Schlick T. Molecular Modeling and Simulation: An Interdisciplinary Guide
Подождите немного. Документ загружается.

130 5. Nucleic Acids Structure Minitutorial
DNA plays a role in life rather like that played by the telephone direc-
tory in the social life of London: you can’t do anything much without
it, but, having it, you need a lot of other things — telephones, wires,
andsoon—aswell.
Review of The Double Helix in The Sunday Times, London (1968).
5.1 DNA, Life’s Blueprint
With the master molecule of heredity, deoxyribonucleic acid (or DNA), so fre-
quently mentioned in the media — in connections ranging from polymerase chain
reaction (PCR) applications (e.g., criminology, medical diagnoses) to cloning
and art [1112] — it is difficult to imagine today that it was only a few decades
ago when Watson and Crick reported their description of the DNA double helix
[1354–1356]! Based on analysis of DNA fiber diffraction patterns and Chargaff’s
rules, they described a spiral image of an orderly helix — two intertwined polynu-
cleotide chains, with a sugar/phosphate backbone on the exterior and pairs of
hydrogen-bonded nitrogenous bases in the center. See [622] for a historical per-
spective of this discovery, including the contribution of all key players (a capsule
of which is given in Chapter 1), and anniversary issues of DNA, for example is-
sued in 2003 in many journals (e.g., Nature Vol. 421 and Science Vol. 300) at the
occasion of DNA’s golden anniversary.
Though the model was imperfect in several respects (e.g., sugar conforma-
tions and base positioning with respect to the helical axis were inexact), Francis
Crick, James Watson, and Maurice Wilkins received the 1962 Nobel Prize in
Chemistry for this monumental discovery. Indeed, soon after the description of the
DNA double helix came a burst of scientific excitement and discovery regarding
the mechanism of heredity. The field of molecular biology gained extraordinary
momentum and, soon after, its tentacles extended to the disciplines of cellular bi-
ology, molecular genetics, genomics, and proteomics, all buttressed by impressive
advances in technology and computing.
5.1.1 The Kindled Field of Molecular Biology
The description of DNA’s three-dimensional (3D) structure provides a classic ex-
ample of the structure/function relationship. The two key functions of DNA —
replication and transcription — were suggested and later proven based on the or-
dered spatial arrangement of the DNA. Replication, or the accurate transmittal of
genetic information from parent to progeny, was explained by base pairing speci-
ficity. Transcription, the transformation of DNA’s genetic information into a form
that directs protein synthesis, was later understood with the deciphering of the

5.1. DNA, Life’s Blueprint 131
genetic code relating triplet nucleotide codons (see Section 5.2 and Figure 5.2 for
a description of the basic building blocks, including the pyrimidine bases T and
C and purine bases A and G) to amino acids.
This genetic code (Table 5.1) reveals interesting patterns:
1. It is highly redundant: as 61 of the 64 triplet codons represent one of the
20 amino acids, and 3 signal ‘stop’ messages for protein synthesis (i.e.,
translation). In particular, Arg, Leu, and Ser are specified by 6 codons; Met
and Trp are each represented by only one; and the rest are specified by 2–4
triplets.
2. Pyrimidine-end commonalities exist: Codons B
1
B
2
TandB
1
B
2
C(where
B
1
and B
2
denote any of the 4 bases) always code for the same amino acid.
3. Purine-end commonalities exist: Codons B
1
B
2
AandB
1
B
2
G code for the
same amino acid except in the cases of AT (ATA = Ile, ATG = Met) and
TG (TGA = ‘
STOP’, TGG = Trp).
Though the genetic code was initially thought to be ‘universal’, DNA se-
quencing in the early 1980s revealed code variations for certain organisms.
Now it is clear that the genetic codes for mammalian and plant mitochondria
and certain protozoa, for instance, exhibit some variations from the code de-
scribed in Table 5.1. In organisms where nonstandard amino acids exist (e.g.,
certain Archaea and eubacteria), such a variation in the code may consist of a
STOP codon that encodes a nonstandard amino acid rather than instruct to halt
translation [68,511,1217].
Recent advances in experimental techniques that allow detailed study of the
components of the translation apparatus are also making possible the study of
how the modern code has evolved from a simpler form [662].
Thus the model for the DNA double helix led the way to establishment of
the first iteration of the central dogma of biology: DNA is self-replicating, DNA
makes messenger ribonucleic acid (mRNA) through transcription,
1
and mRNA
makes protein through translation (DNA → RNA →protein).
2
Although other ar-
rows in the dogma have been suggested or verified (e.g., RNA → RNA in certain
viruses, RNA → DNA in retroviruses), the main components appear known.
1
In transcription, an RNA polymerase glides along one strand of the DNA double helix and
builds an RNA complement by coupling ribonucleotides through dehydration synthesis. The faithful
replicate of one DNA strand then functions as the messenger RNA.
2
In translation, the genetic code of the messenger RNA is read by transfer RNA molecules on
cellular structures called ribosomes. Every transfer RNA molecule carries a specific sequence of three
nucleotides on one end of an L-shaped structure and the corresponding amino acid on the other. The
transfer RNA’s main task is to deposit its amino acids on the ribosomes in proper sequence. In this
process of matching each messenger-RNA codon with the complementary transfer RNA molecule, the
polypeptide chain is assembled. As the amino acids link to one another on the ribosomes, polypeptide
folding is thought to begin.
132 5. Nucleic Acids Structure Minitutorial
Table 5.1. The genetic code in terms of the parental DNA; the mRNA transcript has uracil
(U) instead of thymine (T).
Amino Acid Encoding Triplets
(or Instruction) in Parental DNA
a
Arginine (Arg) CG(*), AG(A,G)
Leucine (Leu) CT(*), TT(A,G),
Serine (Ser) TC(*), AG(T,C)
Alanine (Ala) GC(*)
Glycine (Gly) GG(*)
Proline (Pro) CC(*)
Threonine (Thr) AC(*)
Valine (Val) GT(*)
b
Isoleucine (Ile) AT(T,C,A)
Asparagine (Asp) GA(T,C)
Aspartic acid (Asn) AA(T,C)
Cysteine (Cys) TG(T,C)
Histidine (His) CA(T,C)
Phenylalanine (Phe) TT(T,C)
Tyrosine (Tyr) TA(T,C)
Glutamine (Gln) CA(A,G)
Glutamic acid (Glu) GA(A,G)
Lysine (Lys) AA(A,G)
Methionine (Met) ATG
b
Tryptophan (Trp) TGG
STOP TA(A,G), TGA
a
Short-hand notation is used for the third base of the codon when the first and second bases are
the same: GA(T,C) specifies both GAT and GAC, while GC(*) denotes all four possibilities, namely
GCT, GCC, GCA, and GCG.
b
The codons ATG and (less frequently) GTG form part of the initiation signal in addition to their
coding for Met and Val, respectively.
Much attention has now expanded to new areas of molecular biology, such as
structural molecular biology, computational molecular biology, and offspring of
genomics, such as structural genomics and functional genomics. The functional
relationships among genes and the interaction of genes within the context of the
complete organism are also of great interest.
5.1.2 Fundamental DNA Processes
As the genetic material, DNA carries structural information in the primary se-
quence that not only controls faithful duplication but also regulates expression
5.1. DNA, Life’s Blueprint 133
of the hereditary information [1021]. Genetic variability can result from errors
(or mutations), such as insertions, deletions, or substitutions in the daughters com-
pared to the parental DNA, during the template-copying process; these changes
can lead to altered triplet codes and hence different polypeptide composition.
Since many basic biological processes rely on protein/nucleic-acidinteractions,
the base sequence of polynucleotides also affects profoundly the characteristic 3D
structure of DNA and RNA and hence the nature of fundamental biological pro-
cesses. On a higher level of structure (hundreds and thousands of base pairs),
compact forms of long DNA — such as supercoils, knots, and chromosomes —
are central to fundamental mechanisms for replication, transcription, and recom-
bination [195,1384]. DNA supercoiling, namely the coiling in space of the double
helix axis itself, can condense the DNA by several orders of magnitude and read-
ily store energy for various activities. This supercoiled state is essential for the
storage of DNA in eukaryotic cells, where DNA is wrapped as a left-handed su-
percoil around cores of proteins to form the chromatin fiber. The wide range of
characteristic timescales associated with the configurational rearrangements and
hydrodynamicproperties of supercoiled (or superhelical) DNA represents another
area of intense study.
5.1.3 Challenges in Nucleic Acid Structure
As described above, there are several levels of nucleic acid structure, from the
base-pair level to the cellular level of organization in the chromosomes (see end
of next chapter and [1119] for example). This study of DNA’s rich configurational
levels is particularly challenging to modelers.
Much focus has been placed on protein structure (including protein/DNA com-
plexes) and the protein folding problem — predicting the 3D architecture of a
system from the primary sequence. Yet an analogous folding problem is also rele-
vant to DNA and RNA polymers. In fact, the nucleic-acid analogue of the ‘protein
folding problem’ might be viewed as more challenging than protein structure pre-
diction in the sense that it extends over much larger spatial scales (thousands of
˚
Angstroms) as well as temporal scales (picoseconds to minutes and longer) than
typically associated with proteins. Biologically, elucidating the folding of DNA
in the cell — supercoiling and chromosome condensation, for example — is im-
portant for understanding the regulatory role of DNA in fundamental biological
processes.
For small segments of nucleic acids, X-ray crystallography and NMR data have
been invaluable for providing detailed, atomic-resolution data on single, double,
triple and other forms of nucleic acid oligonucleotides, as well as their complexes
with proteins, other biomolecules, and ligands [126]. The Nucleic Acid Data-
base (NDB, ndbserver.rutgers.edu/)[127] has been beautifully cataloging these
structures and offers many utilities for their analysis.
All-atom molecular dynamics (MD) simulations of nucleic acids have also
shed important insights on DNA sequence/structure relationships, the nature of
the hydration geometries surrounding nucleic acids, nucleic acid flexibility, and
134 5. Nucleic Acids Structure Minitutorial
nucleic-acid protein structures [126, 217, 808, 953, 1026, 1417, 1419, 1421,for
example]. Indeed, simulation improvements and the availability of ultra-high
resolution nucleic-acid crystal structures [234,641,1182] have made possible the
study of fully solvated nucleic acid MD trajectories, with representative ionic
atmospheres [219,239,283,1412, 1416], and even millisecond simulations [987].
The theoretical advances resulted from improvements in long-range electro-
static modeling, force field parameters, representation of the ionic atmosphere,
and novel conformational sampling approaches [217,953,988,1116,1117,1319],
as well as increases in computer memory and speed. The experimental advances
reflect improved methods for crystallization and phase determination, the in-
creased availability of very strong X-ray sources, improvements in algorithms
for model refinement, and innovative approaches such as single-force microscopy
which allows studies of DNA and RNA energetics and dynamics of folding and
unfolding (e.g., [187, 759]) and improved ultra-structural visualization tools for
DNA’s higher levels of structural organization (e.g., chromatin, see next chapter).
Significantly, such modeling and experimental advances have made it possible to
simulate solvated RNA at atomic resolution [86, 502, 525, 526, 1446]. Advances
on both the computational and experimental fronts are ongoing.
Computer scientists also find interest in DNA with the emerging possibility of
using the strands of DNA in vitro for practical applications, such as to produce
electronic devices like nanowires, or as parallel computers to solve very difficult,
combinatorial optimization problems that have non-polynomial complexity [40,
117,162, 825, 1156]. Very recently, a ‘DNA Sudoku’ approach for parallel DNA
sequencing by combinatorial pooling strategies reminiscent of solving sudoku
puzzles has also been reported for analyzing short genome segments associated
with disease markers [366].
Undoubtedly, progress is expected in the bridging between all-atom and macro-
scopic representations of nucleic acids and between experiment and theory. This
unity will enhance our understanding of DNA structure and DNA/protein interac-
tions and, in turn, will likely have important biomedical applications, for example,
in the design of new anti-viral drugs, antibiotics, and anti-cancer agents, some of
which are being designed with DNA or RNA agents such as DNA plasmids and
silencing RNAs.
5.1.4 Chapter Overview
In this chapter, the basic elements of nucleic acids and DNA structure on the base-
pair and helical level will be introduced: the fundamental building blocks and how
they are linked to form polynucleotides, aspects of nucleic acid conformational
flexibility (sugar pseudorotation, torsion angle preferences, and global and local
base-pair parameters), and the three canonical DNA helices (A, B, and Z-DNA).
The next chapter presents further topics regarding the structural diversity of DNA
and RNA, and DNA folding on a higher level, namely supercoiling and chromatin
organization.

5.2. The Basic Building Blocks of Nucleic Acids 135
While RNA is introduced in the next chapter, after the variety of base pairing
arrangements is presented, this in no way means RNA is less fundamentally im-
portant than DNA. In fact, recent discoveries on RNA’s prevalent regulatory roles
in the cell place RNA in the forefront of biology and chemistry.
For basic books on DNA structure, see [100,139,195,269,893,895,1080,1191,
1305]. Lively, less technical introductions can be enjoyed from [272, 420, 1353].
The reader is also well advised to examine some of the rich information on
nucleic acid structure available in the public domain through the NDB [127],
ndbserver.rutgers.edu/.
Throughout this chapter and the next, we abbreviate a base pair as bp and
base pairs as bps. Note that though we mention the three canonical helices be-
fore introducing them formally (using all notation we systematically develop),
novices should be able to follow the material. Students are encouraged to re-read
the chapter after they are more familiar with all the presented material.
5.2 The Basic Building Blocks of Nucleic Acids
In the classic DNA double helix described by Watson and Crick, a flexible ladder-
like structure is formed with the polymer wrapped around an imaginary central
axis (Figure 5.1). The two rails of the ladder consist of alternating sugar (deoxyri-
bose) and phosphate units; the rungs of the ladder consist of nitrogenous bps held
together by hydrogen bonds (see Box 3.2 of Chapter 3 for a definition).
5.2.1 Nitrogenous Bases
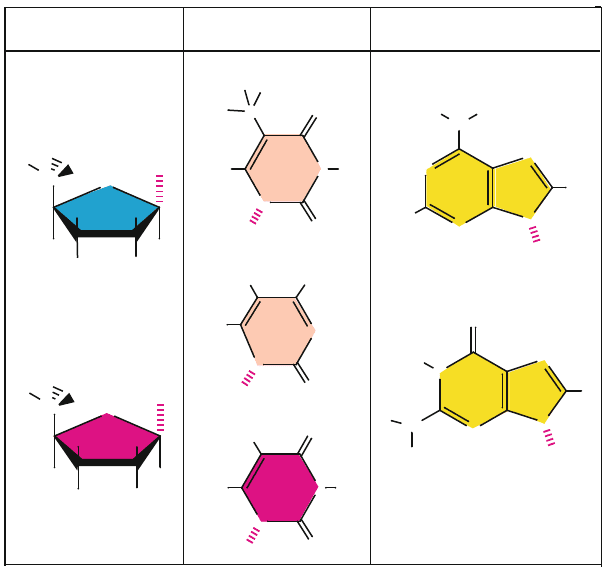
Four nitrogenous bases can be found in DNA: the pyrimidines cytosine (C) and
thymine (T) which are 6-membered rings, and the purines guanine (G) and ade-
nine (A), each of which is a fused system with a 5 and 6-membered ring. In
the single-stranded RNA polynucleotide, ribose replaces deoxyribose and uracil
(U) replaces thymine (Figure 5.2). Some DNAs contain bases that are chemically
modified (e.g., substitutions of a hydrogen by a methyl group).
3
Alternative but
rare tautomeric forms due to proton shifts in aromatic molecules (as introduced in
Chapter 1, subsection on the discovery of DNA structure),
4
are possible but may
not be biologically significant.
3
For example, N6-methyl-dA is a modified adenine base with the N6H
2
(attachment to the ring
carbon C6) moiety replaced by N6HCH
3
; 5-methyl-dC is a modified cytosine where the C5H becomes
C5CH
3
.
4
Two classic tautomerization reactions are keto/enol and amino/imino; the keto and amino forms
are typically favored and are shown in Figure 5.2. Keto/enol tautomerization involves alteration of the
carbonyl group (–C=O) to a hydroxyl group (=C–O–H), shifting the double bond from the carbonyl
group to the nitrogen-carbon bond in the ring (e.g., C6=O of G becomes C6–OH, accompanied by the
change of H–N1–C6 to N1=C6). Similarly, an amino/imino tautomerization involves a change in an
amino nitrogen –NH
2
to an imino form, =NH (e.g., C6–NH
2
of A becomes C6=NH, accompanied
by the change of N1=C6 to H–N1–C6).
136 5. Nucleic Acids Structure Minitutorial
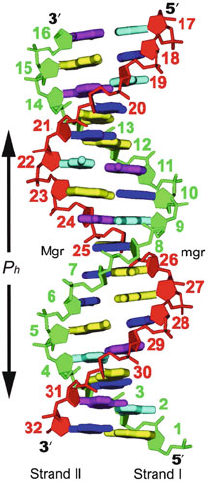
Figure 5.1. The two strands of the classic DNA double helix: alternating subunits of phos-
phates and deoxyriboses held together by hydrogen-bonded bps, normally with adenine
(A) pairing with thymine (T) and guanine (G) pairing with cytosine (C). The two chains of
the DNA double helix run in opposite directions. The chain ending in the lower right ter-
minates with a phosphate group linked to the sugar carbon denoted as C5
, while the chain
ending in the lower left terminates with a free hydroxyl group linked to the sugar carbon
denoted as C3
. Individual bases are numbered 1–16 (or 1 to n where n is the number of
bps) for the sequence strand (Strand I) and 17–32 (n +1to 2n in general) for Strand II,
both in the 5
→ 3
direction; the n base pairs 1–16 are numbered so that they coincide
with the bases of Strand I. The pitch of the helix P
h
is the length along the helix axis for
one complete turn, and n
b
is the number of bps per turn (around 10.5 for DNA in solution).
The unit twist is defined as Ω = 360
o
/n
b
, and the helical rise is h = P
h
/n
b
. Mgr and mgr
denote the major and minor grooves, respectively.
5.2.2 Hydrogen Bonds
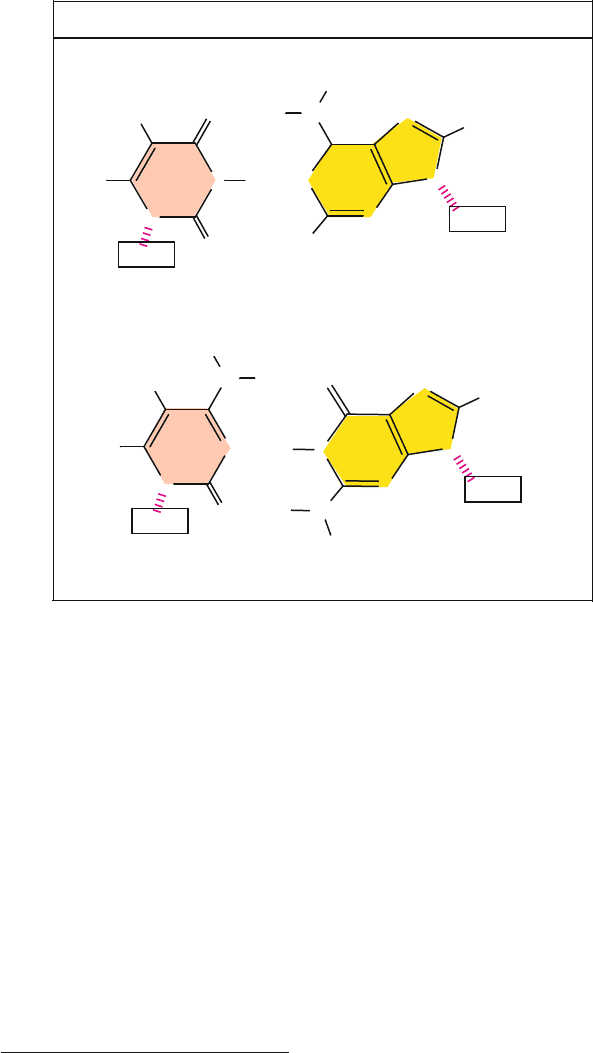
In the Watson-Crick base pairing scheme (termed WC herewith), cytosine pairs
with guanine by forming three hydrogen bonds, and thymine pairs with adenine
by forming two hydrogen bonds (Figure 5.3). This arrangement produces CG and
TA bps whose widths are nearly identical. The approximately uniform width is
significant because any pyrimidine/purine or purine/pyrimidine sequence can be
accumulated on one strand, with a pyrimidine opposing a purine, without much
alteration in overall structure. The discovery of this orderly 3D structure of DNA
5.2. The Basic Building Blocks of Nucleic Acids 137
O
H
HH
HH
O3'H
1'
2'3'
4'
OH
N
N
N
N
N
N
N
N
7
9
8
4
5
6
1
3
2
6
5
4
3
2
1
7
9
8
β−D-2-Deoxyribose
Ribose
Thymine
Cytosine
Uracil
Adenine
Guanine
Sugars Pyrimidine Bases
Purine Bases
N
N
N
N
N
N
4'
5
6
1
2
3
4
54
3
21
6
5
6
4
3
21
T
C
U
A
G
HH
O4
O2
H
HH
H
O2
H
H
O4
HH
O2
H
N6
H
H
O6
H
N2
H
H
H
HH
H
H
C5M
H
H
H
C5'
H
H
O5'H
O
O2'H
HH
HH
O3'H
1'
2'3'
4'
OH
4'
C5'
H
H
O5'H
Figure 5.2. Chemical structures and atomic labels for nucleic acid sugars (deoxyribose in
DNA and ribose in RNA) and nitrogenous bases: T, C, A, G in DNA and U, C, A, G in
RNA. The broken-line pattern in each compound indicates the bond where a link to another
building block is made (see Figure 5.4).
helices explained for the first time how a regular polymer made of repeating
phosphate/sugar/base units (nucleotides) could replicate and encode hereditary
information.
5.2.3 Nucleotides
The basic building block of nucleic acid polymers, the nucleotide, consists of a
5-membered sugar ring — deoxyribose in DNA and ribose in RNA — a phos-
phate, and a purine or pyrimidine base (see Figure 5.4). The unit containing
just the sugar and base is called the nucleoside. Nucleotides are linked together
through the phosphate group to form the polynucleotide chain.
5.2.4 Polynucleotides
When nucleotides are polymerized into nucleic acid chains by the chemi-
cal removal of water molecules, a sugar/phosphate backbone is formed. The
138 5. Nucleic Acids Structure Minitutorial
A
N
N
N
N
O6
N
N
2
3
4
5
6
1
Sugar
Sugar
C
H
Watson - Crick Base Pairs
H
H
H
O2
N2
H
H
N4
H
H
1
2
5
4
6
3
7
9
8
N
N
N
N
Sugar
H
H
1
2
5
4
6
3
7
9
8
N6
H
H
N
N
2
3
4
5
6
1
Sugar
T
H
3
C
H
O2
O4
H
. . .
. . .
. . .
. . .
. . .
G
Figure 5.3. In the classic Watson-Crick base pairing scheme of DNA double helices,
thymine (T) pairs with adenine (A) by forming two hydrogen bonds, and cytosine (C)
pairs with guanine (G) by forming three hydrogen bonds. The hydrogen bonds are often
represented by dots (···). The structural fit (e.g., as measured by C1
–C1
distances)
is such that the difference in width between the two base pairs is less than 3%, allowing
formation of a double helix with a nearly constant diameter.
C3
–hydroxyl group of the nth nucleotide sugar is joined to the C5
–hydroxyl
group of the (n+1)th nucleotide by a phosphodiester bridge.
5
This negative phos-
phate group, located on the exterior of the helix, is readily available for physical
and chemical interactions with solvent water molecules and metal ions present
in the cell (see next chapter). Hydration effects are of great importance for DNA
molecules in solution, since water plays a central role in stabilizing a particular
helical conformation.
5
An ester is an –OR group where R represents an organic chemical group.
5.2. The Basic Building Blocks of Nucleic Acids 139
α
β
O2P
O1P
Base
P
O2P
P
γ
δ
ζ
ε
Base
Nucleotide Unit
C4⬘
O4⬘
HO−C5⬘ − H
2
τ
4
τ
3
τ
2
τ
1
5⬘− end
3⬘− end
τ
0
P
O1P
H
O4⬘
O2P
P
O
N
C
O1P
Base
Base
N9/N1
N9/N1
N9/N1
N9/N1
C3⬘
C5⬘
C4⬘
C3⬘
C2⬘
C1⬘
O3⬘
O5⬘
O5⬘
C4⬘
C4⬘
C3⬘
C3⬘
C2⬘
C2⬘
C1⬘
C1⬘
O3⬘
O3⭈
C5⬘
C5⬘
O5⬘
O4⬘
O4⬘
O3⬘
C2⬘
C1⬘
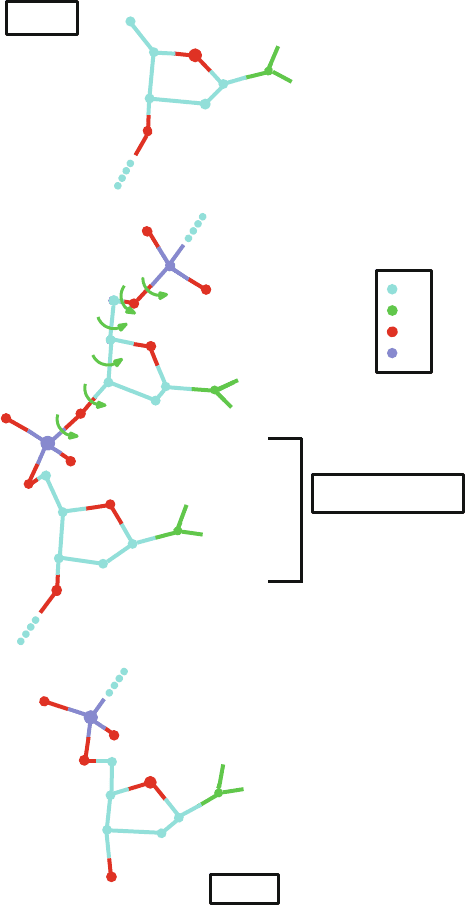
Figure 5.4. The polynucleotide chain, with standard atom labeling shown, runs from the
5
end (of the sugar atom C5
)tothe3
end (of the sugar atom C3
). Nucleotide linkage
is via the 3
to 5
phosphodiester bonds (i.e., C3
–hydroxyl group of one nucleotide sugar
to C5
–hydroxyl group of another). Nucleic acid torsion angles are labeled as α, β, γ, δ,
and ζ along the polynucleotide chain, τ
0
through τ
4
about the endocyclic bonds of the
sugar, and χ about the glycosyl bond, connecting the sugar and base. See Table 5.2 for full
quadruplet sequences of these torsion angles.
