Nof S.Y. Springer Handbook of Automation
Подождите немного. Документ загружается.


Automatic Control in Systems Biology 75.4 Dynamical Models 1345
Two broad classes of methods for SNA have been
developed: metabolic pathway analysis (MPA)andflux
balance analysis (FBA) [75.70–72]. MPA computes and
uses the set of independent pathways-generating rays in
Fig.75.7 – that uniquely describe the entire flux space;
owing to the algorithmic complexity, it can currently
only handle networks of moderate size. FBA, in con-
trast, determines a single flux solution through linear
optimization [75.73], often assuming that cells try to
achieve optimal growth rates. The computational costs
are modest, even for genome-scale models. The ap-
proach was successful, for instance, in predicting the
effects of gene deletions and the outcomes of conver-
gent evolution in microorganisms [75.72,74,75]. FBA,
however, hasto reverse-engineerand operate with an es-
sentially unknown objective function. While maximal
growth has proven to be a reasonable assumption for
lower organisms [75.76], higher cells may tend to min-
imize overall fluxes in the network [75.77]. In general,
FBA has proven effective for simpler organisms, and
when the steady-state assumption is valid. However,
there are many situations where these conditions do
not apply, many of which are biophysically meaningful,
such as the dynamic diauxic shift in E. coli.
Extensions: Dynamics and Control
Stoichiometric constraints restrict the systems dynam-
ics. Thus, the stoichiometric matrix N is fundamental
not only for SNA but also for dynamic processes in re-
action networks, in which the reaction rates r in (75.2)
are time-dependent. For biological systems, the conser-
vation of total amounts of certain molecular subgroups
(conserved moieties such as ATP, ADP and AMP) is
characteristic and can be exploited for systems analy-
sis. Classical work in chemical engineering addressed
this topic for chemical reaction networks. For instance,
Feinberg derived theorems to determine the possible
dynamic regimes, such as multistability and oscilla-
tions, based on network structure alone [75.78, 79].
Challenges posed by biological systems led to re-
newed interest in these approaches and induced further
theory development [75.80, 81]. Application areas in
biology include stability analysis [75.80] and model
discrimination by safely rejecting hypotheses on re-
action mechanisms, thus identifying crucial reaction
steps [75.82]. Algorithms for the identification of de-
pendent species in large biochemical systems – to be
employed, for instance, in model reduction – have re-
cently become available [75.83].
Enabling FBA to deal with dynamics and regulation
proceeded by incorporating additional time-dependent
constraints that reflect knowledge of the operation
of cellular control circuits – an approach termed
regulatory FBA [75.84]. For instance, using superim-
posed Boolean logic models to capture transcriptional
regulatory events has extended the validity of the
methodology for a number of complex dynamic sys-
tem responses [75.84] and for data integration [75.85].
Other dynamic extensions of the FBA algorithm have
been proposed in [75.86]. With these more detailed
models, steady-state analysis suggested that the com-
plex transcriptional control networks operate in a few
dominant states, i. e., generate simple behavior [75.87].
Finally, pathway analysis also allows one to approach
features of intrinsically dynamic systems: for instance,
it helps to identify feedback loops in cellular signal
processing [75.88]. Hence, SNA-related approaches are
about to extend to nonclassical domains, in particular,
through theory development induced by new challenges
in systems biology.
Functional Constraints, Optimality, and Design
In analyzing living systems, one possibility is to start
from the assumptions that they have to fulfill certain
functions and that cells have been organized over evo-
lutionary time-scales to optimize their operations in
a manner consistent with mathematical principles of
optimality. FBA demonstrates the utility of this assump-
tion; note that its implicit functional constraint, i. e.,
steady-state operation of metabolic networks, is not
self-explanatory. Similarly, other approaches invoking
principles of optimal control theory have opened new
avenues for systems analysis in biology.
The cybernetic approach developed by Kompala
et al. [75.89]andVa rn er and Ramkrishna [75.90]is
based on a simple principle: evolution has programmed
or conditioned biological systems to optimally achieve
physiological objectives. This straightforward concept
can be translated into a set of optimal resource allo-
cation problems that are solved at every time-step in
parallel with the model mass balances (basic metabolic
network model). Thus, at every instant in time, gene ex-
pression and enzyme activity are rationalized as choice
between sets of competing alternatives, each with a rel-
ative cost and benefit for the organism. Mathematically,
this can be translated into an instantaneous objective
function. The researchers in this area have defined sev-
eral postulates for specific pathway architectures, and
the result is a computationally tractable(i.e., analytical)
model structure. The potential shortcoming is a limited
handling of more flexible objective functions that are
commonly observed in biological systems [75.91–95].
Part H 75.4

1346 Part H Automation in Medical and Healthcare Systems
Instead of focusing on a single objective func-
tion, mathematical models and experimental data can
be used to test hypotheses on optimality principles,
given a specific cellular function to be fulfilled. For
instance, extensions of FBA suggested that E. coli opti-
mizes the tradeoff between achieving high growth rates
and maintaining wild-type metabolic fluxes after gene
deletions [75.96]. MPA showed that the interplay be-
tween the metabolic network (the controlled plant) and
gene regulation (the controller) in E. coli might be de-
signed to achieve optimal tradeoffs between long-term
objectives, such as metabolic flexibility, and short-term
adjustment for metabolic efficiency [75.97]. Optimal
production pipelines for biomass components, with fast
responses to environmental changes and minimal addi-
tional efforts for enzyme synthesis, were predicted in
detail to employ wave-like gene expression programs,
which was later confirmed experimentally [75.77, 98].
Hence, at least certain cellular design principles can be
revealed by evaluating assumptions on cellular optimal-
ity principles.
Finally, without assuming optimality, we can ask
how functions in biological systems could be es-
tablished in principle. Among others, drawing from
analogies with engineered systems helps to understand
more general design principles in biology. From non-
linear dynamics, for instance, it is well-known that
functions such as oscillators and switches require some
source of nonlinearity. Establishing sucha functionwith
biological building blocks, thus, allows only for cer-
tain circuit designs [75.99,100]. Similar ideas can prove
powerful at different levels of abstraction. For instance,
highly structured bow-ties with multiple inputs, chan-
neled through a core with standardized components and
protocols to multiple outputs, could be the common
organizational principles to establish complex produc-
tion systems in engineering and biology [75.53]. On
the other hand, El-Samad and colleagues studied the
bacterial heat-shock response, pointing out that the in-
tertwined feedback and feedforward loops present can
be assigned individual functions parallel to those loops
in designed control circuits that have to yield fast re-
sponses in highly fluctuating environments [75.101].
Notably, most of the examples discussed here involved
newdevelopmentsin theory toaddress challenges posed
by biology.
75.5 Network Identification
Model development involves the translation of identi-
fied biological processes to coupled dynamical equa-
tions, which are amenable to numerical simulation
and analysis. These equations describe the interac-
tions between various constituentsand the environment,
and involve multiple feedback loops responsible for
system regulation and noise attenuation and amplifica-
tion. Currently, however, our knowledge of essentially
all biological systems is incomplete. Despite genome
projects that allow enumeration and, to a certain ex-
tent, characterization of all genes in a system, this
does not imply knowledge about all network compo-
nents (for instance, all protein variants that can be
derived from a single gene), interactions, and properties
thereof [75.22]. An important task in systems biology
consists of specifying network interactions, which can
concern qualitative or quantitative properties (existence
and strength of couplings), or detailed reaction mecha-
nisms, for genome-based inventories of components.
Essentially, this is a systems identification prob-
lem. Given a set of experimental data and prior
knowledge, the network generating the data is to be
determined [75.102]. Alternatively termed reverse en-
gineering [75.103], network reconstruction [75.104]or
network inference [75.105], the general network iden-
tification problem provides a key interface between
science and engineering. Several qualitatively different
approaches for biological systems have been proposed,
which can be roughly classified into three categories:
data-driven, approximative and mechanistic.
75.5.1 Data-Driven Methods
Empirical or data-driven methods rely on large-scale
datasets that can be generated, for instance, through
microarray analysis for gene regulatory networks.
The relevant methods include singular value decom-
position analysis of microarray data [75.106, 107],
self-organizing maps [75.108], k-means clustering or
hierarchical clustering [75.109], protein correlation and
dynamic deviation factors [75.110], and robust statis-
tics approaches [75.111, 112]. For instance, clustering
methods are routinely applied for identifying groups of
coregulated genes from microarray data. The interpre-
tation of clustering results employs (implicit) models
such as coexpressed genes that are likely to have a com-
Part H 75.5

Automatic Control in Systems Biology 75.5 Network Identification 1347
mon regulator. Data quality andalgorithmic choices (for
instance, of distance measures) critically influence the
clustering results; in addition, validation of clustering
results and techniques is an open issue [75.113–115].
In contrast to the mechanistic approaches discussed
later, most empirical approaches employ discrete-time
gray box models [75.116–119]. For instance, infer-
ence methods based on probabilistic graphical (e.g.
Bayesian) models help to elucidate causal couplings
between the network components [75.120]. Their scal-
ability for large systems and the ability to integrate
heterogeneous datasets make them attractive [75.5,
121]. Yet, these approaches deliver only qualitative
descriptions of network function, and have inherent lim-
itations. For instance, Bayesian models cannot cope
with the ubiquitous feedback in cellular networks, since
causal relationships have to be represented by directed
acyclic graphs [75.120].
A number of challenges are present in treating ex-
perimental data for such problems: (1) the sampling rate
is rarely uniform and may be exponentially spaced by
design, and (2) data from multiple research groups are
often combined (e.g. from WWW-posted data) to yield
data records with inconsistent sampling, experimen-
tal bias, etc. From a systems engineering perspective,
another critical point is the potentially divergent qual-
itative behavior between continuous-time and discrete-
time models of corresponding order [75.122]. Recent
work has shown the promise of continuous-time formu-
lations of empirical models using modulating function
approaches [75.40].
More generally, correctly identifying network
topologies (corresponding to the model structure)
clearly does not suffice for establishing predictive
mathematical models. Experiences with engineered
genetic circuits illustrate this point: with identical topol-
ogy, qualitatively different behavior can result and
vice versa [75.123]. Hence, quantitative characteris-
tics, which are usually incorporated through parameters
in deterministic models, are also required. Corre-
sponding identification methods are rooted in systems
and information theory and, thereby, also provide the
largest intersection among biology, other sciences, and
engineering.
75.5.2 Linear Approximations
The identification of dynamically changing interac-
tions requires corresponding dynamic models. In a first
approximation, we can consider linear systems, i.e.,
systems with additive responses to perturbations. In
systems engineering, a standard form for linear time-
invariant (where the shapeof the output does not change
with a delay in the input) systemswith n states andm in-
puts is given by (75.3) with n ×1 state vector x(t), n ×n
system matrix A, n ×m input matrix B,andm ×1 in-
put vector u(t). Linearization of the general dynamic
system dx(t)/dt = f (x, p, t) with parameter vector p
provides first approximations to the network dynam-
ics, even for highly nonlinear systems such as those
encountered in biological networks. Linear models cap-
ture the local dynamics, for instance, in the vicinity of
a steady state, instead of aiming at more complicated
global behaviors.
dx(t)
dt
= f(x, p, t) ≈Ax(t)+Bu(t)
(75.3)
Mathematically, most methods reconstruct the system
matrix A, which corresponds to the Jacobian matrix
J =∂ f(x, p)/∂x, from the measured effects of (suffi-
ciently small) perturbations. However, direct recovery
of the system matrix A will be unreliable with noisy
data and inputs. In a study using linear models and
perturbation experiments to identify the structure of
genetic networks, Tegner et al. [75.103] therefore pro-
posed an iterative algorithm that uses rational choices
of perturbations to improve the identification quality.
For a developmental circuit, despite high nonlinearities
in the system, the reverse engineering algorithm, which
involves building and refining an average connectivity
matrix in successive steps, recovered all genetic inter-
actions [75.103]. A related approach that uses linear
models and multiple linear regressions showed simi-
lar performance. The algorithm attempts to exploit the
sparsity of systems matrices for biological networks
owing to, for example, (estimated) upper bounds on the
number of connections per node [75.105, 124]. Both
algorithms are scalable – a central concept in engineer-
ing, but until recently considered of less importance in
biology.
Newer approaches to systems identification aim
at exploiting modularity in biological networks. For
a modular system with one output per module, the
method employsinversion of the global response matrix
for identification of network connectivities and of lo-
cal responses from perturbation experiments [75.125].
It requires a reduced number of measurements com-
pared with other methods because only changes in
so-called communicating intermediates have to be
recorded. Apparently, some simplifying assumptions
have to be made; for example, modules are coupled
by information flow only, and mass flow is negligi-
Part H 75.5

1348 Part H Automation in Medical and Healthcare Systems
ble [75.125]. An important result of extending the
modular identification to time-series data is that, for
identifying all connections of a node, it is not nec-
essary to perturb this node directly – inference can
rely on detecting the network responses to remote
perturbations [75.126]. Extensions to include the ef-
fects of uncertainties in experimental data and prior
knowledge [75.127], and the possibility of a unified
mathematical framework [75.128] make modular iden-
tification methods particularly promising.
75.5.3 Mechanistic Models, Identifiability
and Experimental Design
Mechanistic models, owing to effects such as saturation
in enzymatic reactions, pose particular challenges be-
cause they involve identification of nonlinear systems.
Depending on whether model structure and parame-
ters, or only the parameters have to be identified, the
problems fall into the classes of mixed-integer nonlin-
ear programs or nonlinear programs, respectively. As
a clear limitation, finding a unique global optimum in
the estimation, or convergence of the algorithms cannot
be guaranteed. In addition, model identification comes
at high computational costs owing to numerous model
simulations [75.129].
In terms of parameter estimation, which is a com-
mon problem in different scientific domains [75.102],
realistic modeling of complex, nonlinear dynamics of
biological networks has given new impulses for the
evaluation of existing methods and development of new
methods. For instance, though stochastic algorithms
show superior performance over deterministic meth-
ods for parameter optimization in these systems, they
are computationally expensive [75.130]. Novel hybrid
methods try to exploit synergies between both ap-
proaches in order to increase robustness and efficiency
(e.g. [75.131, and references therein]).
More fundamentally, identifiability and design of
informative experiments need to be addressed. Unstruc-
tured approaches to model identification are completely
ill-posed when faced with, for instance, modeling
a yeast cell with 6200 genes and four possible states
per gene; we obtain an overall expression state dimen-
sioninexcessof10
15
[75.132]. Clearly a number of
a priori constraints and correlations must be exploited.
For discrete models, usage of the experimentally ob-
served upper bound on the number of interactions per
species brings the amount of data needed for identi-
fication into realistic dimensions [75.133]. However,
mere extrapolation of current high-throughput technol-
ogy will not solve these high dimensional data issues.
Several recent studies have highlighted the importance
of proper design of perturbations to reveal the logical
connectivity of gene networks [75.104, 134]. Systems
engineering concepts of experimental design to pro-
vide rich datasets can be exploited to develop predictive
mechanistic models.
Parameter estimation accuracies are central to mea-
suring identifiability of mechanistic models. Low accu-
racies mean that the corresponding parameters may be
varied to a greater extent – and still describe the data –
than it is possible for parameters with high estimation
accuracy (low associated error). They combine infor-
mation on model sensitivities with experimental data
(Fig.75.8). More specifically, the Fisher information
matrix (FIM) F(p) [75.135], for a point in parameter
space p, links model and experiment via state sensitiv-
ities S(t) =∂x/∂ p and measurement covariance matrix
for a discrete sampling time t
i
, C(t
i
). For an unbiased
estimator, the Cramér–Rao theorem then gives a lower
bound for the estimation error (minimum variance).
FIM-based approaches, for instance, yielded insight
into the importance of suitable design of input per-
turbations for signaling networks [75.40], optimality
criteria for the design of such inputs [75.136], and al-
gorithms for the optimization of sampling times for
dynamic experiments [75.137]. New hybrid parameter
estimation methods [75.131] and closed-loop (i.e., inte-
grating iterations between estimation, evaluation of the
identifiability, and experimental design) optimal identi-
fication procedures [75.138] rely on the FIM formalism.
Note, however, that while these proof-of-concept stud-
ies with small models and synthetic data are valuable,
the performance for real biological problems awaits
assessment.
Mathematical model
Parameter sensitivities
Fisher information matrix: F(p) =
Quality of identification: σ
j
≥
S(t
i
)
T
C
–1
(t
i
) S(t
i
)
Experimental data
Measurement
covariance matrix C
S(t) =
∂x(t)
∂p
x = x(t,p), p = p
0
Σ
[
F(p)
–1
j,
j
[
[
[
N
i=1
Fig. 75.8 FIM and identification quality. The lower bound
for the estimation error of parameter j, σ
j
, is derived from
the inverse of the main diagonal of the FIM (after [75.4])
Part H 75.5

Automatic Control in Systems Biology 75.6 Quantitative Performance Metrics 1349
Information-rich datasetsfor integrative models will
have to be derived from sources across all levels
of biological regulation, such as the transcriptome,
proteome, and metabolic fluxes. Concomitantly, we
need novel statistical frameworks for data integra-
tion [75.139]. Systems identification would greatly
benefit from the direct in vivo determination of ki-
netic parameters; the work by Ronen et al. [75.140]
for transcriptional control is a first step in this direc-
tion. As a complement, synthetic genetic circuits could
provide means for controlled excitation of the system,
for instance, by inducible genetic switches [75.103],
or through genetic oscillators to incorporate analysis
methods inthe frequencydomain [75.102].Novel meth-
ods could also take known uncertainties associated with
measurements, suchas experimentally determined char-
acteristics of stochastic noise, explicitly into account.
Finally, identification depends on adequate specifica-
tion of the system and model (e.g., [75.141]). While
models are currently either set up ad hoc, or through
manual comparison of few alternative structures (in-
cluding kinetic terms in the equations), uncertainties in
biology pose a major challenge for systems sciences:
deriving advanced approaches to model discrimination
for the simultaneous identification of model structures
and parameters.
75.6 Quantitative Performance Metrics
In biology, as in engineering, robust performance refers
to the attainment of a particular behavior or response by
a system in the presence of uncertainty. This appears to
be a ubiquitous property of biological processes that are
subject to constant uncertainty in the form of stochas-
tic phenomena [75.142], fluctuating environment, and
genetic variation (reviewed in [75.15]). Biology has
adapted a number of approaches for coping with these
sources of uncertainty, which include:
1. Back-up systems (redundancy)
2. Disturbance attenuation through feedback and feed
forward control
3. Structuring of networked systems into semi-
autonomous functional units (modularity)
4. Reliable coordination of network elements through
hierarchies and protocols.
The robustness problems in systems biology have
only begun to yield, in recent years, formal quanti-
tative analyses, owing largely to their nonlinear (and
nonstationary) nature. As with engineering systems, ro-
bust performance requires the precise specification of
both a performance metric and the type/size of uncer-
tainty. When both these elements are specified, it may
be possible to analyze biological systems with estab-
lished engineering tools. It is important to note that
the performance metric is often difficult to be defined
precisely in biology, as it is an implicit element of an
evolved entity.
The last 10 years have demonstrated that sensi-
tivity analysis can provide unique insights into the
functioning of complex biophysical networks [75.4,
143–145]. Of particular interest is the behaviour of
biological circuits that exhibit oscillations (e.g., circa-
dian rhythm, cell cycle, neuron firing, cardiac cycles,
etc.). In [75.146], a novel set of sensitivity metrics
was introduced for performance that were based on
a number of different phase-measures: period, phase,
corrected phase, and relative phase. The motivation
was that phase appears to be the biological imperative,
rather than period, for optimal regulation of circadian
rhythms. Both state-based and phase-based tools were
applied to free-running (absence of light/dark cycles)
Drosophila melanogaster and Mus musculus circadian
models. Each metric produced unique relative sensitiv-
ity measures used to rank parameters from least to most
sensitive. Similarities among the resulting rank distri-
butions strongly suggested a conservation of sensitivity
with respect to parameter function and type. A con-
sistent result, for instance, is that model performance
of biological oscillators is more sensitive to global pa-
rameters than local (i.e., circadian specific) parameters.
Differences across the metrics revealed that the conclu-
sions about robustness were dependent on the metric
employed for performance.
In [75.147], a novel sensitivity measure, the para-
metric impulse phase response curve (pIPRC), was
derived to both characterize the phase behavior of an
oscillator and provide the means for computing the
response to an arbitrary signal (in the form of paramet-
ric perturbation). The pIPRC builds on the knowledge
that biologists have collected for decades in the form
of phase response curves (PRCs), to more general
classes of input perturbations. The PRCs and infinitesi-
mal PRCs presented in that study provided quantifiable
measures of robustness for oscillators acting as pace-
Part H 75.6

1350 Part H Automation in Medical and Healthcare Systems
makers. In these systems, robust performance involves
proper maintenance of phasebehavior. In the case ofthe
circadian clock, this means that the PRC to light must
have notonly theproper shape, but alsothe correct mag-
nitude. Zeilinger et al. [75.148] were able to invalidate
a model of the circadian clock in the plant Arabidop-
sis thaliana, because the pIPRC had neither the proper
shape nor the proper magnitude.
In [75.149], sensitivity methods were employed to
predict the likelihood that noise propagates in stochas-
tic models of the circadian network. The results showed
that noise introduced into a sensitive point in the clock
propagates very well, while noise introduced into an
insensitive (or robust) point is undetectable elsewhere.
The noise propagates without regard to distance from
point of introduction to point of measurement. It was
concluded that the sensitive global parameters are the
sites of effective noise propagation in the clock. The hy-
pothesis was that global parameters govern reactions at
critical points in the network and therefore may suggest
those parts of other systems most worthy of investiga-
tion.
The previous studies focused on single cell models,
but network properties must also be analyzed at higher
levels in the system. Bagheri et al. [75.150] demon-
strated that computational techniques applied to single
cell data are fundamental for tuning and predicting the
behavior of oscillatory phenomena at the population
level [75.151], since the results of such investigations
point to the coupling mechanisms that give rise to
spontaneously synchronized networks of stochastic bio-
physical nodes. Without such insight, it would not have
been possible to reproduce the synchrony observed in
the SCN. As a result, it is important for experimental
biologists to adopt the tools necessary to analyze the
structure of both in vitro and in vivo systems.
75.6.1 State-Based Sensitivity Metrics
Parametric state sensitivity is captured in an m by
n matrix consisting of individual state performance
values with respect to isolated parametric perturba-
tions. The direct method [75.152, 153] is a means
to determine exact parametric state sensitivity mea-
sures, S
ij
(t) = dx
i
(t)/dρ
j
. This approach relies on the
continuity of f[x(t), ρ] with respect to the parameter
vector ρ. Applying the chain rule results in partial
derivatives of the function with respect to states and pa-
rameters produces an ordinary differential equation of
sensitivity dynamics (75.4). The initial conditions for
the sensitivities are zero unless the system initial condi-
tions x
0
depend on the parameters.
˙
S(t) =
∂ f[x(t), ρ]
∂x
•S(t)+
∂ f[x(t), ρ]
∂ρ
(75.4)
In oscillatory systems, coefficients of raw state sensitiv-
ity, S(t) = dx(t)/ dρ, rely on multiple coupled outputs
(such as period and phase) and grow unbounded in
time for parameters whose period sensitivities are
nonzero [75.154–156]. The secular term is due to com-
putation of sensitivities involving a nonuniformly valid
expansion of a periodic system [75.154]. Tomovic and
Vukobratovic [75.155] demonstrate that state sensitiv-
ity, evaluated at the constant nominal period τ, provides
a specific measure corresponding to limit cycle be-
havior. This measure is unbiased to changes in the
period/frequency of oscillation. The resulting m by n
matrix is referred to as the cleaned-out or shape sensi-
tivity measure S
C
(t) =(∂x(t)/∂ρ)
τ
; S
C
(t) is periodic in
time and describes how parametric perturbations affect
the shape of state trajectories [75.154]. In its raw form,
state sensitivity for oscillatory systems may be decom-
posed intoa combination of shape andperiod sensitivity
measures (75.5) [75.155]. This decomposition of raw
state sensitivity highlights its linear time growth t/τ
while isolating period sensitivity, S
τ
= dτ/dρ. The de-
composition is generally accomplished by calculating
state and period sensitivities, and then solving for the
cleaned-out shape sensitivity matrix.
S(t) = S
C
(t)−
t
τ
f[x(t),ρ]
dτ
dρ
(75.5)
Zak et al. [75.156] proposed a method to determine pe-
riod sensitivity S
τ
by making use of the decomposition
(75.5) and evaluating state sensitivities at a large time
t
1
*τ. At this time, the second term on the right-hand
side of (75.5) dominates the cleaned-out sensitivity ma-
trix. Singular value decomposition of state sensitivity
produces S(t
1
) =UV
, where is an m by n di-
agonal matrix of nonnegative singular values σ and
matrices U and V contain the eigenvectors of SS
and S
S, respectively. Hence, period sensitivity of any
given state may be approximated by (75.6) where σ
1
is
the largest singular value in , f[x(t
1
)], ρ is the vec-
tor norm of the state matrix evaluated at time t
1
,and
V
1
is the first column vector of V. This approximation
holds true at any large times t *τ, when the system is
nonzero, and when the period of oscillation is sensitive
to at least one parameter [75.156].
S
τ
≈±
σ
1
τ
f[x(t
1
)], ρt
1
V
1
(75.6)
Part H 75.6
Automatic Control in Systems Biology 75.6 Quantitative Performance Metrics 1351
Amplitude sensitivity S
∧
describes how maximum state
values vary due to independent parametric perturba-
tions. As discussed, cleaned-out sensitivity defines how
parametric perturbations affect the shape of state trajec-
tories at every time in the cycle; at peak concentration
times this measure relates directly to the state’s maxi-
mum concentrations. Thus, amplitude sensitivity values
are calculated from shape sensitivity by evaluating the
time-dependent measure (75.7) at peak concentrations
times t
peak
S
∧
=S
C
(t
peak
) . (75.7)
75.6.2 Phase-Based Sensitivity Metrics
In cases involving oscillatory networks and often noisy
state amplitude data, phase may be employed as a met-
ric for model development and analysis. Standard (raw)
phase and decoupled (corrected) phase are among
the set of numerical performance measures introduced
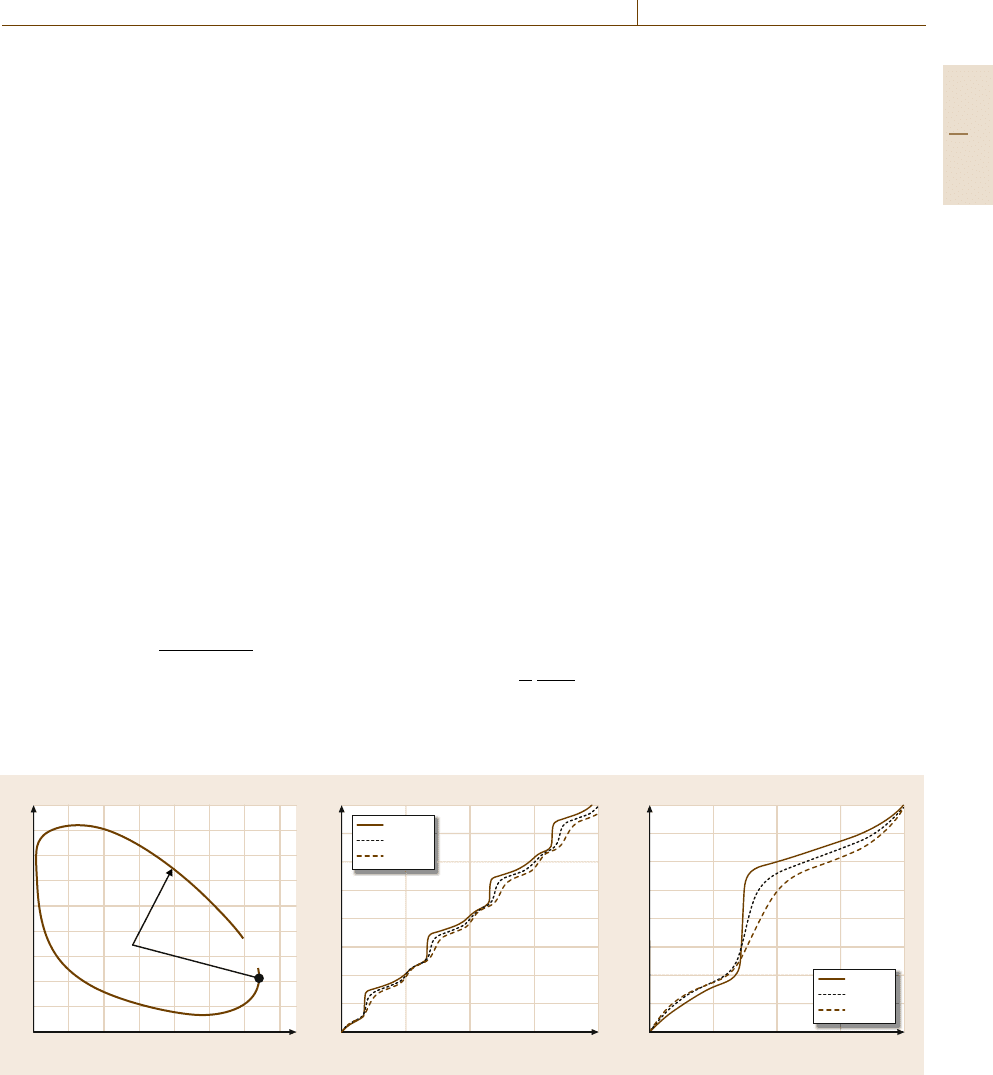
in [75.146]. Radian-based phase angles θ(t,ρ) are ex-
tracted from the system’s limit cycle (Fig.75.9a) using
the cosine rule [75.158]. Resulting phase dynamics re-
flect the oscillator’s real-time position, or concentration
(75.8), with respect to a static reference r
cos[θ(t, ρ)]≈±
x(t,ρ)
r
x(t, ρ)r
.
(75.8)
Phase measures are recorded under varying parameter
sets, θ(t,
˜
ρ), where
˜
ρ indicates measurements with re-
spect to a perturbation of magnitude δ affecting a single
0
a) Protein concentration (nM)
mRNA concentration (nM)
0.4 0.8 1.2 1.6 2
θ (t)
2.4 2.8
2
1.6
1.2
0.8
0.4
0
b) Phase (rad/π)
Time (h)
96724824
8
7
6
5
4
3
2
1
0
0
c) Phase (rad/π)
Percent of a single cycle
100755025
2
7/4
3/2
5/4
1
3/4
1/2
1/4
0
1.15 v
sp
1 v
sp
0.85 v
sp
1.15 v
sp
1 v
sp
0.85 v
sp
Reference
vector
Real-time
state vector
Fig. 75.9a–c Time-dependent phase dynamics. (a) Real-time phase dynamics captured in a 2-D limit cycle. Phase is the radian-
based measure θ(t, ρ) that describes the angular relationship between the state vector x(t, ρ) and some predetermined reference r.
Phase measures are recorded as a function of time under various parameter sets.
(b) Coupling of period and phase as trajectories
diverge in time.
(c) Decoupled measure of phase behavior as circadian phase dynamics are normalized with respect to their
perturbation-induced periods (after [75.146,157] by permission of Oxford University Press)
parameter ρ
j
. This perturbation changes the nominal
parameter vector from ρ to a perturbed parameter vec-
tor ρ
j
˜
ρ =ρ +δe
j
, ∀j ∈[1, n]. (75.9)
The collected phase trajectories capture the oscillator’s
position and map them onto nominal time (Fig. 75.9b).
If the perturbation strength and length are the same, the
information contained in these trajectories is analogous
to thatcontained in phase transitioncurves [75.159]. Di-
rect evaluation of phase trajectories (Fig.75.9b) yields
two types of sensitivity measures: period and phase,
because the change in period (or period sensitivity)
is merely an accumulation in the change in phase (or
phase sensitivity) evaluated over an entire cycle. In
the case of period sensitivity, phase trajectories are
evaluated at a 2π L-radian interval, where the integer
interval L is chosen to balance the numerical error
with computational expense. The difference between
the perturbed 2π L-radian crossing time
˜
k and the nom-
inal 2π L-radian crossing time k yields the system’s
periodic performance. A normalized time difference be-
tween perturbed and nominal phase trajectories defines
the system’s period sensitivity, S
θ
τ
(75.10): a series of
measurements denoting the quantitative change in pe-
riod with respect to a change in the j-th parameter
S
θ
τ
j
=
1
L
˜
k−k
δρ
j
θ(k,ρ)=θ(
˜
k,
˜
ρ)=2π L
,
∀j ∈[1, n], L ∈[1, ∞) .
(75.10)
Part H 75.6
1352 Part H Automation in Medical and Healthcare Systems
In the case of strict phase sensitivity, one can evalu-
ate phase trajectories at specific times t =t
k
. The radian
phase difference between perturbed and nominal trajec-
tories describes the raw phase sensitivity, S
θ
(75.11):
a series of measures reflecting the induced change in
phase with respect to a change in the j-th parameter
t
k
-hours after the perturbation
S
θ
j
(t
k
)=
θ(t,
˜
ρ)−θ(t, ρ)
δρ
j
t=t
k
, ∀j∈[1, n]. (75.11)
10
–2
a)
d) e)
(Phase) Period sensitivity (h/nM)
(State) Period sensitivity (h/nM)
10
0
10
2
10
1
10
0
10
–1
10
–2
10
2
Autoinhibition
Transcription
Translation
mRNA degradation
Protein degradation
Complex degradation
Unspecified degradation
(De)phosphorylation
(Dis)association
Transport
0 10 20 30
b) Relative phase ranking
Corrected phase ranking
30
20
10
0
Autoinhibition
Transcription
Translation
mRNA degradation
Protein degradation
Complex degradation
Unspecified degradation
(De)phosphorylation
(Dis)association
Transport
Most
sensitive
Degradation
Degradation, complex
Degradation, mRNA
Degradation, protein
Transport
Translation
Transcription
Expression
(Dis)association
Phosphorylation
Dephosphorylation
Global
State
Amplitude
Shape sensitivity
Period (state)
Period (phase)
Corrected phase
Phase
Relative phase
Mixed
Local
Least
sensitive
Most
sensitive
Least
sensitive
c) State ranking
Amplitude ranking
Autoinhibition
Transcription
Translation
mRNA degradation
Protein degradation
Complex degradation
Unspecified degradation
(De)phosphorylation
(Dis)association
Transport
Most
sensitive
Degradation
Degradation, complex
Degradation, mRNA
Degradation, protein
Transport
Translation
Transcription
Expression
(Dis)association
Phosphorylation
Dephosphorylation
Global
State
Amplitude
Shape sensitivity
Period (state)
Period (phase)
Corrected phase
Phase
Relative phase
Mixed
Local
Least
sensitive
Most
sensitive
Least
sensitive
0102030
30
20
10
0
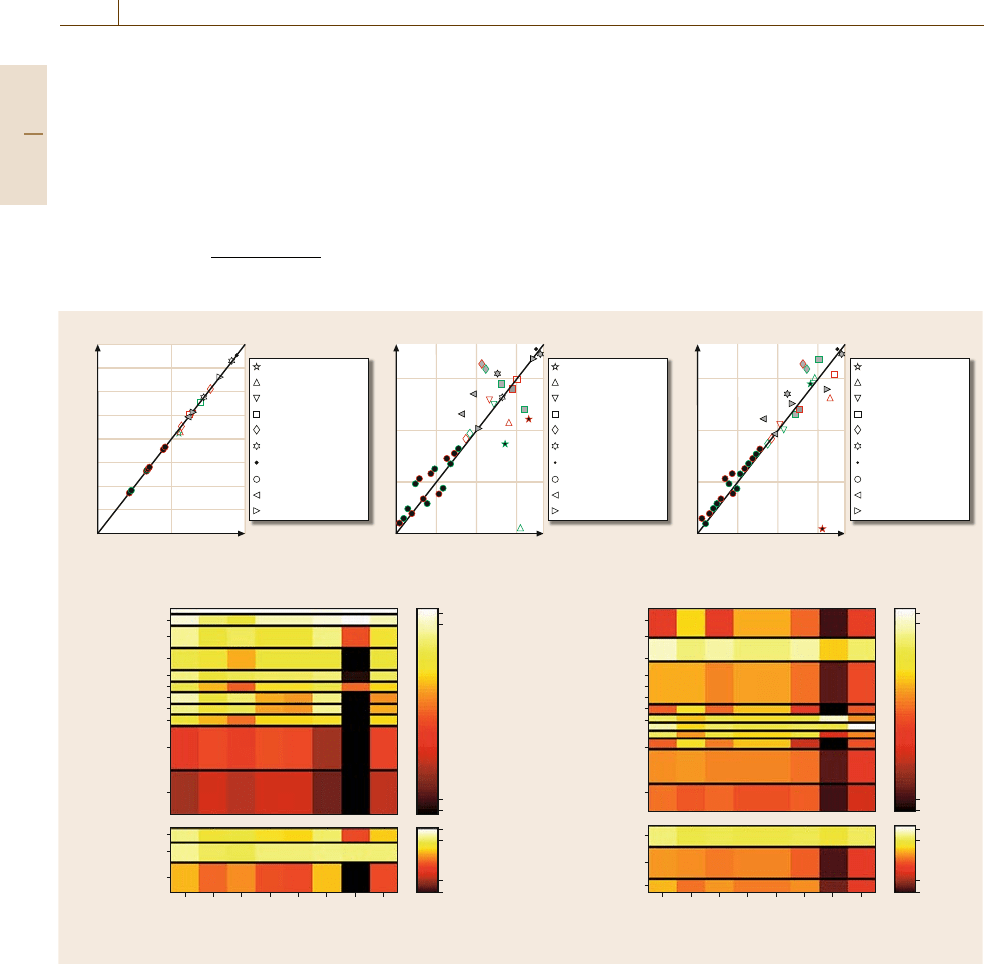
Fig. 75.10a–e Metric evaluation. (a–c) Parametric sensitivity metrics ordered from least to greatest absolute value for the
Drosophila model:
(a) state-based and phase-based period sensitivity, (b) corrected phase (decoupled angular phase trajecto-
ries) and relative phase (time interval between peak per mRNA and nuclear PER/TIM protein concentrations) sensitivity ranks
and
(c) amplitude-based and state-based metrics. Spearman rank correlation coefficients for these pairs of metrics are 1.00, 0.64
and 0.85, respectively. Legends describe the parameters’particular biological processes as the shading of each data point describes
their type: global (open), mixed (gray) and local (black) parameters. Data points outlined in red reflect per gene dynamics, and
those in green reflect tim gene dynamics.
(d,e), Color-coded average sensitivities (values are scaled between 10
−4
and 1) among
parameter function (upper subplots) and parameter type (lower subplots) for each metric in the fly
(d) and the mammalian (e)
model. See the supplementary material for alternate correlation diagrams and ranking plots (after [75.146,157] by permission of
Oxford Univ. Press)
Just as state sensitivity diverges over time, phase
sensitivity grows unbounded due to the nonuniform
expansion of a periodic system. To correct for the inte-
grated response of perturbation effects (in this case, the
coupling of period and phase), each phase trajectory is
normalized with respect to the period of the system af-
ter parametric perturbation
˜
τ: dividing each time series
by
˜
τ decouples the system’s phase from its period. As
a result, normalized datasets begin and end at the same
relative time points (0 and 100% of their respective cy-
Part H 75.6
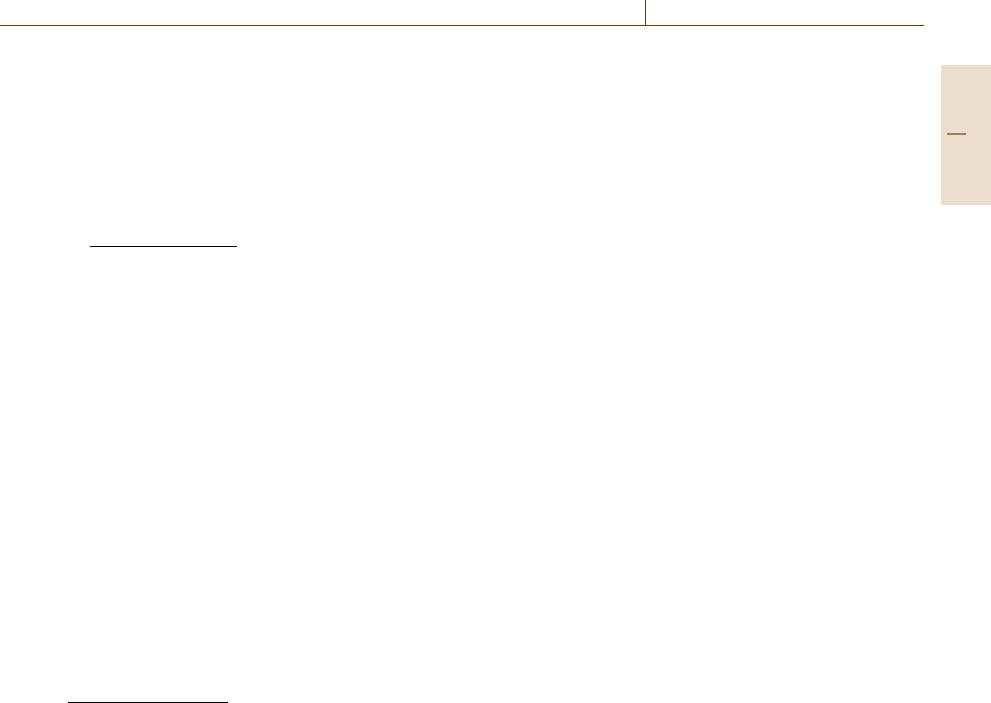
Automatic Control in Systems Biology 75.7 Bio-inspired Control and Design 1353
cles). These modified datasets allow for a comparison
between nominal and perturbed phase at every point
in the cycle (Fig.75.9c). This corrected phase sensi-
tivity assumes a linear scaling of raw phase measures
resulting in a time-dependent performance quantity that
identifies specific points of the cycle most susceptibleto
uncertainty (75.12)
S
%
Θ
j
(t
k
)=
θ(t/
˜
τ,
˜
ρ)−θ(t/τ, ρ)
δρ
j
, ∀j∈[1, n]. (75.12)
Phase-based period, phase, and corrected phase sensi-
tivity analysis examine the biological network relative
to a static reference. In some cases, a relative analysis
that studies relationships within the perturbed network
may be more useful. The timing effects relating tran-
scription, translation, phosphorylation,and transport are
governed byglobal cellularprocesses. Variationof these
specific time intervals as a result of parameter ma-
nipulation indicates a degree of sensitivity. Relative
sensitivity S
φ
investigates the time interval relating the
hour-difference between the occurrence of particular
events; for instance, the time interval between peak
mRNA concentrations and their corresponding protein
concentrations. This time interval φ[x(t), ρ], is a func-
tion of the system’s state and parameter vectors. It
explores how a system’s individual components change
relative to one another due to parametric perturba-
tion (75.13)
S
φ
j
=
φ[x(t),
˜
ρ]−φ[x(t), ρ]
δρ
j
, ∀j∈[1, n]. (75.13)
75.6.3 Global Versus Local Parameters
The investigated performance metrics depict classes of
sensitivity that associate with parameter function, prov-
ing a certain conservation of robustness for specific
biochemical processes.A similar sensitivity distribution
was found when parameters were separated into three
types: global, mixed, and local [75.15]. Global param-
eters are involved in core cellular reactions nonspecific
to the circadian rhythm;they encompass properties con-
sistent with the entirety of the cellular network. Local
parameters are primarily attributed to the circadian sys-
tem; their processes and/or elements are not shared with
many other cellular circuits. Global parameters include
transcription rates, various mRNA and protein degrada-
tion rates, and translation rates.
Figure 75.10a–c depict phase-based and state-based
period sensitivities, relative and corrected phase sen-
sitivities, and amplitude and state sensitivities. Global
parameters were consistently found in the first (up-
per right) quadrants of these plots. Local parameters,
including protein phosphorylation and dephosphoryla-
tion, were consistently in the lower left (less sensitive)
corners. Figure 75.10d and e (lower subplots) empha-
size this conservation of sensitivity apparent in both
models by assigning a color reflective of the average
sensitivity ranking within each parameter type.
The results demonstrated that every defined per-
formance metric was more sensitive to perturbations
involving global and mixed parameters than it was
to perturbations involving local parameters. Grouping
parametric sensitivity based on parameter type provided
a more consistent and distinct distribution of sensitiv-
ity measures among various metrics than the grouping
of sensitivity by function. This outcome agreed with
a previous study suggesting that circadian performance
is greatly affected by changes in global parameters and
less susceptible to changes in local parameters [75.13,
15].
75.7 Bio-inspired Control and Design
Biological networks offer a number of opportunities
for inspired design of engineering networks. Aside
from the overlapping computational toolkit (e.g., sim-
ulation methods for high dimensional, stochastic, stiff,
multiscale systems), there are numerous behaviors in
biological networks that offer promise for improved
communications and sensors networks. Given the space
constraints, we highlight only two of them here, but re-
fer the reader to the thorough NRC report on network
science for additional details [75.160].
Using control engineering parlance, one may refer
to the elements that influence or entrain the circadian
oscillator (zeitgeber: time giver) as manipulations, and
the elements that exhibit quantifiable circadian rhythms
as measurements. The use of this terminology is re-
flective of the spirit in looking for ways to influence
rhythms to finetune physiological performance.Contin-
uing the control analogy, the open-loop characteristics
(i.e., with no intervention) of the typical human are to
adjust at arate ofapproximately 60–90min ofphase per
day [75.161].In otherwords, jet lag accommodation oc-
curs at a rate of approximately 2–3days for a 3 h time
zone change. Westward travel is slightly easier, as the
natural free-running human circadian clock (i. e., in ab-
Part H 75.7

1354 Part H Automation in Medical and Healthcare Systems
sence of light) has a period of approximately 25h. From
a control perspective, the two important attributes of the
resetting response characteristics are phase shift (cap-
tured by the phase response curve), and the associated
transient, which we define via the transient response
curve.
Sensitivity analysis can be used to develop insight
on the parts of a network that are most sensitive and,
consequently, the most susceptible to intervention such
as the targeting of a drug. However, a typical applica-
tion will require a temporal forcing of a node (or nodes)
in a network to elicit an optimal response. In recent
work, model predictive control (MPC) algorithms have
been employed to generate the optimal forcing proto-
col that will reset the circadian clock from a condition
of phase offset (i.e., jet lag) [75.157, 162]. Through
parametric state sensitivity analysis, key driving mech-
anisms for optimal manipulation of the large complex
circadian network were identified. Importantly, the use
of nonphotic control inputs outperforms light-based
phase resetting dynamics. Aside from targeting indi-
vidual parameters as control inputs, Fisher information
matrix based parametric sensitivity analyses identified
combinations of parameters for control (i. e., vector
strategies). The derived MPC algorithm is found to be
robust to model mismatch and outperforms the open-
loop 24h sun cycle based phase recovery strategy by
nearly threefold.
75.8 Emerging Trends
Systems biology has an exciting future. Opportunities
for research exist in the areas of network identifica-
tion, constraints and optimality, stochastic systems, and
robustness [75.4], as well as synthetic approaches to
network reconstruction:
1. Knowledge of biological systems is incomplete,
and codification of the interactions among network
components, interactions, and properties requires
continued elaboration. This systems identification
problem can be approached via empirical, approx-
imative, and mechanistic methods.
2. An understanding of biological systems may pro-
ceed from an appreciation of constraints, which
can be divided into three classes: (1) Functional
constraints exist because biological systems per-
form specified functions arising from a finite set
of components. Since biological systems evolve to
a performance metric, it is sensible to presume op-
timality. This leads to a smaller design space with
highly structured, but rarely composed, reliably
networks.(2) Analysis ofexperimentaldata can pro-
vide data on the possible arrangements for network
structure. (3) The laws of physics and chemistry,
must, of course, be observed.
3. Developments in systems biology will continue
to depend upon algorithmic innovations to ap-
proach stochasticity, particularly for distributed
models.
4. There exist intrinsic scaling problems with those
engineering methods that provide a formal frame-
work for robustness analysis. For large, complex,
nonlinear systems, the methods are conservative
and computational tractability is questionable. Con-
sequently, the scalability of robustness analysis
methodologies provides interesting future chal-
lenges.
One promising application of a systems approach
to biology lies in healthcare [75.163]. Unlike today’s
medicine, with its emphasis on therapies for existing
disease, tomorrow’s medicine will predict and prevent
illness. Multiparameter diagnostics will be used rou-
tinely to assess health status. Network science will be
used to ascertain drug targets, leading to true preventive
medicine.
The emerging bio-inspired automation and au-
tomatic control mechanisms, techniques and mod-
els will continue to offer rich sources of innova-
tion. We will continue to search how the benefits
of systems biology knowledge in living organisms
can help us solve complex, critical problems in in-
creasingly interrelated systems and service infrastruc-
tures.
References
75.1 J. Hasty, D. McMillen, F. Isaacs, J.J. Collins: Com-
putational studies of gene regulatory networks:
In numero molecular biology, Nat. Rev. Genet. 2,
268–279 (2001)
Part H 75
