Schlick T. Molecular Modeling and Simulation: An Interdisciplinary Guide
Подождите немного. Документ загружается.

100 3. Protein Structure Introduction
ALIPHATIC SIDE CHAINS
ALIPHATIC HYDROXYL SIDE CHAINS
SECONDARY AMINO GROUP
ACIDIC SIDE CHAINS AND THEIR AMIDE DERIVITIVES
SULFUR-CONTAINING SIDE CHAINS
BASIC SIDE CHAINS
AROMATIC SIDE CHAINS
Val (V)
H
2
C
+
H
2
N
C H
α
CH
2
H
2
C
C
α
CH
2
CH
2
C
C
α
CH
2
CH
2
C NH
2
O
CH
2
C
CH
2
C NH
2
O
C
α
C
α
CH
2
OH
C
α
CH
C
α
C
α
C
α
CH
2
CH
3
CH
3
CH
3
CH
3
CH
3
CH
2
CH
3
C
α
C
CH
3
OH
H
C
H
CH
C
α
CH
2
C
α
CH
2
C
α
CH
2
C
α
CH
2
C
α
CH
2
C
α
CH
2
SH
CH
2
S CH
3
CH
2
CH
2
CH
2
NH
3
+
CH
2
CH
2
H
N C NH
2
NH
2
+
C
+
HN
C
H
NH
CHCH
2
C
α
C
HC
N
H
OH
CH
2
C
α
χ
1
χ
2
χ3
χ4
O
O
O
O
_
_
Leu (L)
Ser (S) Thr (T) Pro (P)
Asp (D)
Asn (N)
Glu (E)
Gln (Q)
Met (M) Cys (C)
Lys (K)
Arg (R)
His (H)
Phe (F)
)W(prT)Y(ryT
Ile (I)
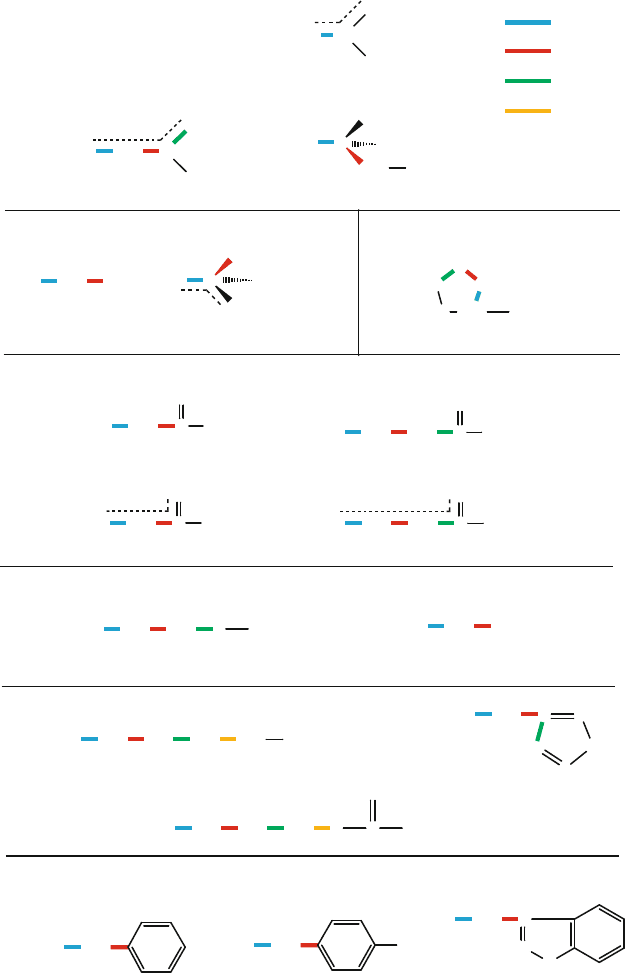
Figure 3.17. Rotameric notation used for 18 of the 20 amino acids is illustrated using
different colors for χ
1
–χ
4
, as shown in the top right key.
3.4. Protein Conformation Framework 101
−90 0 90 180
−90
0
90
180
−180
φ (degrees)
Ψ
(degrees)
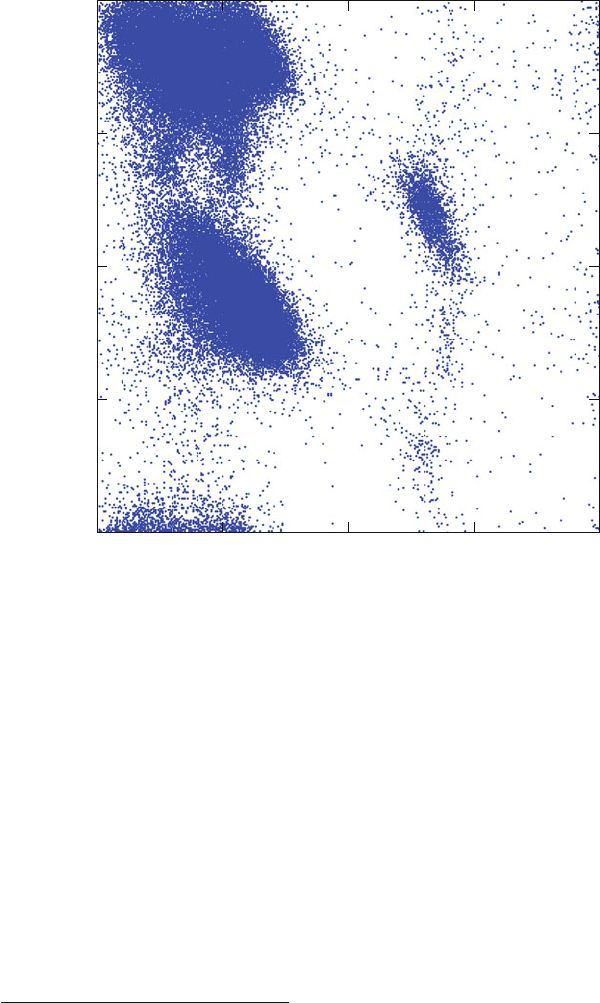
Figure 3.18. Ramachandran plots, obtained from a subset of the PDB40 dataset [959],
corresponding to X-ray protein structures with resolution of 2.5
˚
A or better (470 proteins,
95778 total residues plotted, with proline and glycine excluded).
G.N. Ramachandran
4
and coworkers in 1963, after which Ramachandran plots
are called. Around the same time, John Schellman and coworkers were working
independently along the same lines of mapping the energetically favorable and
excluded regions for protein conformations [1098].
These diagrams in the {φ,ψ} space, as shown in Figure 3.18,areused
to describe this {φ,ψ} flexibility (actually inflexibility) in polypeptides and
proteins. See also Figure 3.19 for a comparative view of Ramachandran plots de-
rived from the moderate-resolution X-ray structures shown in Figure 3.18 versus
high-resolution X-ray as well as NMR-derived structures.
Often, Ramachandran diagrams are presented by plotting the backbone dihedral
angles of all nonterminal residues in a protein for a large group of known protein
4
For lovers of scientific history, the 1998 biography of this renowned Indian molecular biophysi-
cist (1922–2001) is recommended [1091]. His peppered poetry is an added bonus. (My favorite poem
is number 9, on Superhelical Twisting and Replication of D N A [1091, p. 159].)
102 3. Protein Structure Introduction
180
0
−180
180
0
−180
180
0
−180
1800 0 1000500
100
50
0
50
25
0
100
50
0
Ψ (degrees)
Counts
Length (residues)φ (degrees)
X−ray, mod. res.
X−ray, higher res.
NMR
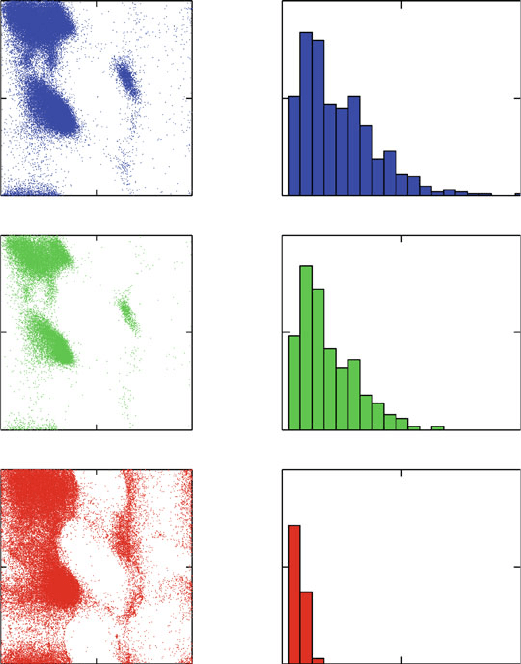
Figure 3.19. Three sets of Ramachandran plots based on the PDB40 dataset [959], corre-
sponding to: (top) X-ray protein structures with resolution of 2.5
˚
A or better (470 proteins,
95,778 total residues plotted, with proline and glycine excluded); (middle) X-ray protein
structures with resolution of 1.8
˚
A or better (183 proteins, 29,758 total residues plotted,
proline and glycine excluded); and (bottom) NMR-derived structures (113 proteins, 84,719
total residues plotted, proline and glycine excluded). For each subset of structures, the
length distribution is also shown.
structures. This superimposed view, averaged over many residues, approximates
protein conformational tendencies. The favorable regions correspond to common
secondary-structure elements, such as helices and sheets, with finer motifs also
noted.
In addition to favorable combinations of φ and ψ in polypeptides, the side-
chain dihedral angle χ
1
has been found to cluster around one of three conformers

3.4. Protein Conformation Framework 103
known as gauche
+
(or g
+
, χ
1
=+60
o
), gauche
−
(or g
−
, χ
1
= −60
o
), and
trans (or t, χ
1
= 180
o
). These are the favored orientations about tetrahedral atoms
(Figure 3.13). Some dependence of χ
1
on the residue’s φ and ψ values has also
been noted.
3.4.4 Conformational Hierarchy
Most natural proteins adopt specific 3D structures that are associated with their bi-
ological activity. Of course, proteins are dynamic, but typical thermal fluctuations
and local configurational arrangements revolve around a specific globally-folded
structure. The majority of proteins is believed to be unknotted in a topolog-
ical sense, though the polypeptide chain is frequently covalently bonded via
disulphide links and noncovalently held together by hydrogen bonds [1254].
One of the hallmarks of biomolecular structure is that the amino acid sequence
determines the 3D structure of a protein. This was first shown by Christian B.
Anfinsen and his colleagues in the early 1960s [51]. Anfinsen shared the Nobel
Prize in Chemistry in 1972 for his work on ribonuclease — connecting the amino
acid sequence to the biologically active conformation — with Stanford Moore
and William H. Stein — who connected ribonuclease’s chemical structure to its
catalytic activity.
5
In Anfinsen’s work, the protein ribonuclease was denatured by
destroying its hydrogen bonding network as well as intrinsic disulfide bonds. The
researchers observed that the protein spontaneously refolded into its native state
in a short time, regaining all its enzymatic activity. Of course, we recognize now
that accessory chaperone molecules may be necessary to assist in the folding of
many large proteins in vivo,asdiscussedinChapter2.
Four basic levels are used to describe protein structure:
• primary structure — the sequence of amino acids;
• secondary structure — regular local structural patterns such as α-helices
and β-sheets, or combination motifs thereof (supersecondary structure);
• tertiary structure — the 3D arrangement of all atoms in the polypeptide
chain in space; and
• quaternary structure (used for large proteins with independent subunits) —
the complete 3D interaction network among the different subunits.
The next chapter describes in turn the secondary and supersecondary, tertiary,
and quaternary structure of proteins.
5
Readers are invited to browse the electronic museum of Profiles in Science at
www.profiles.nlm.nih.gov/ for a glimpse not only of Anfinsen’s scientific activities but also of his
other hobbies and interests.
104 3. Protein Structure Introduction

4
Protein Structure Hierarchy
Chapter 4 Notation
S
YMBOL DEFINITION
α
R
classic right-handed α-helix
α/β
protein class
α + β
protein class
β-sheet
aggregating amino-acid strands
β
2
hairpin motif
β
4
Greek key motif
C
α
α-Carbon
π
helix form looser than α
R
φ {N–C
α
} rotation about peptide bond
ψ
{C
α
–C}=O rotation about peptide bond
3
10
helix form tighter than α
R
Try to learn something about everything and everything about
something.
Thomas Henry Huxley (1825–1895).
T. Schlick, Molecular Modeling and Simulation: An Interdisciplinary Guide, 105
Interdisciplinary Applied Mathematics 21, DOI 10.1007/978-1-4419-6351-2
4,
c
Springer Science+Business Media, LLC 2010
106 4. Protein Structure Hierarchy
4.1 Structure Hierarchy
The complexity of protein structures requires a description of their structural
components. This chapter describes the elements of protein secondary struc-
ture — regular local structural patterns — such as helices, sheets, turns, and
loops. Helices and sheets tend to fall into specific regions in the {φ,ψ} space
of the Ramachandran plot (see Figures 3.18 and 3.19). The corresponding width
and shape of each region reflects the spread of that motif as found in proteins.
Following this description of each secondary structural element, we discuss
the basic four classes of protein supersecondary or tertiary structure (the 3D spa-
tial architecture of a protein), namely α-proteins, β-proteins, α/β-proteins, and
α + β-proteins. This is followed by a presentation of the fold motifs for each such
class. Classes and folds are at the top of protein structure classification, as intro-
duced in the last section. Describing these folds and structural motifs is far from
an exact science, so variations in some of these aspects are common.
4.2 Helices: A Common Secondary Structural Element
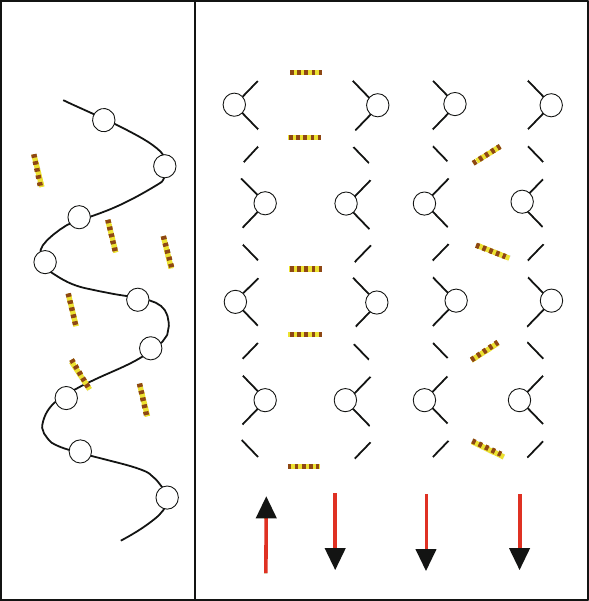
4.2.1 Classic α-Helix
In the classic, right-handed α-helix (α
R
), a hydrogen bonding network connects
each backbone carbonyl (C=O) oxygen of residue i to the backbone hydrogen of
the NH group of residue i +4(see Figure 4.1). This hydrogen bonding provides
substantial stabilization energy.
The regular spiral network of the α-helix is ubiquitous in proteins. It is as-
sociated with a {φ,ψ} pair of about {−60
o
, −50
o
}. The resulting helix has
3.6 residues per turn, and each residue occupies approximately 1.5
˚
A in length.
The helix may be curved or kinked depending on the amino acid sequence, as
well as on solvation and overall packing effects. Such distortions are reflected
by the {φ,ψ} distribution around the α
R
region in typical Ramachandran plots.
Hemoglobin, myoglobin, bacteriorhodopsin, human lysozyme, T4 lysozyme,
Trp repressor,andrepressor-of-primer (Rop) are all examples of proteins that
are virtually entirely α-helical. See Figures 4.2 and 4.3 for illustrations of such
α-proteins (see below) and Figure 3.10 for Rop.
An α-helix is associated with a dipole moment: the amino terminus of the helix
has a positive charge and the carboxyl end has a negative charge clustered about it.
Thus, residues that are negatively charged on the amino end and positively-
charged on the carboxyl end stabilize the helix; residues with the opposite charge
allocation destabilize the helix.
Experimental and theoretical work has shown that both intrinsic and extrinsic
(inter-residue interactions) factors are important for helix formation in proteins.
Residues with restricted sidechain conformations, due to long or bulky groups,
are poorer α-helix participants than other residues. Glutamine, methionine,
4.2. Helices: A Common Secondary Structural Element 107
C
α
C
α
C=O
N−H
C
α
C
α
C
α
C
α
C
α
C
α
C
α
C
α
C
α
O=C
C=O
H−N
C
α
H
|
N
N
−H
C
O
C
O
C
O
C
O
C
O
C
O
C
O
C
O
C
O
H
|
N
H
|
N
H
|
N
H
|
N
H
|
N
H
|
N
H
|
N
C
α
C
α
C
α
C
α
C
α
C
α
C
α
C
α
N−H
O=C
H−N
C=O
C=O
H−N H−N
O=C
O=C
N−H
N−H
C=O C=O
H−NH−N
O=C O=C
N−H N−H
C=O
C=O
H−N
H−N
=
=
=
=
=
=
=
=
=
H−N
C
α
C
α
C
α
C
α
H−N
N−H
O=C
H−N
C=O
N−H
O=C
C
α
α
R
antiparallel β−sheet
parallel β−sheet
Figure 4.1. Hydrogen bonding patterns in the classic α-helix (α
R
), with the ribbon tracing
the α-carbons (left), anti-parallel β-sheet (middle), and parallel β-sheet (right).
and leucine favor α-helix formation, while valine, serine, aspartic acid, and
asparagine tend to destabilize α-helices (e.g., due to steric and electrostatic
considerations).
4.2.2 3
10
and π Helices
There are more common variants of the α-helix motif that are typically not sta-
ble in solution but can play a part in overall protein structure. These include the
tighter 3
10
and looser π helices, with {φ, ψ} angles around {−50
o
, −25
o
} and
{−60
o
, −70
o
}, respectively.
The tighter 3
10
helix of three residues per turn (instead of 3.6 in the classic
α-helix) involves hydrogen bonds between residues i and i+3 instead of i and i+4
as in α
R
. There are 10 atoms within the hydrogen bond; hence the nomenclature
3
10
. The more loosely coiled π helix has hydrogen bonds between residues i and
i +5of the polypeptide.
108 4. Protein Structure Hierarchy
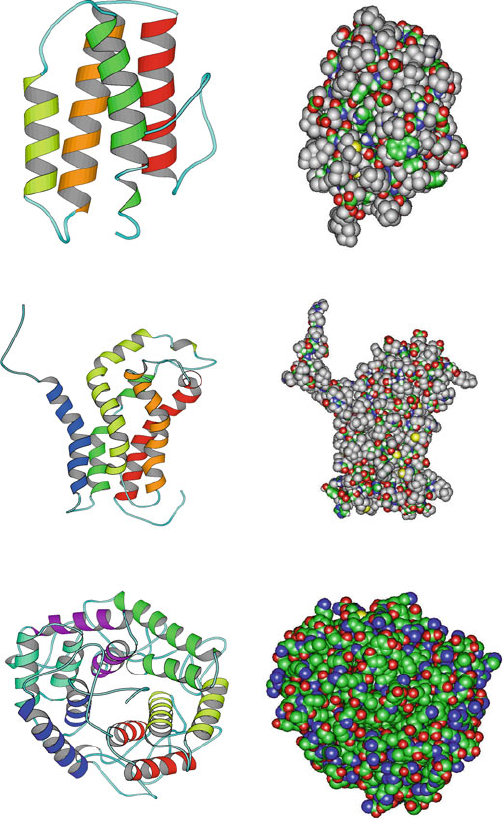
Pix (1BY1, 209 residues, five−helix bundle)
Myohemerythrin (2MHR, 118 residues, four−helix bundle)
Cellulase Cela (1CEM, 363 residues, six−alpha hairpins)
Figure 4.2. Examples of α-proteins: myohemerythrin, pix,andcellulase cela.
Because of their close packing, 3
10
helices generally form for a few residues
only, often at the C-terminus end of classic α-helices where the helix tends to
tighten. Similarly, the π helix occurs rarely since the backbone atoms are so
loosely packed that they leave a hole.
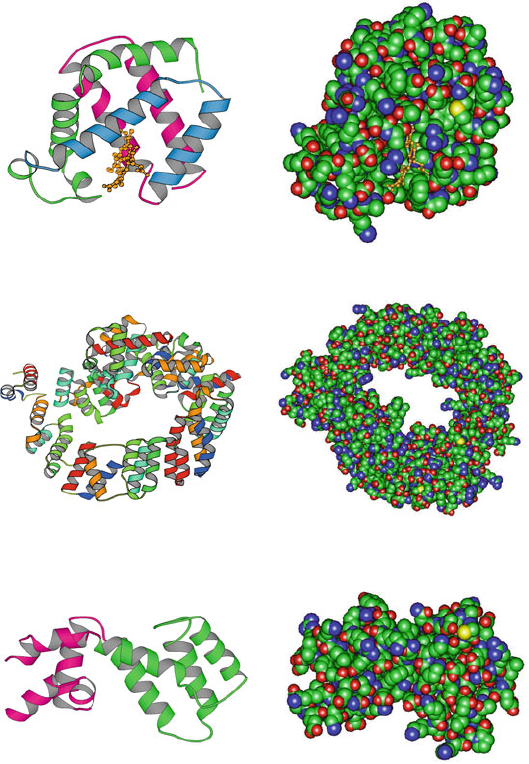
4.2. Helices: A Common Secondary Structural Element 109
Guanine Nucleotide−Binding Protein G(I)
(1AGR, 205 residues, two all−alpha domains)
Soluble Lytic Transglycosylase Slt70
(1QSA, 618 residues, folded leaf superhelix)
Myoglobin (5MBA, 147 residues, folded leaf)
Figure 4.3. Examples of α-proteins: myoglobin; soluble lytic transglycosylate protein of
bacterial muramidase, in the N-terminal region of the enzyme muramidase in bacterial cell
walls; and guanine nucleotide-binding protein, an irregular α-helical protein with a fold
containing a 4-helix bundle with left-handed twist.
4.2.3 Left-Handed α-Helix
A left-handed α-helix is theoretically possible, with {φ, ψ}= {+60
o
, +60
o
}.
However, this motif is generally unstable. The chirality preference for α-helices
follows the chirality of L-amino acids.
