Schlick T. Molecular Modeling and Simulation: An Interdisciplinary Guide
Подождите немного. Документ загружается.

230 7. Topics in Nucleic Acids Structure: Noncanonical Helices and RNA Structure
junction
bulge
loop
stem
Tree
Dual
edge vertex
vertex
edge
edge
edge
edge
edge
edge
edge
vertex
vertex
junction
bulge
loop
stem
vertex
vertex
vertex
vertex
stem
2bp
1bp
2b
bulge
loop
We consider:
Translating RNA motifs to
graph theory objects
B0
G
C
G
A
C
C
G
A
G
C
C
A
G
C
G
A
A
A
G
U
U
G
G
G
A
G
U
C
G
C
5'
10
20
3'
S1
S2
B1
B2
Leadzyme
Tree Graphs
Dual Graph
B2
B1
S1
S2
G
C
G
G
A
U
U
U
A
G
C
U
C
A
G
U
U
G
G
G
A
G
A
G
C
G
C
C
A
G
A
C
U
G
AA
G
A
U
C
U
G
G
A
G
G
U
C
C
U
G
U
G
U
U
C
G
AU
C
C
A
C
A
G
A
A
U
U
C
G
C
A
C
C
A
5'
3'
10
20
30
40
50
60
70
B3
B1
B2
B0
B4
S1
S2
S3
S4
B2
B1B3 B4
S2
S1
S3
S4
B2
B1
B3
S1
S2
S3
S4
B0
B4
B2
B1
B0
S1
S2
tRNA
>
>
>
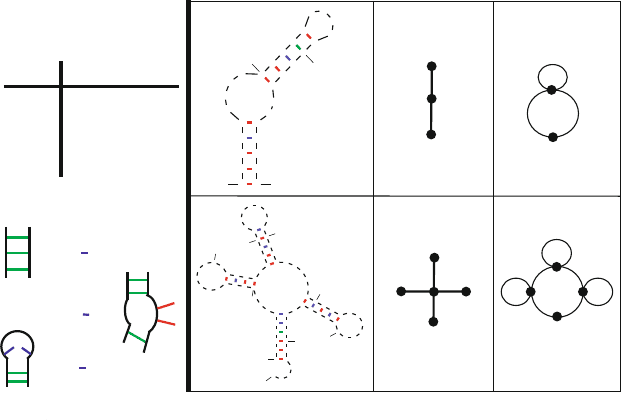
Figure 7.11. Graphic Representations of RNA Secondary Structures shown for two RNAs,
using Tree and Dual Graphs, using the definitions at right [388, 438].
holds promise in the area of RNA structure analysis. Graph theory is suitable for
RNA because this field of mathematics is widely used for analyzing networks
and enumerating structural possibilities, including chemical structures, genetic
and biochemical networks, monetary transactions, terrorist organizations, social
networks, and the Internet [33,900]; see also the 24 July 24 2009 issue of Science,
volume 325, devoted to networks.
Graph theory can be used to represent RNA secondary structures as two types
of graphical objects: trees and dual graphs according to specified rules for defin-
ing RNA stems, bulges, junctions, and loops (Fig. 7.11). All possible RNA 2D
structure motifs, as simplified by this approach, can then be cataloged by the
graph vertex number, as well as ranked by topological complexity according to
the second-smallest eigenvalue of the Laplacian matrix of the graph [388,438].
7.5.2 RNA-As-Graphs (RAG) Resource
One such RNA topology resource, RAG (RNA-As-Graphs) (http://monod.
biomath.nyu.edu/rna), is being used to classify/analyze topological character-
istics of existing RNAs [438, 645](seeFig7.12 for graph enumeration segments
from RAG) and to design and predict novel RNA motifs [643–646]. See also some
RNA reviews commenting on graph theory applications to RNA [542,736].
Specifically, RAG and the associated graph theory framework for RNA [388,
435] have been used to classify and catalog RNA motifs [438, 645], esti-
mate the size of RNA’s structural/functional repertoire [645], detect structural
7.5. Application of Graph Theory to Studies of RNA Structureand Function 231
and functional similarity among existing RNAs [965], identify RNA motifs of
antibiotic-binding aptamers (found synthetically) in genomes [702–704], analyze
the structural diversity of random pools used for in vitro selection of RNAs [450],
simulate aspects of the process of in vitro selection in silico [643, 644, 646],
and analyze RNA thermodynamics landscapes to better understand riboswitch
mechanisms to ultimately enhance their design [1028].
Other applications of RAG in the community which include graph theory ex-
tensions involve classification and prediction of ncRNAs [503,633,801,901,902,
1181], various RNA structure analyses [50,78,148,170,533,534,538,982,1041],
and various applications of graph theory [82,416,470,471,538,761,1324,1431].
Examples of graph extensions include labeled dual graphs [633] and directed
tree graphs [503]. Our application of spectral theory to catalog RNA graphs
has also been extended to other biological and physical systems [82, 416, 470,
471,761,1324,1431]. See Kim’s thesis for a more detailed descriptions on these
applications [642].
RNA Structure Enumeration
Cataloging based on graph theory enumeration suggests that the RNA structure
universe is dominated (more than 90%) by pseudoknots, in agreement with avail-
able data [645], as also discussed in [78,170,982,1041]. Significantly, the existing
RNA classes represent only a small subset of possible 2D RNA motifs as enumer-
ated by graph index; some of these motifs may be natural while others may be
possible to generate in the laboratory. Still, others may not exist.
RNA-Like Motifs
The usage of clustering techniques to separate graphs that are ‘RNA-like’ from
those that do not resemble natural RNAs also led to predictions of many new
RNA-like motifs, including ten specific examples of sequences that might lead
to novel-like RNA topologies [645], as shown in Figure 7.13. Some of these
motifs predicted in 2004 have since been solved: C1 in mammalian CPEB3 ri-
bozyme [1084], C2 in a purine riboswitch [819], C3 in the tymovirus/Pomovirus
tRNA-like 3
UTR element [840], C4 in the tombusvirus 3
UTR region IV [1463],
and C7 in the flavivirus DB element [235]. Significantly, the predicted and actual
sequences have between 45 to 51% homology.
Graph theory tools are also natural for comparing RNA structures to find ex-
isting RNA motifs within large RNAs based on graph isomorphisms [965]. This
idea was applied to identify topological similarities among existing RNA classes
and to define motifs of RNA within larger RNA topologies for major RNA classes
(e.g., tRNA, tmRNA, hepatitis delta virus RNA, 5S, 16S, 23S rRNAs).
Furthermore, the representation of RNAs as graphs led to identification of RNA
motifs in genomes. Since natural aptamers exist in many bacterial genomes and
other organisms, it appeared likely that natural counterparts of synthetic motifs
exist in vivo. This led to development of an efficient search tool for identifying
small RNA motifs in genomes by exploiting many artificial motifs derived from
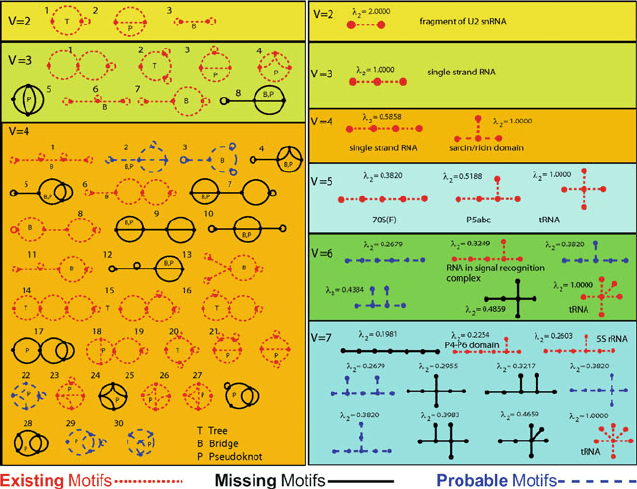
232 7. Topics in Nucleic Acids Structure: Noncanonical Helices and RNA Structure
Figure 7.12. Segments from RAG showing some enumeration of graphs for tree and dual
segments of low V numbers. The graphs are coded according to motifs found in nature
(red), motifs not yet found that are RNA-like (blue), as determined by clustering analysis,
and remaining motifs (black) [645]. See RAG website for details and updates.
RNA in vitro selection experiments [702–704]. The search for antibiotic-binding
aptamers produced 37 candidate sequences from bacterial and archaeal genomes.
RNA Design
Finally, graph theory tools can also advance the design of novel RNAs by
mimicking the process of in vitro selection in silico. As a first step, understand-
ing the structural diversity of random pools was important for improving in vitro
technology. It is simple to generate random pools computationally, but graph
theory allowed rapid analysis of the resulting 2D motifs using 2D folding al-
gorithms [450]. By characterizing the distribution of secondary structure motifs
in computer-generated random pools using sets of possible RNA tree structures
from graph theory [450], we showed that random pools do not have a uniform
distribution of possible topologies and instead favor simple topological motifs.
This was expected from experimental observations that typically yield simple mo-
tifs. Further studies also found that the proportion of multiply branched structures
increases with sequence length, in agreement with other studies.
7.5. Application of Graph Theory to Studies of RNA Structureand Function 233
single strand RNA
(NDB:PR0055)
bulged hairpin
(Rfam:CopA)
DsrA RNA
(Rfam:DsrA)
single strand RNA
(NDB:PR0055)
bulged hairpin
(Rfam:CopA)
single strand RNA
(NDB:PR0055)
C1
C2
C3
C4
C5
C6
C7
C8
C9
C10
DsrA RNA
(Rfam:DsrA)
bulged hairpin
(Rfam:CopA)
single strand RNA
(NDB:PR0037)
DsrA RNA
(Rfam:DsrA)
Flavivirus DB element
(Rfam:RF00525)
Purine Riboswitch
(Rfam: RF00167)
Tymovirus tRNA-like
3' UTR element
(Rfam: RF00233)
Candidates Discovered After 2004
RNA Secondary Structure With
Natural Submotif
Graph Representation
With Natural Submotif
Tombuvirus
3' UTR region IV
(Rfam: RF00176)
Mammalian CPEB3 Ribozyme
(Rfam: RF00622)
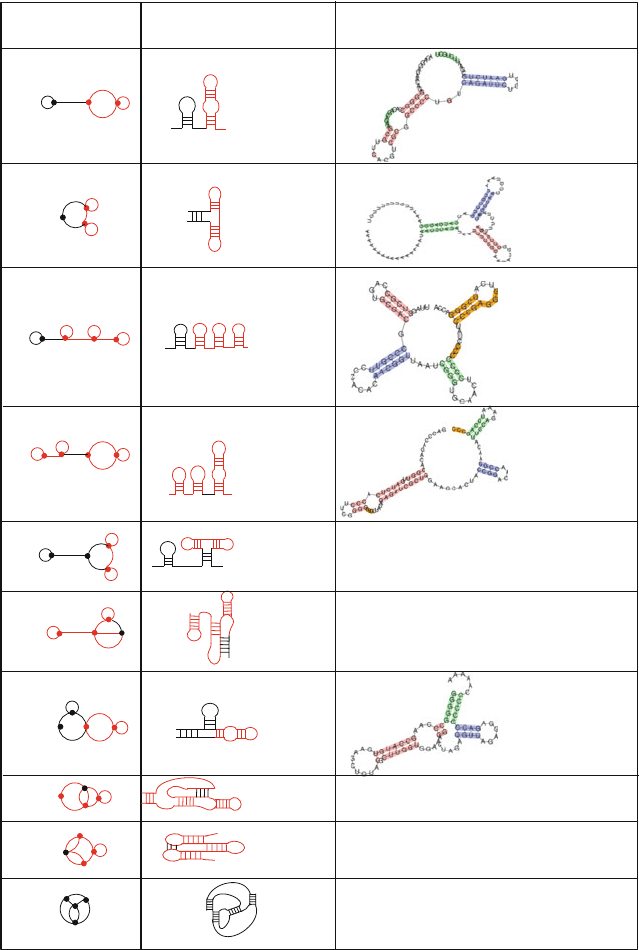
Figure 7.13. Ten candidates predicted to have RNA-like topologies, as determined by clus-
tering analysis, represented as dual graphs; the red submotif occurs in natural RNAs [645].
Of those ten, five RNAs were discovered since the published predictions, as shown in the
third column.
234 7. Topics in Nucleic Acids Structure: Noncanonical Helices and RNA Structure
4x4 mixing matrix M
RNA sequences
A
C
G
U
A
C
G
U
A
C
G
U
A
C
G
U
ACGUAGGGACCGUUU
UAAUAGGGACUACUU
UCCCUAGGGUACUAG
UACUGUAGUACUUAC
UCAGUUUUGGGGUAA
AAUAAUGAACUGACG
ACUGACCCUACGUAC
CAUGAUUUACGAUAC
~ 10
15
M
MMM
MMMM
M
MM
M
M
M
MM
AA
CA
GA
UA
AC
CC
GC
UC
AG
CG
GG
UG
AU
CU
GU
UU
Aptamers
Task
1. Specifying
sequence
mutations
Chemical probing
with functional assays
2. Generating
nucleotide
pools
3. Characterizing
structure and
function of
sequences
4 vials containing
A, C, G, and U
2D folding of sequences,
analysis of graph motifs
Initial
Structure
Candidate aptamer
sequences
Experiment Theory
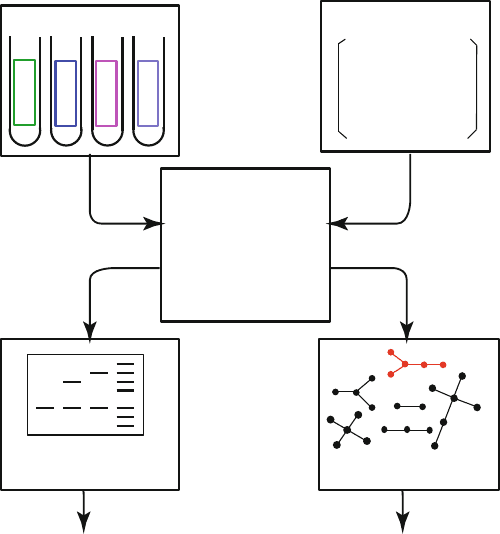
Figure 7.14. Experimental RNA pool synthesis and an in silico approach to mimic the pro-
cess [643,646]. Pool generation can be simulated using the mixing matrix, which specifies
nucleotide mixtures in synthesis port or mutation rates for all nucleotide bases and can be
defined to mimic specific biological situations [646]. The resulting sequences are “folded”
into 2D structures using existing algorithms and analyzed further to screen and filter the
candidates.
Mimicking the experimental process in silico can be done on the basis of
a “mixing matrix” framework (see Fig. 7.14), which led to development of
RAGPOOLS, a publicly-available web server for pool design [643]. Such mixing
matrices specify the mixing ratios of nucleotides in synthesis ports (see Fig. 7.14).
When applied to a starting sequence, each mixing matrix will generate a pool of
specific sequences via mutations of the given sequence. Thus, different design
strategies (based on covariance mutations like AU to CG or conversion of AU to
CG base pairs) produce different sequence pools, and the main idea is to design
these pools to yield the desired structure and/or function.
Using this new tool and other computational and experimental resources,
very large pools of nucleotides (up to 10
14
) can be generated, screened, and
filtered according to various 2D-structure similarity and flanking sequence
7.5. Application of Graph Theory to Studies of RNA Structureand Function 235
analyses [644]. Such computational and theoretical yields agree for simple RNA
motifs. For real aptamer targets, the in silico procedure overestimates the yields
found experimentally, as expected, because experimental yields represent lower
bounds and the screening does not yet involve 3D structural aspects.
Advances on all these fronts are ongoing.
236 7. Topics in Nucleic Acids Structure: Noncanonical Helices and RNA Structure
2 3 4 5 6 7 8 9
−0.2
−0.1
0
0.1
0.2
0.3
0.4
0.5
50 100 150 200 250 300 350
0
0.5
1
1.5
2
2.5
3
50 100 150 200 250 300 350
0
1
2
3
4
0 1 2 3 4 5
0
50
100
150
200
Morse
Quartic
(special)
Cubic
...........
Quartic
Harmonic
r (bond length) [A]
Bond energy [kcal/mol]
2 fold
3 fold
sum
.........
τ
(dihedral angle) [deg.]
Dihedral angle energy [kcal/mol]
O3'
C3'
C2'
O2'
O
P
O5'
C5'
Coulombic energy [kcal/mol]
LJ Energy [kcal/mol]
C...C
O...H
110 115 120 125 130
0
0.2
0.4
0.6
0.8
1
1.2
1.4
θ
(bond angle) [deg.]
Bond Angle energy [kcal/mol]
Harmonic
Trig.
τ
(dihedral angle) [deg.]
0 0.1 0.2 0.3 0.4 0.5
−5000
0
5000
r (distance) [A] r (distance) [A]
C...C
O...H
Dihedral angle energy [kcal/mol]
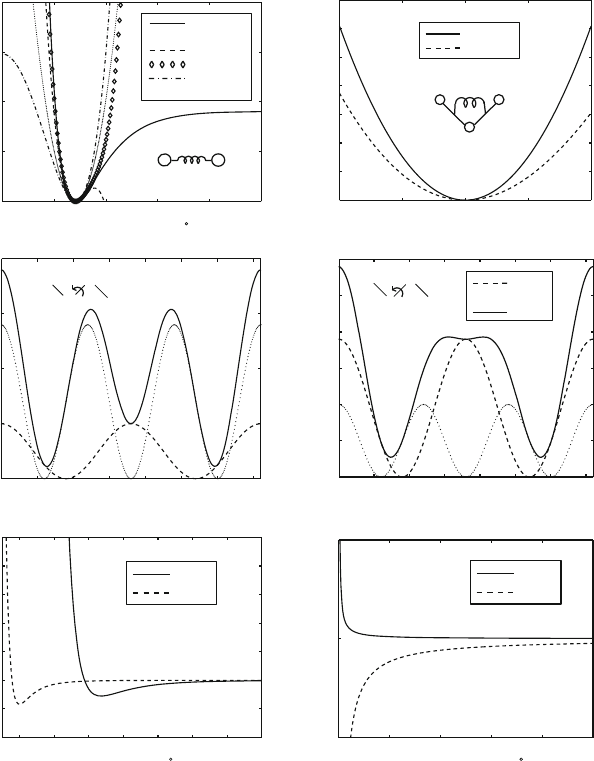
8
Theoretical and Computational
Approaches to Biomolecular Structure
Chapter 8 Notation
S
YMBOL DEFINITION
Vectors
a, b, c interatomic distance vectors (in definition of τ)
n
ab
, n
bc
unit normals
r
ij
interatomic distance vector (x
j
− x
i
)
x
i
position vector of vector i, components x
i1
,x
i2
,x
i3
P
collective momentum (for nuclei and electrons)
X
collective position (for nuclei and electrons)
X
collective position (nuclei only), components
x
1
,...,x
N
∈R
3N
V collective velocity (nuclei only)
ψ
n
eigenfunctions of the Hamiltonian operator
Scalars & Functions
b
i
bond length i
q
i
Coulomb partial charge for atom i
r
ij
interatomic distance (between atoms i and j)
t
0
initial time
A
ij
,B
ij
Lennard-Jones coefficients
H
Hamiltonian operator (E
k
+ E
p
)
E
bond
bond length energy
E
bang
bond angle energy
E
coul
Coulomb energy
E
k
kinetic energy
E
local
local (short-range) energy
E
n
eigenvalues of the Hamiltonian operator (quantum states)
E
nonlocal
nonlocal (long-range) energy
E
p
potential energy (also E)
T. Schlick, Molecular Modeling and Simulation: An Interdisciplinary Guide, 237
Interdisciplinary Applied Mathematics 21, DOI 10.1007/978-1-4419-6351-2
8,
c
Springer Science+Business Media, LLC 2010
238 8. Theoretical and Computational Approaches to Biomolecular Structure
Chapter 8 Notation Table (continued)
S
YMBOL DEFINITION
E
tor
torsional (dihedral) angle energy
E
LJ
Lennard-Jones energy
K
ijk
bond angle force constant
M
F
dimension of the Fock matrix
N
number of atoms
N
e
number of electrons
R
n
Euclidean space of dimension n
S
ij
bond length force constant
S
B
set of bonds
S
BA
set of bond angles
S
DA
set of dihedral angles
S
NB
set of atom pairs computed in the nonbonded energy
T
temperature
V
n
torsional barrier height of periodicity n
dielectric function
θ
i
or θ
ijk
bond angle (e.g., θ
dha
for donor–hydrogen···acceptor
sequence in a hydrogen bond)
τ
i
or τ
ijkl
dihedral or torsion angle
Δt
timestep
Science says the first word on everything, and the last word on
nothing.
Victor Hugo (1802–1885).
8.1 The Merging of Theory and Experiment
8.1.1 Exciting Times for Computationalists!
Computational techniques for exploring three-dimensional (3D) structures of nu-
cleic acids and proteins are now well recognized as invaluable tools for revealing
details of molecular conformation, motion, and associated biological function.
Over a decade ago, the theoretical chemist Henry Schaefer declared: “It is clear
that theoretical chemistry has entered a new stage ... with the goal of being no
less than full partner with experiment”[1092].
Commenting on the two 1998 Chemistry Nobel Prize awardees in quantum
chemistry — Walter Kohn of the University of California, Santa Barbara, and
John Pople of Northwestern University — a reporter in The Economist wrote: “In
the real world, this could eventually mean that most chemical experiments are
8.1. The Merging of Theory and Experiment 239
conducted inside the silicon of chips instead of in the glassware of laboratories.
Turn off that Bunsen burner; it will not be wanted these ten years” (17 October
1998)!
It remains to be seen how effective “in silico biology” will be, but clearly a
new enthusiasm is stirring in the molecular biophysics community in the wake of
many methodological and technological improvements. The following categories
reflect improvements that lend computation a stronger basis than ever before:
1. Improvements in instrumentational and experimental techniques
[1043]. Rapid advances in sequencing and mapping genomes are being
made, as discussed in Chapter 1, making available an enormous amount
of biological data requiring analysis [83]. Many procedures in biomo-
lecular NMR and electron spectroscopy [361, 908, 1184], including 4D
NMR [1433] and X-ray crystallography [110, 994, 1224], are accelerating
the rate and improving both the accuracy and scope of structure determi-
nation; much of this progress was stimulated by the structural genomics
initiatives. And newer microscopic techniques such as scanning tunnel-
ing and cryo [61, 189, 336, 361, 685, 1077, 1286] are being applied. With
such advances, structures that were considered unconquerable a decade
ago are being resolved — RNAs, nucleosomes, ribosomes, ion channels,
and membrane proteins, for example — although limitations still remain
for macromolecular complexes. Theoreticians can use these structures
with other experimental data and biological information as solid bases for
simulations and computer analyses.
2. New models and algorithms for molecular simulations.Improvedforce
fields are being developed, some simpler (e.g., coarse-grained [1117])
and others more complex (e.g., polarizable, more variables [223]) that
are sophisticated versions of early coarse-grained (e.g., [749]) or po-
larizable (e.g., [1344]) protein folding models. Innovative models for
protein and DNA folding and dynamics and for macromolecular com-
plexes are shedding insights into important biological processes. Faster
and more advanced dynamic simulations of complex systems with full
account of long-range solvation and ionic effects are being used. Quantum-
mechanical and classical/quantum hybrid studies of biomolecules are
becoming routine. And many enhanced sampling methods are making pos-
sible simulation of a wide range of conformational states of biomolecules,
including computation of reaction rates [1116, 1117].
3. The increasing speed and availability of supercomputers, parallel
processors, and distributed computing. Faster, cheaper, and smaller com-
puting platforms and graphics workstations are entering the laboratory
at reduced costs, though computer memory requirements are explod-
ing. Supercomputing centers are making available very fast computing
platforms, and specialty hardware like Anton [1169] hold promise for
advancing macromolecular modeling and simulation.
