Latchman. Eukariotic Transciption Factors
Подождите немного. Документ загружается.


80 EUKARYOTIC TRANSCRIPTION FACTORS
Figure 4.2
Amino acid sequences
of several Drosophila
homeodomains,
showing the
conserved helical
motifs. Differences
between the
sequences of the Ubx
and Ftz
homeodomains from
that of Antp are
indicated; a blank
denotes identity in the
sequence. The helix-
turn-helix region is
indicated.

in the next section (for reviews see Hayashi and Scott, 1990; Gehring et al.,
1994).
4.2.3 DNA BINDING BY THE HELIX-TURN-HELIX MOTIF IN THE
HOMEOBOX
The first indication that the homeobox proteins were indeed transcription
factors came from the finding that the homeobox was also present in the yeast
mating type a and gene products which are known to be transcription
factors that regulate the activity of a and -specific genes (for review see
Dolan and Fields, 1991), hence suggesting, by analogy, that the Drosophila
proteins also fulfilled such a role.
Direct evidence that this is the case is available from a number of different
approaches. Thus it has been shown that many of these proteins bind to DNA
in a sequence specific manner as expected for transcription factors (Hoey and
Levine, 1988). Moreover, binding of a specific homeobox protein to the
promoter of a particular gene correlates with the genetic evidence that the
protein regulates expression of that particular gene. For example, the
Ultrabithorax (Ubx) protein has been shown to bind to specific DNA
sequences within its own promoter and in the promoter of the
Antennapedia gene, in agreement with the genetic evidence that Ubx
represses Antennapedia expression (Fig. 4.3).
The ability of the homeobox-containing proteins to bind to DNA is directly
mediated by the homeobox itself. Thus if the homeobox of the Antennapedia
protein is synthesized in isolation either in bacteria or by chemical synthesis,
it is capable of binding to DNA in the identical sequence specific manner
characteristic of the intact protein.
This ability to define the sixty amino acid homeodomain as the region
binding to DNA has led to intensive study of its structure in the hope of
elucidating how the protein binds to DNA in a sequence specific manner
(for reviews see Kornberg, 1993; Gehring et al., 1994b). In particular, the
crystal structure of the Antennapedia (Antp) homeodomain bound to DNA
has been determined by nuclear magnetic resonance spectroscopy (NMR)
while similar structural studies of the engrailed (eng) and the yeast MAT2
homeodomains bound to DNA have been carried out by X-ray crystallogra-
phy.
By this means the Antp homeodomain was shown to contain a short N-
terminal arm of six residues followed by four alpha helical regions (Fig. 4.4).
The first two helices are virtually anti-parallel to each other with the other two
helices arranged at right angles to the first. Most interestingly, helices II and
III are separated by a beta turn forming a helix-turn-helix motif (Fig. 4.5). The
FAMILIES OF DNA BINDING TRANSCRIPTION FACTORS 81
eng and MAT2 homeodomains also have a similar structure with an N-term-
inal arm and a subsequent helix-turn-helix motif. In this case, however, the
third and fourth helices observed in Antp form a single helical region.
Interestingly, the helix-turn-helix structure typical of the homeodomain is
very similar to the DNA binding motif of several bacteriophage regulatory
proteins such as the lambda cro protein or the phage 434 repressor which
have also been crystallized and subjected to intensive structural study.
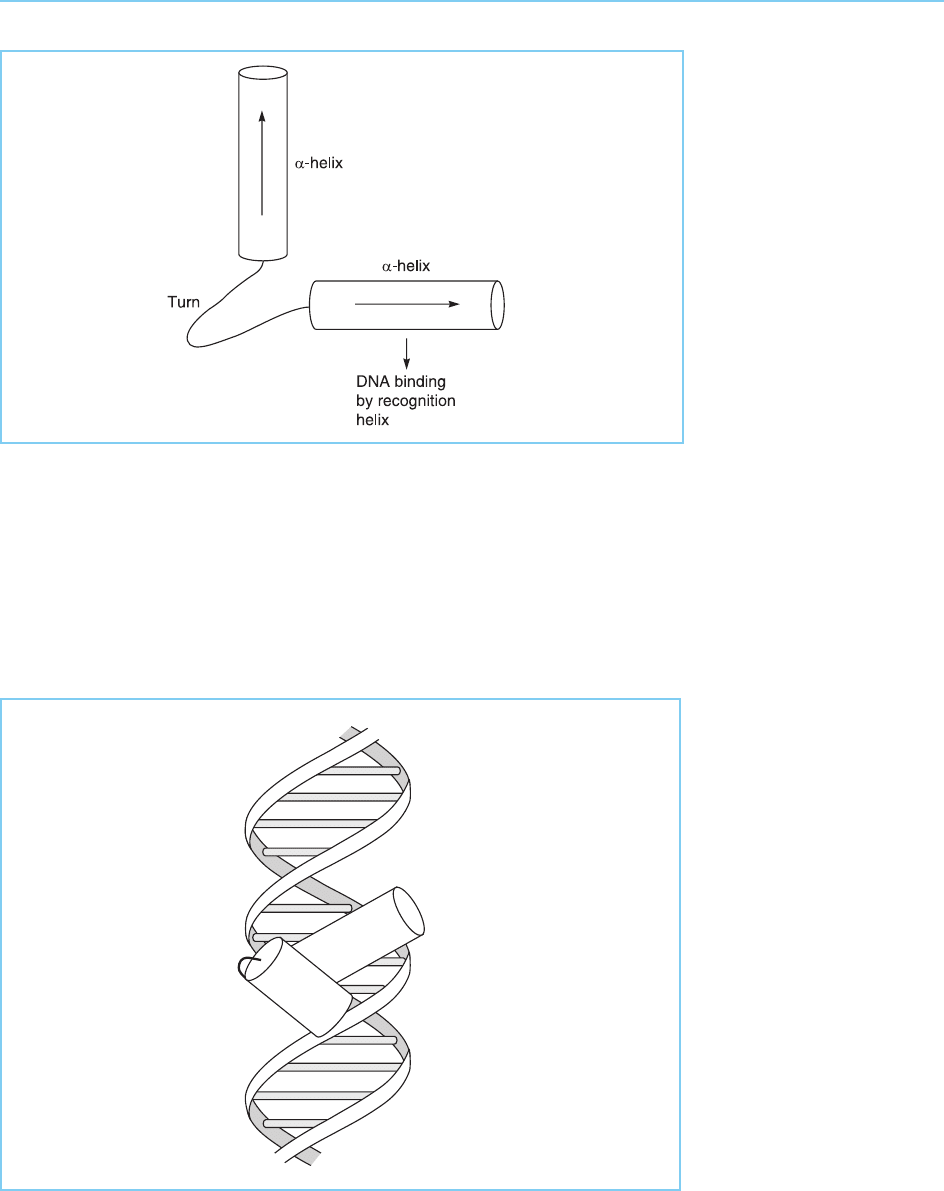
82 EUKARYOTIC TRANSCRIPTION FACTORS
Figure 4.4
Structure of the
Antennapedia
homeodomain as
determined by nuclear
magnetic resonance
spectroscopy. Note the
four alpha-helical regions
(I–IV) represented as
cylinders with the amino
acids at their ends
indicated by numbers and
the one letter amino acid
code.
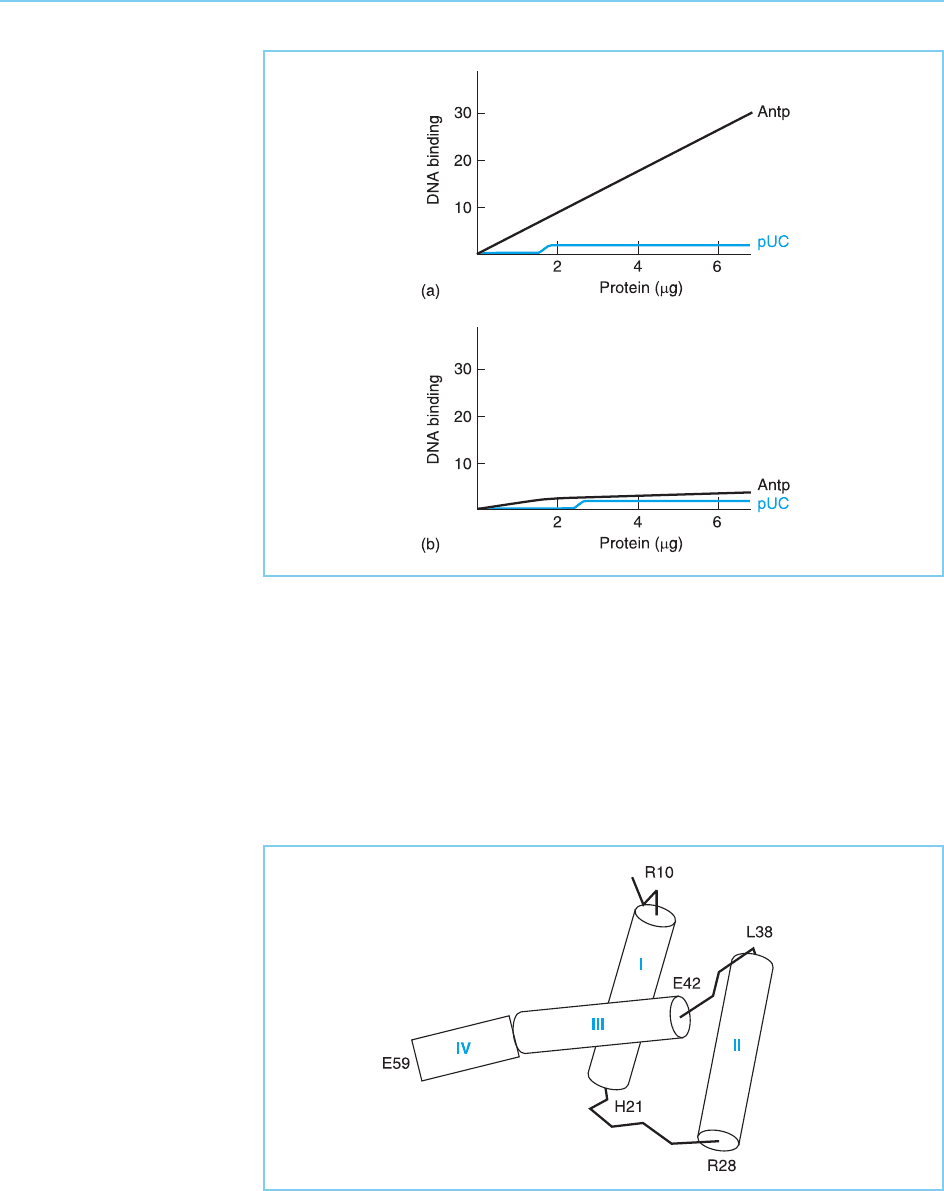
Figure 4.3
Assay of protein binding
to a DNA fragment from
the Antennapedia gene
promoter (Antp) or a
control fragment of
plasmid DNA (pUC) using
protein extracts from E.
coli which have been
genetically engineered to
express the Drosophila
Ubx protein (a) or protein
extracts from control E.
coli not expressing Ubx
(b). Note the specific
binding of Ubx protein to
the Antennapedia DNA
fragment.
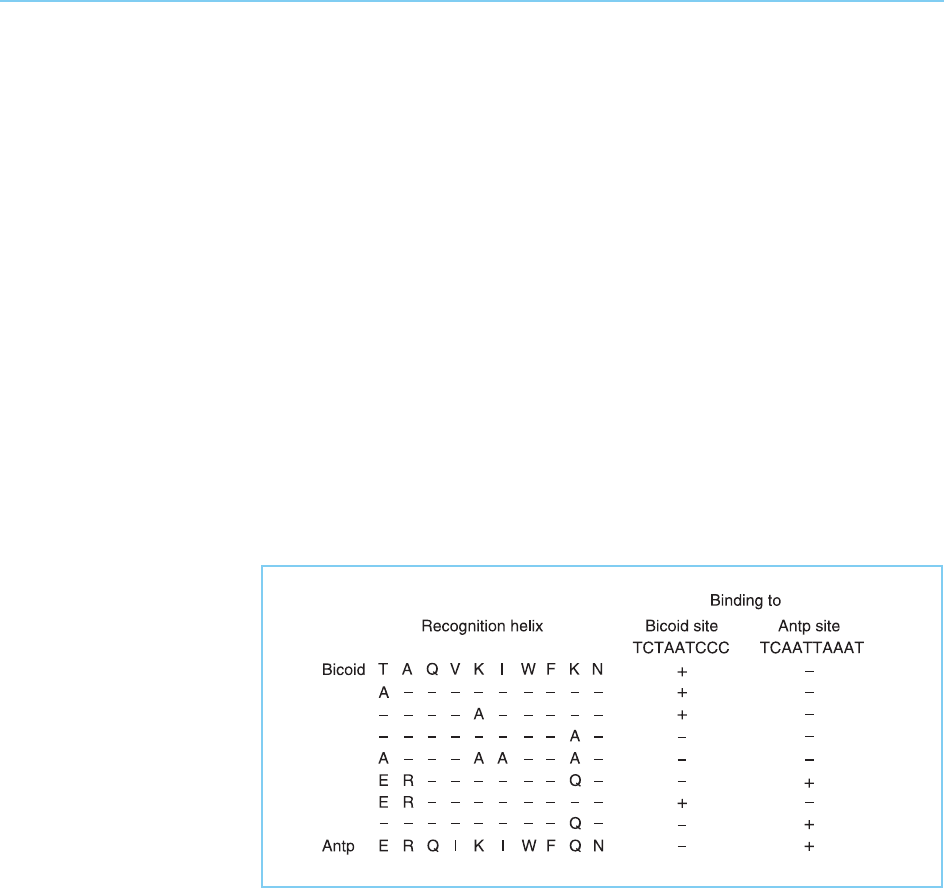
In these bacteriophage proteins X-ray crystallographic studies have shown
that the helix-turn-helix motif does indeed contact DNA. One of the two
helices lies across the major groove of the DNA while the other lies partly
within the major groove where it can make sequence specific contacts with the
bases of DNA. It is this second helix (known as the recognition helix) that
therefore controls the sequence specific DNA binding activity of these
proteins (Fig. 4.6).
FAMILIES OF DNA BINDING TRANSCRIPTION FACTORS 83
Figure 4.6
Binding of the helix-turn-
helix motif to DNA with
the recognition helix in
the major groove of the
DNA.
Figure 4.5
The helix-turn-helix motif.
The similarity in structure of helices II and III in the eukaryotic homeo-
domains to the two helices of the bacteriophage proteins led to the suggestion
that these two helices in the homeodomain are similarly aligned relative to the
DNA with helix III constituting the recognition helix responsible for sequence
specific DNA binding. Hence the precise amino acid sequence in the recogni-
tion helix in different homeodomain proteins would determine which DNA
sequence they bound (for review see Treisman et al., 1992).
In agreement with this idea, exchanging the recognition helix in the Bicoid
(Bcd) homeodomain for that of Antp resulted in a protein with the DNA
binding specificity of Antp and not that of Bicoid. Most interestingly a Bcd
protein with the DNA binding specificity of Antp could also be obtained by
exchanging only the ninth amino acid in the recognition helix, replacing the
lysine residue in Bcd with the glutamine residue found in the Antp protein
(Fig. 4.7), whereas the exchange of other residues which differ between the
two proteins has no effect on the DNA binding specificity. Hence the ninth
amino acid within the recognition helix of the homeodomain plays a critical
role in determining DNA binding specificity.
It is likely that the amino group of lysine found at the ninth position in the
Bcd protein makes hydrogen bonds with the N6 and N7 positions of a gua-
nine residue in the Bcd-specific DNA binding site whereas the amide group of
glutamine found at the corresponding position in the Antp recognition helix
forms hydrogen bonds with the N6 and N7 positions of an adenine residue at
the equivalent position within the Antp-specific DNA binding site. Hence the
replacement of lysine with glutamine results in the loss of two potential
hydrogen bonds to a Bcd site and the gain of two potential hydrogen bonds
to an Antp site explaining the observed change in DNA binding specificity
(Fig. 4.8).
84 EUKARYOTIC TRANSCRIPTION FACTORS
Figure 4.7
Effect of changing the
amino acid sequence in
the recognition helix of
the Bicoid protein on its
binding to its normal
recognition site and that
of the Antennapedia
(Antp) protein. Note the
critical effect of changing
the ninth amino acid in
the helix which
completely changes the
specificity of the Bicoid
protein.
A similar critical role for the ninth amino acid in determining the precise
DNA sequence which is recognized is also seen in other homeobox-containing
proteins, replacement of the serine found at this position in the paired pro-
tein with the lysine found in Bicoid or the glutamine found in Antp, allowing
the paired protein to recognize respectively Bcd or Antp-specific DNA
sequences. Hence the DNA sequence recognized by a homeobox-containing
protein appears to be primarily determined by the ninth amino acid in the
recognition helix, proteins with different amino acids at this position recog-
nizing different DNA sequences whereas proteins such as Antp and fushi-
FAMILIES OF DNA BINDING TRANSCRIPTION FACTORS 85
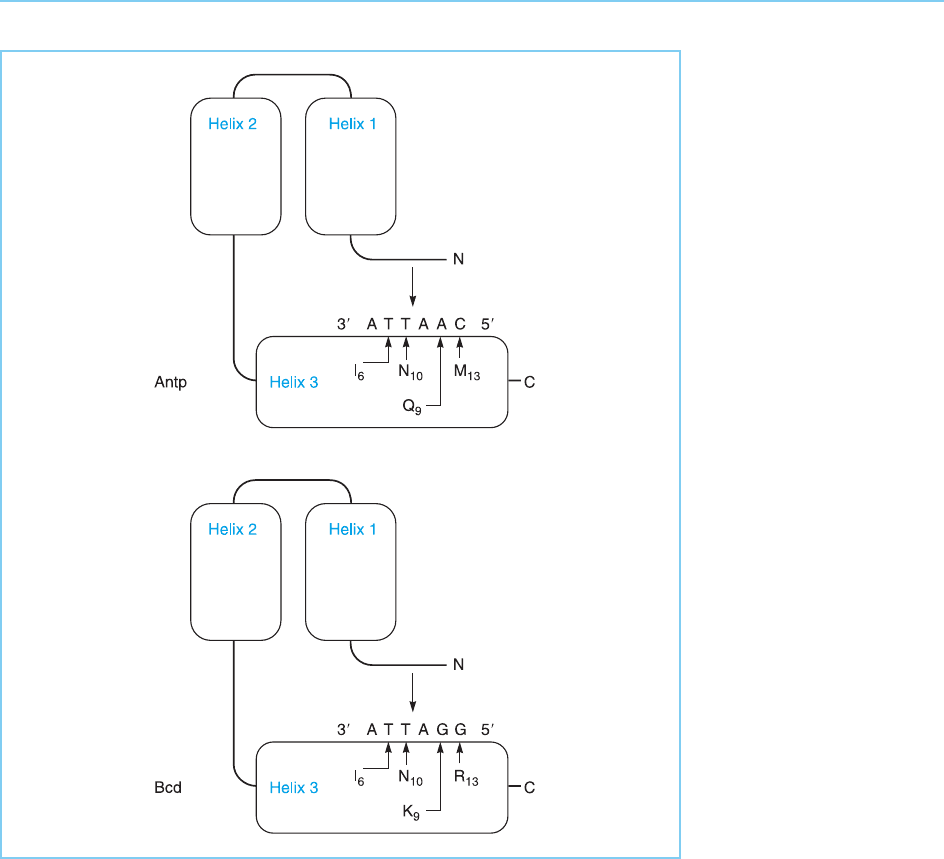
Figure 4.8
Contacts between DNA
and the Antp or Bcd
homeodomains. Note
that the change in the
ninth amino acid of the
recognition helix (helix 3)
alters the base that is
preferentially bound from
a G for Bcd to an A for
Antp, as discussed in the
text, while the N-terminal
arm of the homeodomain
contacts the ATTA
sequence common to the
recognition site of both
proteins.

tarazu which have the same amino acid at this position recognize the same
DNA sequence.
This critical role of the ninth amino acid is in contrast to the situation in the
bacteriophage proteins in which the helix-turn-helix motif was originally
defined. In these proteins, the most N terminal residues (1–3) in the recogni-
tion helix play a critical role in determining DNA binding specificity (for
review see Pabo and Sauer, 1992). As shown in Figure 4.7, however, these
amino acids appear to play little or no role in determining the DNA binding
specificity of eukaryotic helix-turn-helix proteins suggesting, therefore, that
the recognition helix of these proteins is oriented differently in the major
groove of the DNA.
This idea is in agreement with the structural studies of the eukaryotic
homeodomains bound to DNA which have identified the actual protein–
DNA contacts. These studies have shown that as in the bacteriophage
proteins, the recognition helix directly contacts the bases of DNA in the
major groove. However, in the eukaryotic homeobox proteins this helix is
oriented within the major groove somewhat differently such that the critical
base-specific contacts are, as predicted, made by the C terminal end of the
helix which contains residue nine (see Fig. 4.8).
It is clear therefore that the helix-turn-helix motif in the homeobox med-
iates both the DNA binding of the protein and also, via the recognition helix,
controls the precise DNA sequence that is recognized. Interestingly, however,
the short N-terminal arm of the homeodomain also contacts the bases of the
DNA, although it makes contact in the minor groove rather than the major
groove. Removal of this short N-terminal arm dramatically reduces the DNA
binding affinity of the homeodomain indicating that this region contributes
significantly to the DNA binding ability of the homeodomain probably by
contacting the ATTA bases common to the DNA binding sites of several
homeodomain proteins (see Fig. 4.8).
Although DNA binding is important for the modulation of transcription, it
is necessary to demonstrate that the homeobox proteins do actually affect
transcription following such binding. In the case of the Ubx protein, this
was achieved by showing that co-transfection of a plasmid expressing Ubx
with a plasmid in which the Antennapedia promoter drives a marker gene
resulted in the repression of gene expression driven by the Antennapedia
promoter (Fig. 4.9). Hence the observed binding of Ubx to the Antp promoter
(see above) results in down regulation of its activity in agreement with the
results of genetic experiments.
Most interestingly, the Ubx expression plasmid was able to up regulate
activity of its own promoter in co-transfection experiments, this ability
being dependent on the previously defined binding sites for Ubx within its
86 EUKARYOTIC TRANSCRIPTION FACTORS
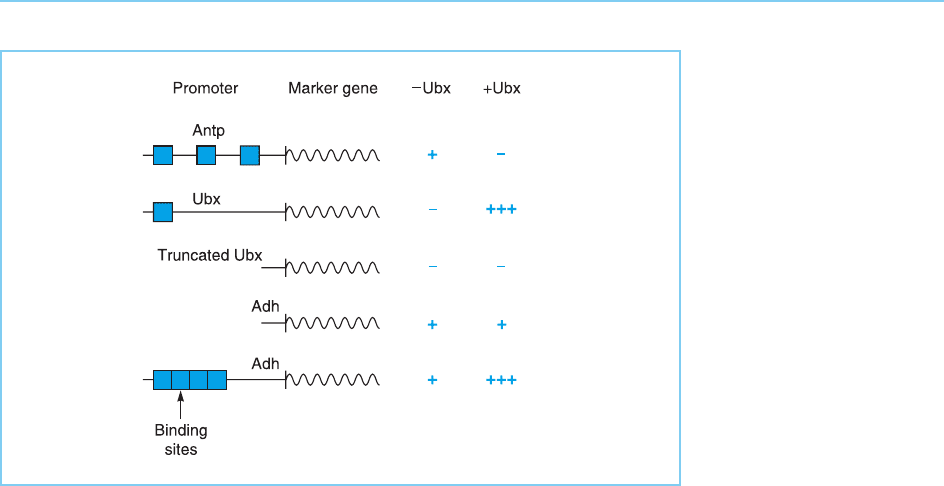
own promoter. Similarly, although Ubx normally has no effect on expression
of the alcohol dehydrogenase (Adh) gene it can stimulate the Adh promotor
following linkage of the promoter to a DNA sequence containing multiple
binding sites for Ubx. Hence a homeobox protein can produce distinct effects
following binding, Ubx activating its own promoter and a hybrid promoter
containing Ubx binding sites but repressing the activity of the Antp promoter
(Fig. 4.9).
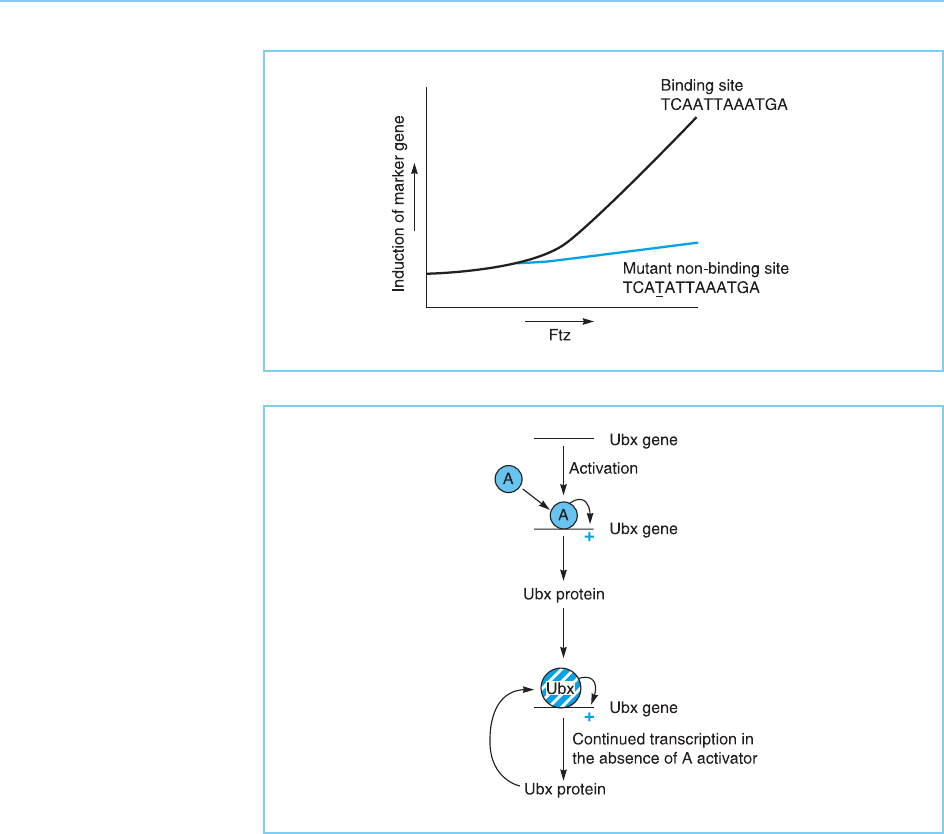
A similar transcriptional activation effect of DNA binding has been demon-
strated for the Fushi-tarazu (Ftz) protein. This protein binds specifically to the
sequence TCAATTAAATGA. As with Ubx, linkage of this sequence to a mar-
ker gene confers responsivity to activation by Ftz, such activation being depen-
dent upon binding of Ftz to its target sequence, a one base pair change which
abolishes binding also abolishing the induction of transcription (Fig. 4.10).
Interestingly, the ability of Ubx to induce its own transcription provides a
mechanism for the long-term maintenance of Ubx gene expression during
development since once expression has been switched on and some Ubx
protein made, it will induce further transcription of the gene via a simple
positive feedback loop even if the factors which originally stimulated its
expression are no longer present (Fig. 4.11). This long-term maintenance of
Ubx expression is essential since, if the Ubx gene is mutated within the larval
imaginal disc cells, which eventually produce the adult fly, the cells that would
normally produce the haltere (balancer) will produce a wing instead. Thus,
although these cells are known to already be committed to form the adult
FAMILIES OF DNA BINDING TRANSCRIPTION FACTORS 87
Figure 4.9
Effect of Ubx on various
marker genes with or
without binding sites
(hatched boxes) for the
Ubx protein. Note that
Ubx can stimulate its own
promoter which contains
a Ubx binding site and
this effect is abolished by
deleting the Ubx binding
site. Similarly, the alcohol
dehydrogenase (Adh)
gene which is normally
unaffected by Ubx, is
rendered responsive to
Ubx stimulation by
addition of Ubx binding
sites. In contrast the
Antennapedia promoter,
which also contains Ubx
binding sites, is
repressed by Ubx. Hence
binding of Ubx can
activate or repress
different promoters.
haltere at the larval stage, the continued expression of the Ubx gene is essen-
tial to maintain this commitment and allow eventual overt differentiation (see
Hadorn, 1968 for a review of imaginal discs and their role in Drosophila
development).
4.2.4 REGULATION OF DNA BINDING SPECIFICITY BY
INTERACTIONS BETWEEN DIFFERENT HOMEOBOX PROTEINS
Although we have previously described the DNA binding specificity of indivi-
dual homeobox proteins, it is possible for the DNA binding specificity of one
factor to be altered in the presence of another factor. Thus several homeobox
88 EUKARYOTIC TRANSCRIPTION FACTORS
Figure 4.11
The stimulatory effect of
the Ubx protein on the
transcription of its own
gene, ensures that once
Ubx gene transcription is
initially switched on by an
activator protein (A),
transcription will continue
even if the activator
protein is removed.
Figure 4.10
Effect of expression of
the Ftz protein on the
expression of a gene
containing its binding site,
or a mutated binding site
containing a single base
pair change which
abolishes binding of Ftz.
proteins, such as Ubx and Antp, bind to the same DNA sequences when tested
in isolation in vitro (Hoey and Levine, 1988) yet, paradoxically, the effects of
mutations which inactivate the genes encoding each of these proteins are
different indicating that they cannot substitute for one another. Similarly,
in vivo Ubx can bind to a site in the promoter of the decapentaplegic (dpp)
gene and activate its expression whereas Antp cannot do so.
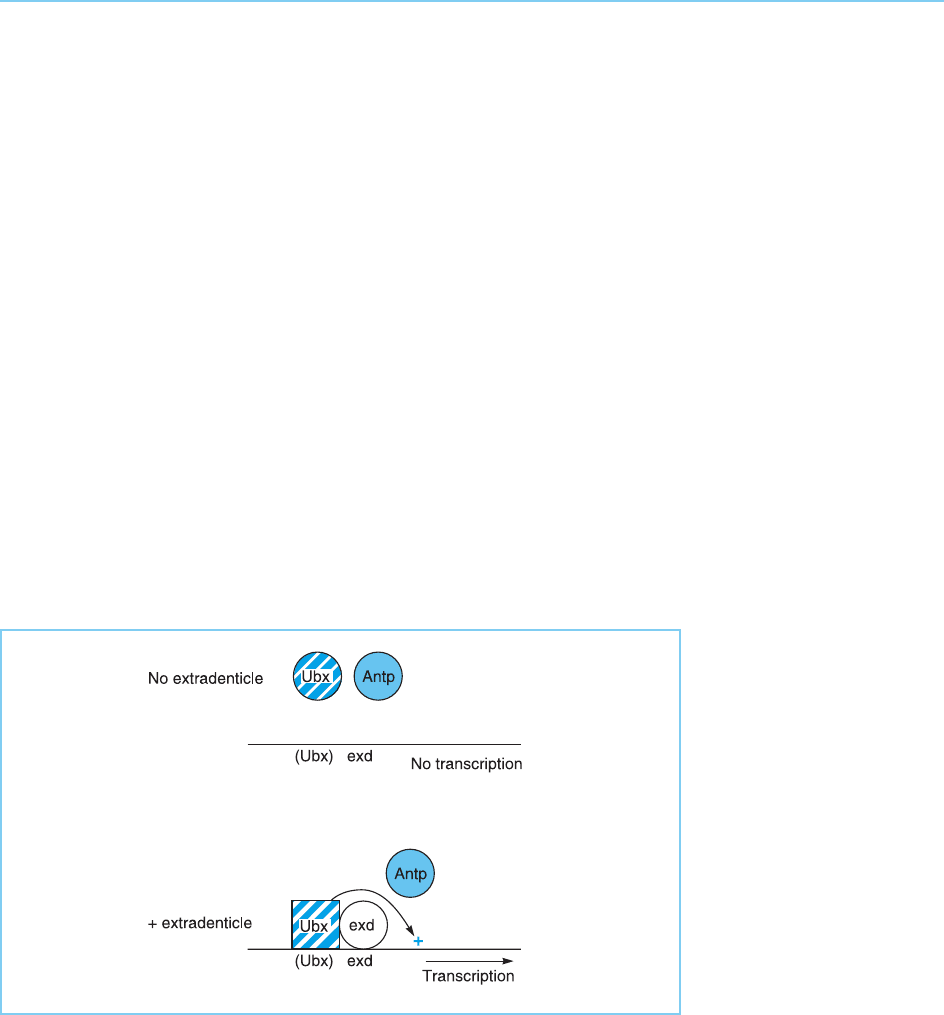
This paradox is explained by the presence in the dpp promoter of a bind-
ing site for another homeobox protein extradenticle (Exd) which lies adjacent
to the site to which Ubx binds. The Exd protein interacts with the Ubx protein
and both enhances its DNA binding affinity and modifies its DNA binding
specificity so it can bind strongly to the dpp gene promoter and activate its
expression (Fig. 4.12) (for review see Mann and Chan, 1996). As Antp does
not interact with Exd, its specificity is not modified in this way. Hence, it does
not bind to the dpp gene promoter and therefore cannot activate this pro-
moter. Interestingly, structural studies have shown that Ubx and Exd bind to
opposite sides of the DNA and that a short region of Ubx N-terminal to the
homeodomain extends round the DNA and inserts into a cleft in the Exd
homeodomain resulting in interaction of the proteins and enhanced DNA
binding by the complex (Passner et al., 1999; for review see Scott, 1999).
A similar interaction is observed in the case of the yeast homeodomain
proteins a1 and 2, which control the mating type in this organism (for review
see Dolan and Fields, 1991). Thus, in the absence of a1, the 2 protein has a
weak DNA binding ability. However, in the presence of a1, an a1/2 hetero-
dimer forms and binds to specific gene promoters. As the 2 protein is a
FAMILIES OF DNA BINDING TRANSCRIPTION FACTORS 89
Figure 4.12
The Exd protein interacts
with the Ubx protein to
allow it to bind with high
affinity to its potential
binding site in the dpp
promoter (indicated as
(UBX)) and activate its
expression. In contrast,
the Antp protein cannot
interact with Exd and so
does not bind to the dpp
promoter.
