Schlick T. Molecular Modeling and Simulation: An Interdisciplinary Guide
Подождите немного. Документ загружается.

90 3. Protein Structure Introduction
3.3.1 Globular Proteins
In most proteins, the twenty amino acids occur at roughly similar frequencies.
Notable exceptions occur for certain amino acids like methionine, which is fre-
quently found at the N-terminus of the peptide since it serves as the amino acid
initiator of synthesis, or special groups of proteins, such as membrane or fibrous
proteins.
Table 3.1 shows the frequency of occurrence of amino acid residues in the
PDB40 dataset of 971 domains of unrelated proteins with a sequence identity
of 40% or less [959], and Figure 3.8 displays the data as histograms. We see that
nonpolar Ala and Leu (boldfaced entries in the table) have the highest percentages
(above 8%) within the representative protein database. The lowest frequencies
(4% and below) occur for Trp (aromatic sidechain), Cys (sulfur-containing side-
chain), His and Met (the other sulfur-containing sidechain), Tyr and Phe (aromatic
sidechains also), and Gln.
Table 3.1. Amino acid frequencies in proteins based on the data of [615] which analyzed
45,137 proteins from 15 taxa. Bold and italics types are used, respectively, for the highest
(>8%) and lowest (≤ 2.5%) frequencies.
Amino Acid Freq. [%]
Alanine (Ala, A) 8.1
Arginine (Arg, R) 5.1
Asparagine (Asp, D) 5.2
Aspartic acid (Asn, N) 4.0
Cysteine (Cys, C) 1.2
Glutamine (Gln, Q) 3.8
Glutamic acid (Glu, E) 6.5
Glycine (Gly, G) 7.2
Histidine (His, H) 2.2
Isoleucine (Ile, I) 6.8
Leucine (Leu, L) 10.3
Lysine (Lys, K) 5.9
Methionine (Met, M) 2.5
Phenylalanine (Phe, F) 4.2
Proline (Pro, P) 4.3
Serine (Ser, S) 6.2
Threonine (Thr, T) 5.1
Tryptophan (Trp, W) 1.1
Tyrosine (Tyr, Y) 3.2
Valine (Val, V) 6.9
3.3.2 Membrane and Fibrous Proteins
Membrane proteins are embedded in a dynamic lipid bilayer environment, where
mobility is more restricted. They therefore have more hydrophobic residues than
3.3. Sequence Variations in Proteins 91
globular proteins, which favor polar groups on the exterior surface. Since mem-
brane proteins are particularly difficult to crystallize, simulation work is especially
important in this area to understand their detailed function.
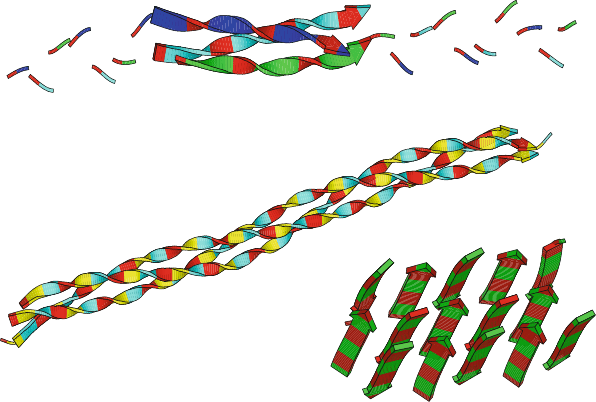
Fibrous, or structural, proteins tend to have repetitive sequences. The triple-
stranded collagen helix, for example, is composed of repeating triplets which
include glycine as the first residue and often proline as one or both of the other
residues of the triplet. A model of collagen is shown in Figure 3.9.
Since collagen is needed to rebuild joint cartilage, there are important prac-
tical applications to skin and bone ailments. For example, a gelatin-containing
(glucosamine and calcium-enriched) powdered drink mix called Knox NutraJoint
is being touted as a dietary supplement that helps maintain healthy joints and
bones. (‘Juice Your Joints’ touts an ad featuring an athletic sexagenarian water
skier). Gelatin is rich in two amino acids, glycine and proline, that make up
collagen.) Even though our bodies make these two amino acids, manufacturers
claim that this gelatin-containing supplement may be helpful in decreasing the
progression of osteoarthritis, a condition caused by cartilage deterioration.
Collagen model (1CLG) (Gly−Pro−Pro) units
12
3
Silk fibroin model (2SLK) (Gly−Ala) units
Triple−helical collagen−like peptide (1BKV)
Ile−Thr−
Gly−Ala−Arg−Gly−Leu−Ala−Gly−Pro−Hyp−Gly
[capped by (Pro−Hyp−Gly) on both ends]
3
Figure 3.9. Models of the fibrous proteins collagen (triple helix) and silk, along with a
crystallographically-determined collagen-like peptide (Hyp denotes hydroxyproline).
Another use of collagen is in a skin product used to heal wounds such as from
venous skin ulcers, burns, and skin surgery. In May 1998 the FDA approved
Apilgraf, a product for treating venous skin ulcers made of human skin cells
mixed with collagen cells from cattle.
92 3. Protein Structure Introduction
Silk is another example of a fibrous protein with a repetitive sequence. The
product of many insects and spiders, silk is the polypeptide β-keratin composed
largely of glycine, alanine, and serine residues, with smaller amounts of other
amino acids such as glutamine, tyrosine, leucine, valine, and proline. The softness,
flexibility, and high tensile strength of silk stems from its unique arrangement of
loose hydrogen bonding networks in the form of β-sheets connected by β-turns, a
mixture of both highly-ordered and less densely-packed regions. Figure 3.9 shows
a model of the repetitive β-sheet network of silk (without connecting regions).
3.3.3 Emerging Patterns from Genome Databases
As genome sequencing projects are completed, interesting findings on enzyme
sequences also emerge. For example, the genome of the tuberculosis bacterium
(completed in 1998 by the Wellcome Trust Genome Campus of the Sanger
Institute in collaboration with the Institut Pasteur in Paris) revealed surprisingly
that, unlike other bacteria, repetitive gene families of glycine-rich proteins exist
in M. tuberculosis; these approximately 10% of the enzyme-coding sequences are
associated with gene families involved in anaerobic respiratory functions.
3.3.4 Sequence Similarity
Sequence Similarity Generally Implies Structure Similarity
As mentioned above, sequence similarity generally implies structural, func-
tional, and evolutionary commonality. Thus, for example, if we were to scan
the Protein Databank (PDB) randomly and find two proteins with low se-
quence identity (say less than 20%), we could reasonably propose that they also
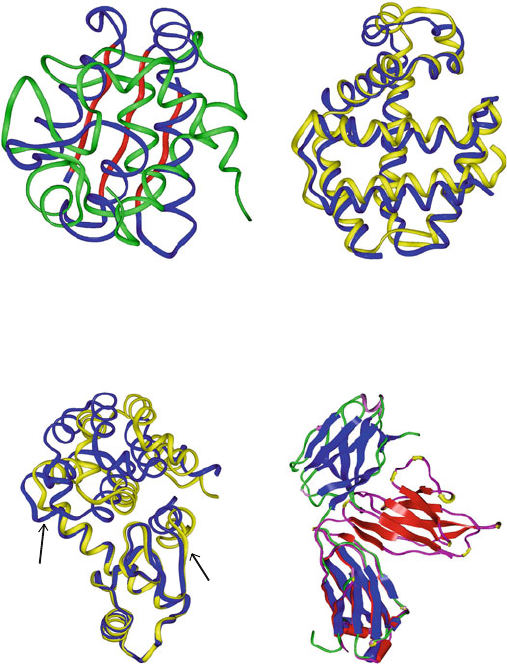
have little structural similarity. Such an example is shown in Figure 3.12 for the
cytochrome/barstar pair. Similarly, large sequence similarity generally implies
structural similarity (see introduction in 2.1.2 of Chapter 2).
In general, small mutations (e.g., single amino acid substitutions) are well
tolerated by the native structure, even when they occur at critical regions of sec-
ondary structure. The small protein Rop (Repressor of primer), which controls
the mechanism of plasmid replication, provides an interesting subject to both this
sequence-implies-structure paradigm, and to exceptions to this rule (discussed
below).
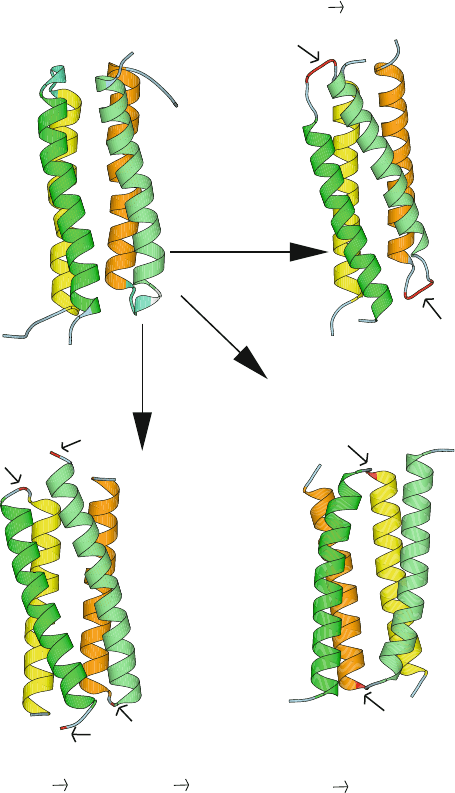
Rop is a dimer, with each monomer consisting of two antiparallel α-helices
connected by a short turn; it dimerizes to form a four-helix bundle as active
form, as shown in Figure 3.10. (Fold details and motifs are discussed in the next
chapter). Recall that Rop was used as the basis for solving Paracelsus challenge
(Chapter 2) because the α-helix motif was thought to be quite stable.
The high stability of Rop emerged surprisingly from experiments of Castagnoli
et al. [205]. When these researchers deleted just a few residues in a key turn region
that produces the overall bundle fold in the native Rop structure, they expected one
long contiguous helix to form. Instead, their tinkering produced a small variation
3.3. Sequence Variations in Proteins 93
N
N
C
C
C
N
C
N
Ala−Asp31−Ala
Ala−Asp31−Ala
C
C
N
N
Gly1
Gly30
Gly30
Gly1
C
C
N
N
Pro31
Pro31
Single mutation:
entire topology altered
Minor mutation
(overall structure
preserved)
Minor insertion
(overall structure
preserved)
ROP wild−type(1RPR)
(left−handed anti−parallel bundle)
ROP mutant (1RPO)
(Asp31 Ala−Asp31−Ala)
ROP mutant (1GTO) ROP mutant (1B6Q)
(Met1 Gly, Asp30 Gly) (Ala31 Pro)
(right−handed mixed bundle)
ab
cd
Figure 3.10. The protein Repressor of Primer (63 residues per monomer) provides in-
teresting examples of the paradigm of structure inference by sequence similarity: the
four-helix bundle motif of the wildtype (a) can be both structurally stable, i.e., resistant
to mutations — as shown by the two variants in (b) and (c) — or structurally fragile and
highly sensitive to mutations, caused by proline substitution at the turn region — as shown
in (d), a mutant with an entirely different topology [462]. In (b), two Ala residues were
inserted at both sides of the amino acid Asp in the loop region. In (c), the Asp residue con-
necting the two α-helices of each monomer was mutated to Gly, and Met1 was changed to
Gly [205].
94 3. Protein Structure Introduction
of the original bundle motif. Apparently, the four-helix bundle motif is so stable
that a new turn was formed from residues that used to be part of the α-helix back-
bone! Thus, the original bundle motif, though slightly smaller, was maintained
in the mutants. This is seen in Figure 3.10, which displays the wildtype enzyme
structure (a) and those of two mutants in the above cited study (b and c).
Though this experiment supports the general notion that protein structures
are remarkably stable to tinkering (mutations), we emphasize that functional
properties of proteins are fragile and quite sensitive to sequence changes.
Exceptions Exist
There are many exceptions, however, to this simple sequence/structure/function
relationship.
Namely, examples exist where despite large sequence similarity there is small
structural and functional similarity. A classic example of this relationship is the
disease sickle-cell anemia, where a minute substitution in sequence leads to al-
tered function with devastating consequences. This abnormality results from the
replacement of the highly-polar glutamate residue in hemoglobin by the nonpo-
lar amino acid valine. This key substitution at the surface of the protein leads to
an entirely different quaternary structure for this multidomain red-blood pigment
protein. This is because the markedly altered structure affects the solubility of
oxygenated hemoglobin and leads to a clumping of the deoxygenated form of the
molecule (HbS instead of HbA).
Conversely, examples exist where despite small sequence similarity there is
large structural, and even functional and evolutionary similarity. A classic ex-
ample for this relationship is the myoglobin/hemoglobin pair of proteins (see
Figure 3.12). These proteins only share 20% of the sequence. However, as
oxygen-carrying molecules, they share structural, functional, and evolutionary
similarity. Proteins in the calmodulin family are also known to display a great
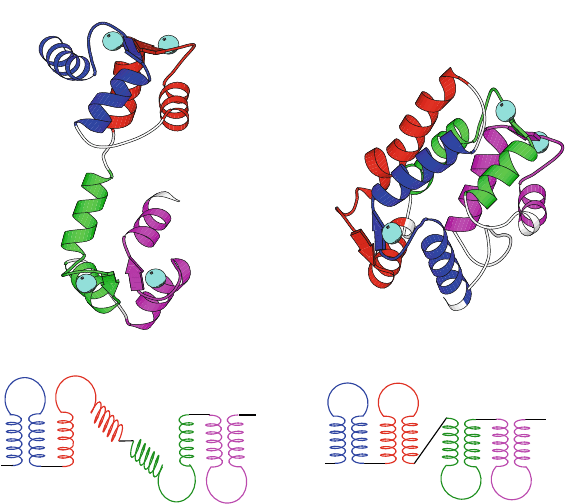
deal of structural variability for similar sequences [664] (see Figure 3.11).
More generally, changes in 3D architecture (despite a nontrivial degree of
sequence similarity) can result from a variety of factors, as follows.
• Mutations in critical regions of the proteins, such as active sites and ligand
binding sites, can change 3D structures dramatically. Such an example is
shown for the pair of immunoglobulins in Figure 3.12.
• Mutations in regions that connect two secondary-structural elements can
also be responsible for structural divergence, as in the helix-loop-helix
motif of the EF-hand family, and the connecting loops in helix bundles.
Figure 3.11 illustrates this principle for the two EF-hand calcium-binding
proteins calmodulin and sarcoplasmic calcium-binding protein: one is
overall extended in shape while the other is more compact [979].
Helix bundles are sensitive to mutations in loop or turn regions that connect
different helices, to the extent that a single amino acid substitution (alanine
3.3. Sequence Variations in Proteins 95
to proline) can change the topology of a homodimeric 4–helical bundle pro-
tein from the canonical left-handed all-antiparallel form to a right-handed
mixed parallel and antiparallel bundle [462]. Figure 3.10(d) shows this dif-
ferent resulting topology of the Rop four-helix bundle subject to the single
mutation Ala31→Pro at the turn region.
• Structural variations can be observed in the same system determined at
different environmental conditions such as solvent or crystal packing. The
same T4-lysozyme mutants in Figure 3.12 (100% sequence similarity) dis-
play intriguing mobility, adopting 5 different crystal conformations [374]
due to a hinge bending motion.
• Multidomain proteins can adapt quaternary structures that depend sensi-
tively on the number of subunits and/or on the sequence.
D
B
C
A
EF hands
EF hands
A
B
C
D
111 118 145 148
1 6 38 45
77
82
C
D
B
A
Calmodulin (3cln),
elongated form
Sarcoplasmic calcium-binding
protein (2scp)
EF hands
EF hands
1 3 38 43 82
89 122 130 159 174
A
B
C
D
Figure 3.11. Structural variability despite large similarity in the protein secondary-struc-
tural elements is illustrated for two calcium-binding proteins — calmodulin (148 residues)
and sarcoplasmic calcium binding protein (174 residues) [979] — due to different over-
all 3D arrangement of the shared motifs. Though sharing only 30% of the sequence, both
proteins are made of two repeating units, each consisting of two EF hand motifs. Each
hand motif contains two helical regions surrounding a calcium binding loop (crystal-bound
calcium atoms are rendered as large spheres; only three are bound to 2scp).
96 3. Protein Structure Introduction
Leu 79
Leu 13
A’
A
B and B’
Immunoglobulins
8FAB:A, strands in red
1DCL:B, strands in blue
All β sandwich, Greek key
Myogloblin (5MBA, yellow)
Hemoglobin (1ASH)
All−α, globin−like, folded leaf
Cytochrome C6 (1CTJ)
all−α, folded leaf
Barstar (1BTA)
α/β, 3 parallel β strands
Mutant T4 lysozymes (150L)
Wild−type like
Hinge angle 32
⬚
, yellow
Mainly α, lysozyme−like
Seq sim 100%, RMS 2 A Seq sim 75%, RMS 9 A
oo
o
Seq sim 3%, RMS 7 A Seq sim 12%, RMS 1.8 A
o
ab
cd
Figure 3.12. Various examples of sequence/structure relationships in proteins: (a) Low
sequence similarity (3% for alignment of 72% of the residues) generally implies low struc-
ture similarity (cytochrome C6 versus barstar). Still, exceptions are found. For example,
in (b), despite low (12%) sequence similarity, there is large structure and function similar-
ity (hemoglobin and myoglobin); conversely, despite high sequence similarity, there can
be structural diversity, due to (c) hinge bending in two lysozyme mutants (Met → Ile in
residue 6) or (d) different orientation of one of the two subunits in two immunoglobulins.
The lysozyme mutant displays 5 different crystal conformations, one similar to the wild-
type (shown in blue) and others overall very similar except for a different hinge-bending
angle (see defining arrows); the form with largest bend (32
o
) is shown in yellow. The two
immunoglobulins differ markedly in tertiary organization due mainly to differences in the
linker domain between the A and B subunits of each protein.
3.4. Protein Conformation Framework 97
gauche
−
gauche
+
τ = 0
o
60
120
−120
−60
180
gauche
−
gauche
+
trans
For n−butane (CH
3
− CH
2
− CH
2
− CH
3
)
CH
3
CH
3
CH
3
CH
3
CH
3
CH
3
CH
3
CH
3
trans
τ
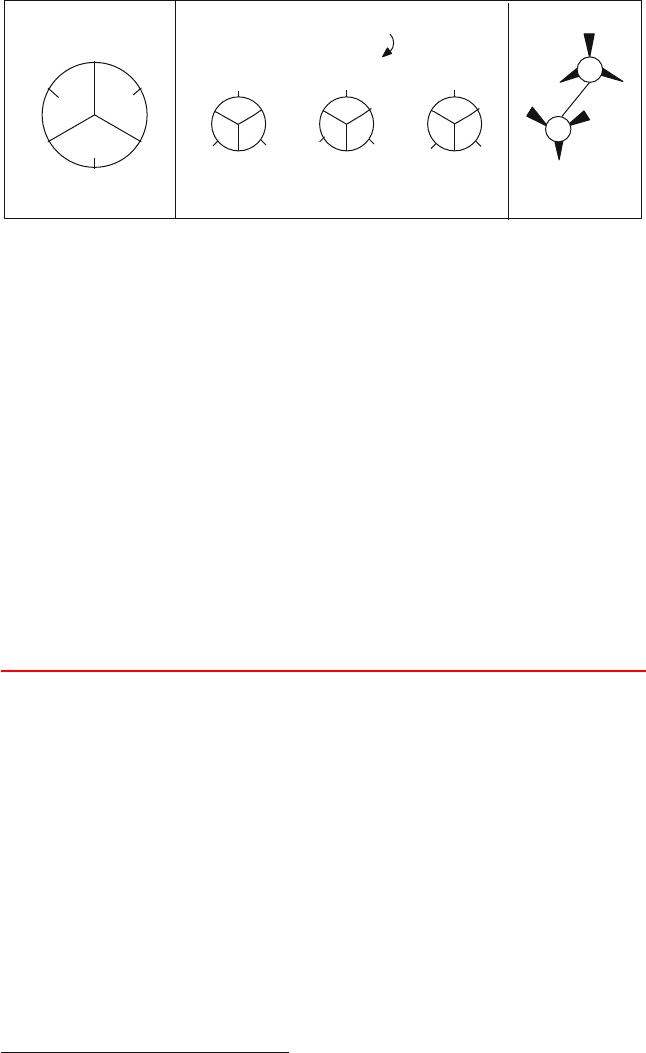
Figure 3.13. Gauche (g) and trans (t) dihedral-angle orientations for n-butane: (left) clas-
sification wheel; (middle) simple Newman projections that illustrate the three favored
orientations of the two end methyl groups about the central C–C bond (perpendicular to the
plane of the paper); and (right) the trans conformation, which has the least steric clashes.
3.4 Protein Conformation Framework
3.4.1 The Flexible φ and ψ and Rigid ω Dihedral Angles
Polypeptides can have a wide variety of conformations, i.e., 3D structures differ-
ing only in rotational orientations about covalent bonds.
2
This type of rotational
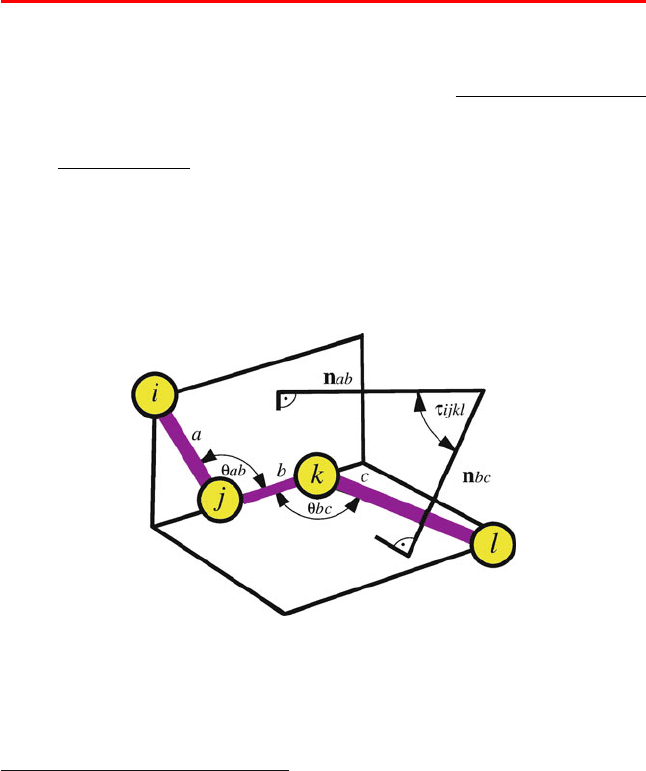
flexibility is characterized by a dihedral angle, which measures the relative ori-
entation of four linked atoms in a molecule, i − j − k − l. A dihedral angle for
a 4-atom sequence that is not necessarily covalently bonded can also be used for
special terms in the potential energy function; see Chapter 9 for examples. See
Box 3.4, Figure 3.14, and the equations in Appendix D, under the Addendum to
Assignment 8 for a definition of a dihedral angle.
Box 3.4: Dihedral Angle
The dihedral angle τ
ijkl
defined for a sequence of linked atoms i–j–k–l (Figure 3.14)is
the angle between the normal to the plane of atoms i–j–k and the normal to the plane of
atoms j–k–l. The sign of τ
ijkl
is determined by the triple product ( a × b) · c,wherea, b
and c are the interatomic distance vectors for atoms i → j, j → k and k → l, respectively.
Strictly defined, the related torsion angle, τ is the angle between the two planes defined
by i–j–k and j–k–l. Thus, τ + τ = π (180
o
). However, the terms torsion and dihedral
angle are often used interchangeably. We will often use dihedral angle to refer to the
numerical value of the angle, and torsion angle or torsional potential when we discuss
general properties of these rotations.
2
see Chapter 8, Subsection 8.4.1, for the related definition of configuration.
98 3. Protein Structure Introduction
When the dihedral angle is 0
o
, the four atoms i–j–k–l are coplanar and atoms i and l coin-
cide in their projections onto the plane normal to the j–k bond; this orientation is defined
as cis or syn. When the dihedral angle is 180
o
, the atoms are coplanar but atoms i and l
lie opposite one another in the projection onto the plane normal to the j–k bond; such an
orientation is defined as trans or anti. More generally, angular regions convenient to de-
scribe protein and nucleic acid conformations are the following: cis (≈0
o
), trans (≈180
o
)
and ± gauche (≈±60
o
). Another common terminology is: syn (≈0
o
), anti (≈180
o
), ±
synclinal (≈±60
o
),and± anticlinal (≈±120
o
). See Figure 3.13 for a simple illustration
for n-butane.
While the peptide group (Figure 3.3) is relatively rigid — it has 40% double-bond
character — there is a great deal of flexibility about each of the single bonds
along the backbone, {N–C
α
} and {C
α
–C}=O. The two dihedral angles φ and ψ
are used to define rotations about the bond between the nitrogen and C
α
of the
mainchain and between C
α
and the carbonyl carbon, respectively (Figure 3.15).
The dihedral angle ω
defines the rotation about the peptide bond, namely for
theatomicsequenceC
α
1
–{C–N}–C
α
2
,whereC
1
and C
2
are the α-carbons of two
adjacent amino acids. Because of the partial double-bond character of the peptide
bond and the steric interactions between adjacent sidechains, ω is typically in the
trans configuration: ω = 180
o
.
3
In this orientation, all four atoms lie in the same
plane, with the distance between C
α
1
and C
α
2
as large as possible (see Figure 3.13
for a definition of various dihedral angle orientations).
Figure 3.14. Definition of a dihedral angle τ
ijkl
=cos
−1
(n
ab
·n
bc
), the angle between the
two normals spanned by atoms i, j, k and j, k, l.
3
Non-trivial deviations from planar peptide bonds can be shown by theory and experiment (e.g., as
reviewed in [348]). A statistical survey of peptide and protein databases verified that the distribution
of rotation angles (or energies associated with peptide bond rotations) follows Boltzmann statistics
[799].
3.4. Protein Conformation Framework 99
O
C
N
H
C
O
H
N
C
i−1
α
C
i
α
R
i−1
H
H
Ri
H
R
i+1
ψ
i−1
ω
φ
i
ψ
i
ω
φ
i−1
i−1
Residue i
i+1
C
i+1
α
Figure 3.15. Rotational flexibility in polypeptides: definition of the φ, ψ and ω dihedral
angles.
3.4.2 Rotameric Structures
Besides the {φ,ψ} flexibility associated with the two backbone bonds involving
C
α
, multiple conformations are possible for 18 of the 20 amino acids when the
sidechain geometries differ (excluded are glycine and alanine). Rotameric struc-
tures of amino acids (and hence proteins) are those that have the same {φ,ψ}
angles but differ in the sidechain conformations. The dihedral angles used to de-
fine sidechain rotations are denoted by χ, with subscripts used as needed (see
Figure 3.17).
C
1
C
2
C
3
C
4
C
α
χ
1
χ
2
χ
3
χ
4
N
H
H
HH
H
H
H
H
H
H
H
H
Figure 3.16. Lysine’s four rotamers defined by torsional variables χ
1
through χ
4
.
For example, in lysine, whose sidechain has four carbons (see Figure 3.16),
dihedral angles χ
1
through χ
4
denote the rotations about bonds C
α
–C
1
,C
1
–C
2
,
C
2
–C
3
,andC
3
–C
4
, respectively (see also Figure 3.17 for other amino acids).
Rotameric structures for polypeptides and proteins depend on the environment of
the polymer and on the secondary and tertiary structures.
3.4.3 Ramachandran Plots
The feasible combinations of the φ and ψ angles are limited due to steric hin-
drance. That is, only certain combinations are typically observed, with some
dependence on residue size and shape. Glycine is unique in its flexibility — it is
therefore a good agent for turns in polypeptides and proteins — but other residues
exhibit a highly limited range of sterically-permissible φ and ψ combinations. In
fact, only roughly one tenth the area of the {φ,ψ} space is generally observed
for polypeptides and proteins. Among the first to note this limitation were
