) to form 1.75 turns of a left-handed supercoil; the linker
histone component H1 is thought to be a key player in regulation through binding
to the outside of this core particle and contacting the linker DNA. The wrapped
DNA has a structure very different from free oligonucleotides as well as DNA
in other protein/DNA complexes [1050]. Distinct features include excess cur-
vature, bending into the minor groove, twist alterations, and DNA stretching.
Five years later, a high-resolution nucleosome structure with tails resolved was
published [287].
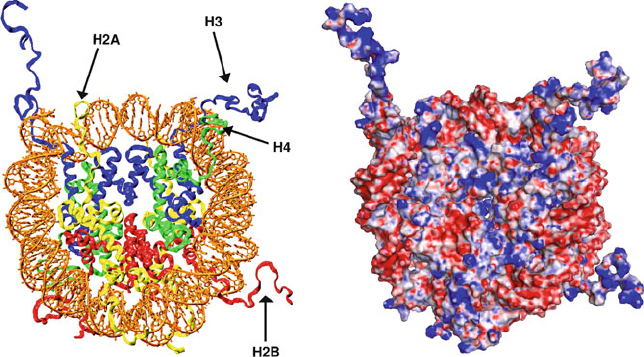
Figure 6.8 illustrates the electrostatic view that emerges from Poisson-
Boltzmann calculations (see discussion of theory and methodology at the end of
Chapter 10) for the high-resolution nucleosome core particle with resolved tails
[287]. Note the positively charged (blue) H3 and H2B tails and histone regions
inside the complex, as well as the negatively charged (red) wrapped DNA.
In 2005, a tetranucleosome was solved crystallographically [1094], and sev-
eral other nucleosome structures have since become available, with modified tails
[786], modified DNA sequences [88], as shown in Figure 6.9, and bound to drugs
like cisplatin [1392]. The nucleosome core structure shows that each histone con-
tains unstructured end regions which make important points of contact between
the protein and the DNA. Specifically, an underwinding (10.2 vs. 10.25 bp/turn)
of the nucleosome-bound DNA superhelix lines up neighboring DNA grooves to
form a channel through which the histone ends can pass. These tails likely play
key roles in regulating biological processes, such as transcription, that require a
conformational change of the complex for initiation. Ongoing investigations of
the transition between the more open and more compact nucleosome structure
will help us understand better transcriptional regulation and DNA packaging.
Polynucleosome Assembly
The nucleosomal packing described above — superhelical DNA around a his-
tone core — represents only a tenfold compression of the DNA. This is because
≈166 bps of DNA (or ≈ 560
˚
A of contour length if stretched) are organized as
a core cylindrical particle of dimensions 110 ×110×55
˚
A, where 110
˚
Aisthe
diameter and 55
˚
A represents the height of this unit. Chromatin fibers within inter-
phase chromosomes possess a hierarchical structure. At low salt concentrations, a
beads-on-a-string model is produced which compacts the DNA further by a fac-
tor of about 40. At physiological ionic strengths, it is believed that a next level of
chromosomal organization is the so called “30 nm” (300
˚
A) chromatin filament
observed by electron microscopy.
Several models for this polynucleosomal level of folding have been sug-
gested based on X-ray crystallography and electron microscopy imaging. Yet,
a consensus has not been reached despite decades of intense research, as reviewed
recently [1271, 1297]. The two broad classes of structures for the chromatin
fiber [330] include two-start zigzag structures, where the nucleosomes criss-
cross one another around the helical axis with straight linker DNA and dominant
interactions between next-nearest neighbors (i
±2), and one-start solenoid struc-
6.5. Cellular Organization of DNA 189
Figure 6.8. The nucleosome core particle (left) [287] and its corresponding electrostatic
surface potential (right) as computed from the Poisson Boltzmann (PB) equation with
the PBEQ module [586]intheCHARMM program [175] (see Chapter 10 for theoret-
ical framework and algorithms). The grid resolution for solving the linear PB equation
was 1.5
˚
A and 1.0
˚
A after focusing; the salt concentration was set to 0.15 M, and par-
tial charges from the CHARMM program were used as input. As customarily done for
macromolecules [903], the unitless (i.e., relative) dielectric constant
r
is set to the numer-
ical value of 2 inside and 80 outside the macromolecule (i.e., 2 and 80 times the vacuum
dielectric constant
0
).
tures, where nucleosomes arrange helically around the fiber axis with bent linker
DNA and dominant interactions between nearest neighbors of nucleosomes (i±1)
(see Figure 6.10).
The solenoid was proposed in 1977 by John Finch, Aaron Klug, and collab-
orators [401] based on electron micrographs that suggested a helical form with
6 nucleosomes per turn stabilized by contacting linker histone molecules. The
zigzag form [330] is evident in the recent crystallographic structure of the tetranu-
cleosome [1094], for example, with short linker DNAs between nucleosome and
without linker histones.
Indeed, this unsettled puzzle for the fiber’s organization is evident in views pre-
sented over 22 years in various editions of the classic Molecular Biology textbook
[17]. The 1986 (second) edition showed chromatin as a zigzag, while the 1994
(third) edition presented a solenoid; eight years later, the solenoid remained in
the fourth edition, but the 2008 (fifth) edition illustrates both zigzag and solenoid
models.
Only recently, have researchers begun to dissect the influence of key inter-
nal and external factors such as length of the connecting linker DNA segments
between nucleosomes (which can vary from 10 to 70 bp), the binding of linker
histones, and the presence of various concentrations of monovalent and divalent
ions on chromatin structure. For example, electron microscopy of reconstituted
