Nof S.Y. Springer Handbook of Automation
Подождите немного. Документ загружается.

1335
Automatic Co
75. Automatic Control in Systems Biology
Henry Mirsky, Jörg Stelling, Rudiyanto Gunawan, Neda Bagheri, Stephanie R. Taylor, Eric Kwei,
Jason E. Shoemaker, Francis J. Doyle III
The reductionist approaches of molecular and
cellular biology have produced revolutionary
advances in our understanding of biologi-
cal function and information processing. The
difficulty associated with relating molecu-
lar components to their systemic function
led to the development of systems biology,
a relatively new field that aims to establish
a bridge between molecular level information
and systems level understanding. The novelty
of systems biology lies in the emphasis on
analyzing complexity in networked biological
systems using integrative rather than reduc-
tionist approaches. By its very nature, systems
biology is a highly interdisciplinary field that re-
quires the effective collaboration of scientists
and engineers with different technical back-
grounds, and the interdisciplinary training of
students to meet the rapidly evolving needs of
academia, industry, and government. This chap-
ter summarizes state-of-the-art developments of
automatic control in systems biology with sub-
stantial theoretical background and illustrative
examples.
75.1 Basics ..................................................1335
75.1.1 Systems Biology ...........................1336
75.1.2 Control Research in Systems Biology1336
75.2 Biophysical Networks ............................1337
75.2.1 Timing and Rhythm:
Circadian Rhythm Networks
and Oscillatory Processes...............1337
75.2.2 Apoptosis: Programmed Cell Death .1338
75.2.3 Signals in Diabetes:
Insulin Signaling Pathway .............1339
75.3 Network Models
for Structural Classification....................1340
75.3.1 Hierarchical Networks...................1341
75.3.2 Boolean Networks, Petri Nets,
and Associated Structures..............1341
75.4 Dynamical Models.................................1342
75.4.1 Stochastic Systems........................1343
75.4.2 Constraints and Optimality
in Modeling Metabolism ...............1344
75.5 Network Identification..........................1346
75.5.1 Data-Driven Methods ...................1346
75.5.2 Linear Approximations..................1347
75.5.3 Mechanistic Models, Identifiability
and Experimental Design ..............1348
75.6 Quantitative Performance Metrics ..........1349
75.6.1 State-Based Sensitivity Metrics ......1350
75.6.2 Phase-Based Sensitivity Metrics .....1351
75.6.3 Global Versus Local Parameters ......1353
75.7 Bio-inspired Control and Design ............1353
75.8 Emerging Trends ..................................1354
References ..................................................1354
75.1 Basics
Advances in molecular biology over the past two
decades have made it possible to probe experimentally
the causal relationships between processes initiated by
individual molecules within a cell and their macro-
scopic phenotypic effects on cells and organisms.
A systematic approach for analyzing complexity in bio-
physical networks was previously untenable owing to
the lack of suitable measurements and the limitations
imposed in simulating complex mathematical models.
Recent studies provide increasingly detailed insights
into the underlying networks, circuits, and pathways re-
sponsible for the basic functionality and robustness of
Part H 75

1336 Part H Automation in Medical and Healthcare Systems
biological systemsand create new andexciting opportu-
nities for the development of quantitative and predictive
modeling and simulation tools [75.1]. The discipline
of systems biology has emerged in response to the
challenges in modeling and understanding complex bi-
ological networks [75.2,3].
75.1.1 Systems Biology
The field of systems biology combines approaches
and methods from systems engineering, computa-
tional biology, statistics, genomics, molecular biology,
biophysics, and other fields [75.5–7] to help create
systems-level understanding of complex biological net-
works. In particular, systems engineering methods are
finding unique opportunities in characterizing the rich
dynamic behavior exhibited by biological systems.
Conversely, the new classes of biological problems are
motivating novel developments in theoretical systems
approaches. Two characteristics of systems biology
are [75.4]: (1) integrative view points towards unravel-
ing complex dynamical systems, and (2) tight iterations
between experiments, modeling, and hypothesis gener-
ation (Fig.75.1).
Although the field of systems biology is relatively
young, one can already point to early successes in
a number of cases. The work of Arkin on λ-phage was
one of the first detailed analyses of a stochastic gene
switch, and showed convincingly that formal stochastic
treatment was requiredto understandthe cell fate switch
between lysis and lysogeny [75.8]. The analysis of per-
fect adaptation in chemotaxis is another example where
multiple groups adopted a systems perspective, and key
insights have been generated [75.9–11]. Notably, the
Experimentation
Experimental
design
Network
identification
Systems modelling
and analysis theory
Fig. 75.1 Systems biology iterations. Interactions between
experimental analysis and theoretical approaches, and the
main tasks for theory at the interfaces (after [75.4])
mechanism for perfect adaptation has been elucidated
and interpreted in classical control engineering terms:
integral feedback [75.11].
The approach of model reduction and systematic
analysis (including requisite modeling assumptions to
yield perfect adaptation) is an excellent example of
an effective systems strategy. This problem continues
to generate new insights, as recent work has shown
that disparate organisms have both overlapping and dis-
tinctive architectures for chemotaxis [75.11]. Another
nice example that has received considerable attention is
the gene network underlying circadian rhythms. Mod-
els have been proposed [75.12], and formal robustness
analysis tools have generated insights on biological de-
sign principles [75.13].
A more detailed case study that might be character-
ized as a success story has also emerged from the work
of Muller et al. on the JAK-STAT pathway [75.14].
They have shown that modeling-experiment iterations
can yield new hypotheses, particularly regarding un-
observable components that can be simulated (but
never measured). One implication, for the JAK-STAT
pathway, involves pharmacological intervention. Cur-
rent practice focuses on the phosphorylation element
of the pathway, but the model shows that a more
effective strategy involves the blocking of nuclear ex-
port.
75.1.2 Control Research in Systems Biology
Natural control systems are paragons of optimality.
Over millennia, these architectures have been honed
to achieve automatic, robust regulation of a myriad
of processes at the levels of genes, proteins, cells,
and entire systems. One of the more challenging
opportunities for systems research is unraveling the
multiscale, hierarchical control that achieves robust per-
formance in the face of stochastic perturbations. These
perturbations arise from both intrinsic sources (e.g., in-
herent variability in the transcription machinery), and
extrinsic sources (e.g., environmental fluctuations). Ro-
bustness in key performance variables to particular
perturbations has been shown to be achieved at the
expense of strong sensitivity (fragility) to other pertur-
bations.
The coexistence of extreme robustness and fragility
constitutes one of the most salient features of highly
evolved or designed complexity [75.15]. Optimally ro-
bust systems are those that balance their robustness to
frequent environmental variations with their coexist-
ing sensitivity to rare events. As a result, robustness
Part H 75.1

Automatic Control in Systems Biology 75.2 Biophysical Networks 1337
and sensitivity analysis are key measures in under-
standing and controlling system performance. Robust
performance reflects a relative insensitivity to perturba-
tions; it is the persistence of a system’s characteristic
behavior under perturbations or conditions of uncer-
tainty. Measuring the robustness of a system determines
the behavior (the output or performance) as a function
of the input (the disturbance). Formal sensitivity ana-
lysis allows the investigation of robustness and fragility
properties of mathematical models, yielding local prop-
erties with respect to a particular choice of parameter
values.
Within the field of systems engineering, control
engineering has had a pervasive influence on the disci-
pline of systems biology. For instance, the chemotaxis
work [75.11] was a paradigm of collaboration between
control engineers (Doyle) and biologists (Simon). Other
examples include the robustness analysis of cellular
function [75.15] and the unraveling of design principles
in circadian rhythm [75.13]. There are many other ex-
amples from the control community: for instance, major
advances in understanding signal transduction [75.16],
and the oscillations underlying a positive feedback gene
switch [75.17].
75.2 Biophysical Networks
Biophysical networks are remarkably diverse, cover
a wide spectrum of scales, and are inevitably character-
ized by a range of rich behaviors. The term complexity
is often invoked in the description of biophysical
networks that underlie gene regulation, protein interac-
tions, and metabolic networks in biological organisms.
There are categorically two distinct characterizations
of complexity: (1) the descriptive or topological no-
tion of a large number of constitutive elements with
nontrivial connectivity (described in Sect.75.3), and
(2) the classical notion of behavior associated with
the mathematical properties of chaos and bifurcations
(described in Sect. 75.4). In both biological and more
general contexts, a key implication of complexity is
that the underlying system is difficult to understand and
verify [75.18]. Simple low-order mathematical models
can be constructed that yield chaotic behavior, and, yet,
rich complex biophysical networks may be designed to
reinforce reliable execution of simple tasks or behav-
iors [75.19].
Biophysical networks have attracted a great deal
of attention at the level of gene regulation, where
dozens of input connections may characterize the reg-
ulatory domain of a single gene in a eukaryote, as
well as the protein level, where literally thousands of
interactions have been mapped in so-called protein in-
teractome diagrams that illustrate the potential coupling
of pairs of proteins [75.20, 21]. Similar networks also
exist at higher levels, including the coupling of indi-
vidual cells via signaling molecules, the coupling of
organs via endocrine signaling, and ultimately the cou-
pling of organisms in ecosystems. To elucidate the
mechanisms employed by these networks, biological
experimentation and intuition are by themselves insuf-
ficient. As noted earlier, the field of systems biology
has laid claim to this class of problems, and engi-
neers, biologists, physicists, chemists, mathematicians,
and many others have united to embrace these prob-
lems with interdisciplinary approaches [75.22]. In this
field, investigators characterize dynamics via mathe-
matical models and apply systems theory with the goal
of guiding further experimentation to better understand
the biological network that gives rise to robust perfor-
mance [75.22].
75.2.1 Timing and Rhythm:
Circadian Rhythm Networks
and Oscillatory Processes
Oscillatory processes are omnipresent in nature, com-
prising the cell cycle, neuron firing, ecological cycles
and others; they govern many organisms’ behaviors.
A well-studied example of a biological oscillator is the
circadian rhythm clock. The term circa- (about) diem
(day) describes a biological event that repeats approx-
imately every 24h. Circadian rhythms are observed at
all cellular levels since oscillations in enzymes and
hormones affect cell function, cell division, and cell
growth [75.23]. They serve to impose internal align-
ments between different biochemical and physiological
oscillations. Their ability to anticipate environmental
changes enables organisms to organize their physiol-
ogy and behavior such that they occur at biologically
advantageous times during the day [75.23]: visual and
mental acuity fluctuate, for instance, affecting complex
behaviors.
The mammalian circadian master clock resides in
the suprachiasmatic nucleus (SCN), located in the
Part H 75.2
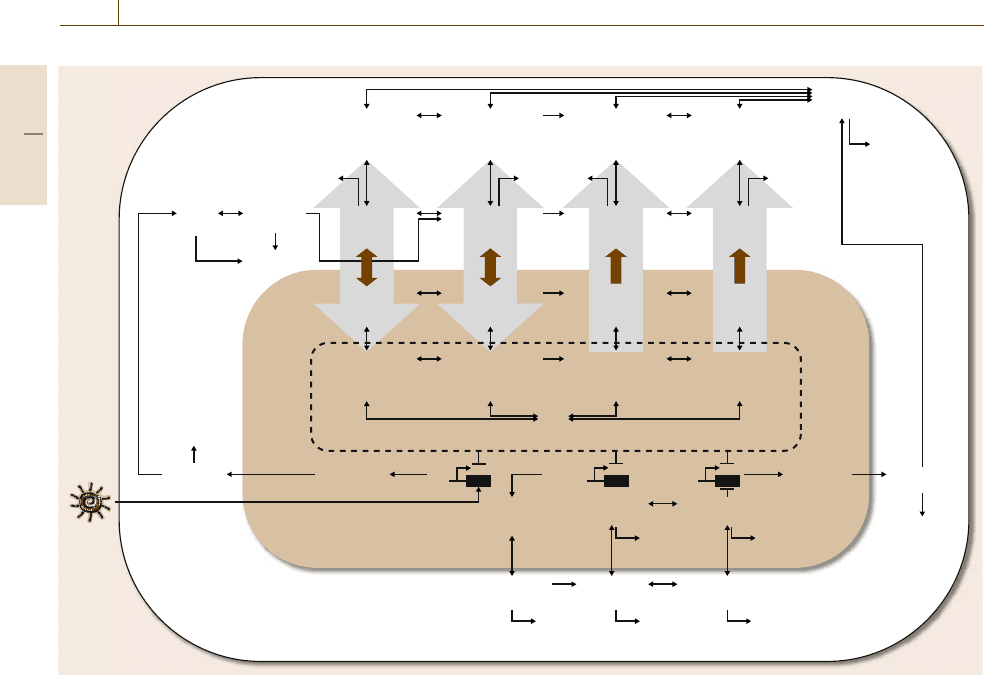
1338 Part H Automation in Medical and Healthcare Systems
1°-Phosphorylated
PER:CRY
(PopRo, PopRt,
PtpRo & PtpRT)
1°-Phosphorylated
PER:CRY:Kinase
(PopCRo, PopCRt,
PtpCRo & PtpC)
2°-Phosphorylated
PER:CRY:Kinase
(PoppCRo, &
PoppCRt)
2°-Phosphorylated
PER:CRY
(PoppRo, & PoppRt)
1°-Phosphorylated
PER
(Ponp & Ptnp)
1°-Phosphorylated
PER:Kinase
(PonpCn, PtnpCn)
2°-Phosphorylated
PER:Kinase
(PonppCn)
2°-Phosphorylated
PER:CRY
(Ponpp)
1°-Phosphorylated
PER:CRY
(PonpRon & PonpRtn,
PtnpRon & PtnpRtn)
1°-Phosphorylated
PER:CRY:Kinase
(PonpCnRon, PonpCnRtn,
PtnpCnRon & PtnpCnRtn)
2°-Phosphorylated
PER:CRY:Kinase
(PonppCnRon &
PonppCnRtn)
2°-Phosphorylated
PER:CRY
(PonppRon &
PonppRtn)
2°-Phosphorylated
PER:CRY
(Popp)
2°-Phosphorylated
PER:Kinase
(PoppC)
1°-Phosphorylated
PER:Kinase
(PopC & PtpC)
1°-Phosphorylated
PER
(Pop & Ptp)
PER
(Po & Pt)
PER:Kinase
(PoC & PtC)
CRY
(Ro & Rt)
CRY
(Ron & Rtn)
Degraded
CRY
Degraded PERDegraded PERDegraded PER
Degraded PER
Degraded
per mRNA
per mRNA
(McPo & McPt)
per mRNA
(MnPo & MnPt)
per rev-erb-α cry
cry pre-mRNA
(MnRo & MnRt)
cry mRNA
(McRo & McRt)
Degraded
cry mRNA
Degraded
REV-ERB-A
Degraded
REV-ERB-A dimer
Degraded
REV-ERB-A dimer
Degraded
REV-ERB-A
REV-ERB-A dimer
(RvRv)
REV-ERB-A
(Rv)
only cry1
rev-erb-α
pre-mRNA
(MnRv)
Degraded
rev-erb-α mRNA
rev-erb-α mRNA
(McRv)
REV-ERB-A
(Rvn)
REV-ERB-A dimer
(RvnRvn)
Fig. 75.2 Gene regulatory network underlying circadian rhythms in neurons in the SCN. Activated complexes of proteins inhibit
the transcription of their corresponding genes, thus leading to time-delayed negative feedback and oscillations (after [75.30])
hypothalamus [75.24]. It is a network of multiple
autonomous noisy (sloppy) oscillators, which com-
municate via neuropeptides to synchronize and form
a coherent oscillator [75.25, 26].Atthecoreofthe
clock is a gene regulatory network in which approxi-
mately six key genes are regulated through an elegant
array of time-delayed and coupled negative and pos-
itive feedback circuits (Fig.75.2). The activity states
of the proteins in this network are modulated (acti-
vated/inactivated) through a series of chemical reactions
including phosphorylation and dimerization. These net-
works exist at the subcellular level. Above this layer
is the signaling that leads to a synchronized response
from the population of thousands of clock neurons in
the SCN. Ultimately, this coherent oscillator then co-
ordinates the timing of daily behaviors, such as the
sleep/wake cycle. Left in constant (dark) conditions, the
clock will run freely with a period of only approxi-
mately 24h such that its internal time, or phase, drifts
away from that of its environment. Thus, the ability to
entrain to external time through environmental factors
is vital to a circadian clock [75.27–29].
75.2.2 Apoptosis: Programmed Cell Death
Another example of biophysical networks is the apop-
tosis network in which an extracellular input controls
the response of the cell as a result of this information
processing network. Apoptosis is the programmed cell
death machinery that is used by nature to strategically
kill off unneeded and infected cells, but this mecha-
nism often becomes impaired in cancer cells, leading
to unchecked proliferation.
The specific example here is triggered by the Fas
ligand. When an activated T-cell contacts a diseased
cell, Fas and its natural ligand bind, resulting in the
formation of the death inducing signalling complex
(DISC). This complex then activates two pathways,
both of which lead to the activation of the so-called
executioner caspase 3. The Fas apoptotic network has
Part H 75.2
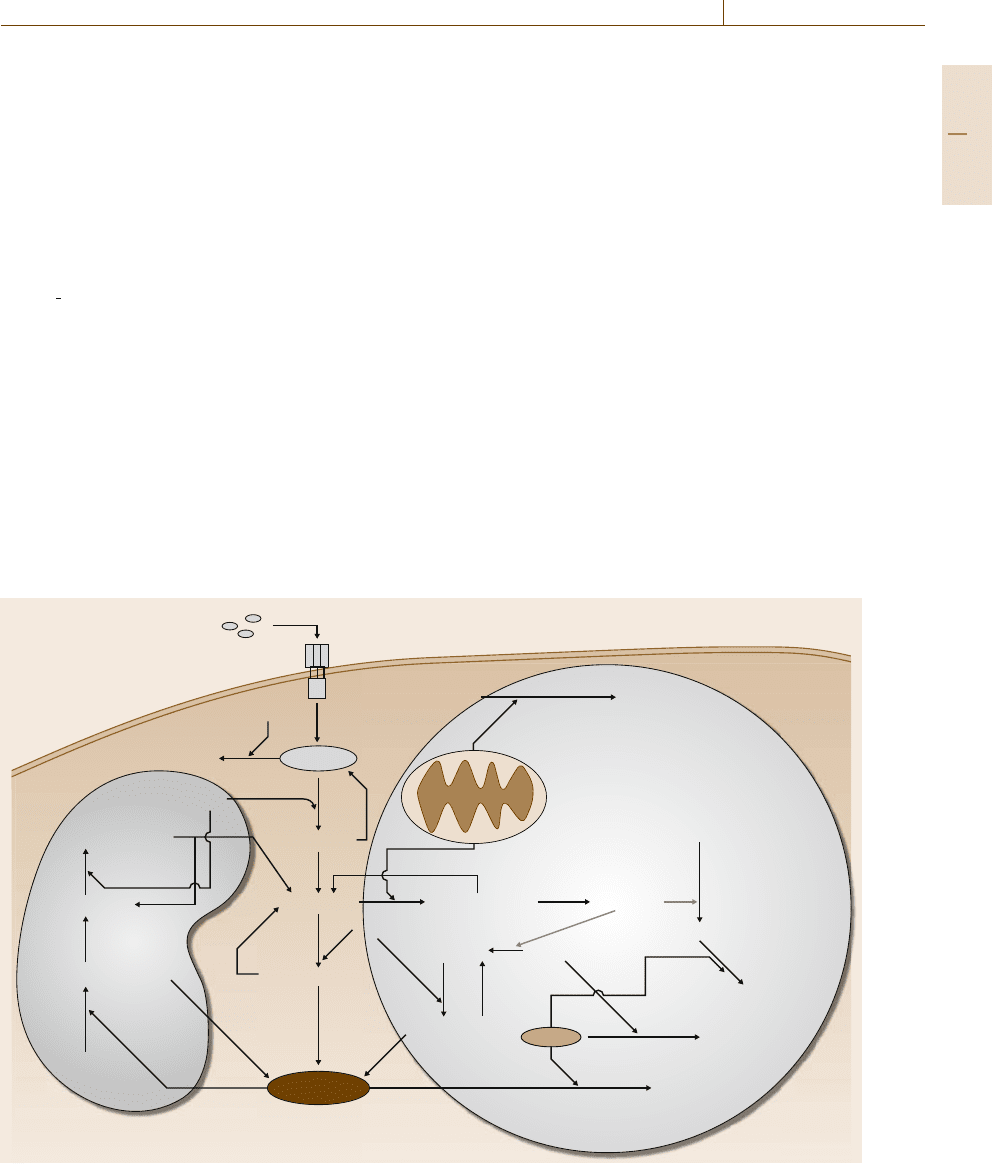
Automatic Control in Systems Biology 75.2 Biophysical Networks 1339
been modeled by [75.31] using ordinary differential
equations and is illustrated in Fig.75.3. In the Type I
pathway, a feedback pathway involving caspase-6 and
caspase-8 regulates the amount of activated caspase-3.
In the Type II pathway, Bcl-2 and active caspase-8 both
interact with the mitochondrial membrane, regulating
mitochondrial permeability. Caspase-8 allows mito-
chondria to become permeable, inducing the activation
of the apoptosome and Smac (second mitochondrial-
activator caspase) [75.32]. The activated apoptosome
(Apopt
active) in turn activates caspase-3, while Smac
can further enhance the activation of the executioner
caspase by removing XIAP (X-linked inhibitor of apop-
tosis protein). FLIP, Bcl-2, and XIAP all antagonize
the apoptotic signal and variations in their quantities
can toggle the apoptotic signal between Type I and
Type II activation or no activation at all. Experimen-
tal evidence has shown that Type I activation requires
a significant amount of caspase-8 to be present in the
cell. Yet, in Type II cells, mitochondrial activity ulti-
mately enhances the death signal, significantly lowering
the amount of caspase-8 that needs to be activated to
induce apoptosis [75.33].
BCL-2: MitoBCL-2
DISC:FLIP
DISC
DISC:Casp8
Casp8_active Casp8_active:Mito
Casp3_active
Casp3_active
Mitochondria
Mitochondria
Type II
Type I
Mito_active
Smac_active
Smac_act:XIAP
Apopt_XIAP
Casp3_active:XIAP
Smac
Casp8_active:Casp3
Casp6_active:Casp8
Casp3_active:Casp6
Casp6_active
Casp6
FLIP
Casp8
Casp3
Apopt_active
Apopt_act:Casp3
Apoptosome
XIAP
FasL
Fas
Fig. 75.3 Network schematic of the Type 1 and Type 2 Fas-induced apoptosis network (after [75.31])
Understanding apoptosis in a broader sense will
lead to a better knowledge of the common platform
for emergence of cancer cells, and perhaps point to
possible cures for certain types of cancer. The com-
plexity of apoptosis, however, makes the understanding
very difficult without a systems level approach using
a mathematical representation of the pathway. Further,
analysis of an apoptosis model can reveal the fragility
points in the mechanism of programmed cell death that
can have physiological implications not only for ex-
plaining the emergence of cancer cells but also for
designing drugs or treatment for reinstating apopto-
sis in these cells. A robust performance analysis of an
apoptotic network identified known network fragilities
as well as new potential targets for therapeutic inter-
vention. Such analyses are crucial to streamlining drug
discovery in highly dynamic networks [75.34].
75.2.3 Signals in Diabetes:
Insulin Signaling Pathway
In healthy cells, the uptake of glucose is regulated by
insulin, which is secreted by β-cells in the pancreas.
Part H 75.2
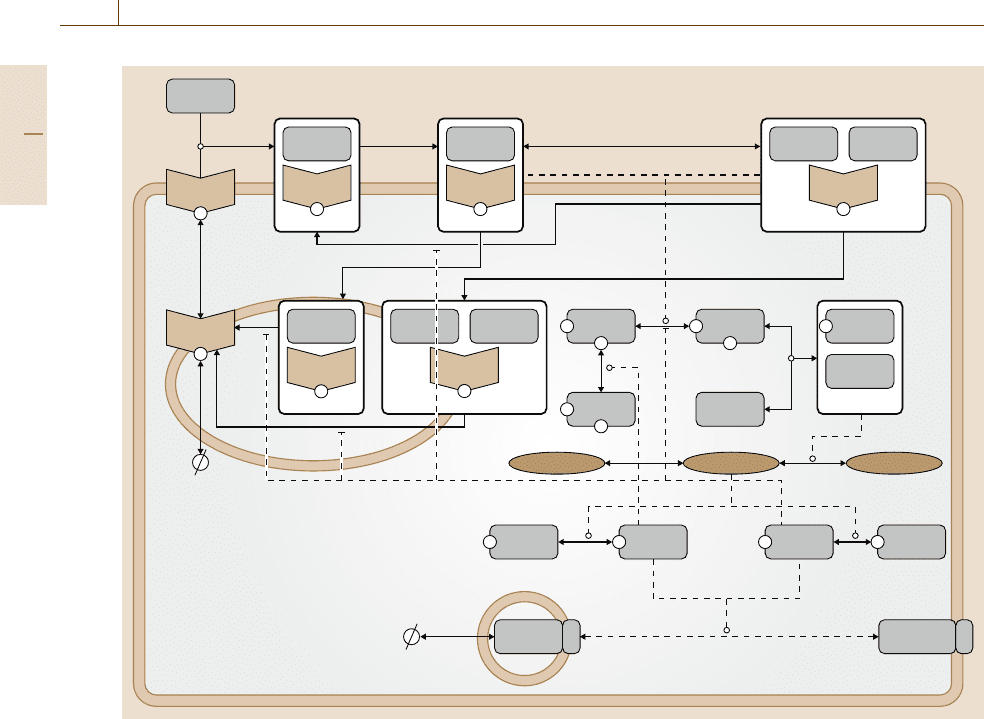
1340 Part H Automation in Medical and Healthcare Systems
Insulin [x
1
]
Insulin
P P
PP
P
Receptor
Receptor [x
2
]
Receptor [x
6
]
x
3
Insulin
Receptor
x
8
IRS1
IRS1 [x
10
]IRS1 [x
9
]
GLUT4 [x
20
] GLUT4 [x
21
]
Vesicle
Vesicle
Cell
PI3K [x
11
]IRS1 [x
10a
]
PI(3,4)P
2
[x
15
]
PKCζ [x
18
] PKCζ [x
19
] Akt [x
17
] Akt [x
16
]
PI3K
x
12
Insulin Insulin
Receptor
x
4
Insulin
Receptor
x
5
PI(3,4,5)P
3
[x
13
] PI(4,5)P
2
[x
14
]
PP
Insulin
Receptor
x
7
Insulin
Fig. 75.4 Insulin signalling pathway model (after [75.35])
Simply stated, in patients with type 1 diabetes, the
pancreas does not produce insulin, whereas in type 2
diabetes, among other consequences, the cells are resis-
tant to the insulin produced by the pancreas. The latter
phenomenon is best understood from detailed consid-
eration of the insulin signalling pathway (illustrated in
Fig.75.4). The sequence of actions occurs as follows:
(1) insulin binds to a receptor on the cell surface, which
causes receptor autophosphorylation and activation;
(2) the activated insulin receptor then phosphorylates
insulin receptor substrate-1 (IRS1), which subsequently
forms a complex with phosphatidylinositol-3-kinase
(PI3K); (3) the IRS1-PI3K complex catalyzes the pro-
duction of phosphatidylinositol triphosphate (PIP
3
),
which then interacts allosterically with phosinositide-
dependent kinase 1 (PDK
1
); (4) the PIP
3
-PDK1 com-
plex phosphorylates protein kinase Akt and protein
kinase C (PKCζ); (5) activated Akt and PKCζ trigger
glucose transporter (GLUT4) translocation from an in-
ternal compartment to the cell membrane. In a healthy
cell, this cascade ultimately leads to uptake of glucose
and helps to regulate glucose levels. In a cell charac-
terized by type 2 diabetes, the cascade is desensitized to
insulin, and the effectivenessof thesignal is diminished.
75.3 Network Models for Structural Classification
An important point in systems biology is the integrative
perspective, i.e., the analysis of the system considered
as a whole and across the different levels (gene, pro-
tein, metabolite, etc.) and not the reductionist analysis
Part H 75.3

Automatic Control in Systems Biology 75.3 Network Models for Structural Classification 1341
of individual components. While it is useful to catego-
rize the elements and levels of a hierarchical regulatory
scheme, it is more useful to analyze such schemes for
behaviors that emerge from combinations of simpler
motifs. Some simple examples of canonical regulatory
constructs (i. e., motifs) that yield specific classes of
behavior in gene networks include [75.36]:
1. Positive feedback: multistability, oscillations, state-
dependent response
2. Integral feedback: robust adaptation
3. Negative feedback: steady-state(homeostasis, adap-
tation)
4. Time delay: complex response, oscillations
5. Protein oligomerization: multistability, oscillations,
resonant stimulus frequency response.
In addition, stochastic fluctuations can induce ran-
dom response to stimuli, random outcomes, as well as
stochastic focusing. Such properties are characteristic
of general networks, including social networks, com-
munication networks, and biological networks [75.37].
Given the wide variety of modeling objectives, as well
as the heterogeneous sources of data, it is not surpris-
ing that many approaches exist for capturing network
interactions in the form of mathematical structures.
75.3.1 Hierarchical Networks
Biophysical networks can be decomposed into modular
components that recur across and within given organ-
isms. One hierarchical classification is to label the top
level as a network, which is comprised of interacting
regulatory motifs consisting of groups of 2–4 compo-
nents such as proteins or genes [75.38–40]. Motifs are
small subnetworks that are over-represented compared
to random networks with the same large-scale proper-
ties. At the lowest level in this hierarchy is the module
that describes transcriptionalregulation, of which a nice
example is given in [75.41]. At the motif level, one can
use pattern searching techniques to determine the fre-
quency of occurrence of these simple motifs [75.39],
leading to the postulation that these are basic build-
ing blocks in biological networks. Of relevance to the
present discussion is the fact that many of these com-
ponents have direct analogs in systems engineering
architectures. Consider the three dominant network mo-
tifs found in Escherichia coli [75.39]:
1. Coherent feedforward loop: in this, one transcrip-
tion factor regulates another factor, and, in turn, the
pair jointly regulates a third transcription factor
2. Single input module (SIM): in systems terminology,
a single-input multiple output block architecture
3. Densely overlapping regulons: in systems termi-
nology, a multiple-input multiple output block
architecture.
Similar studies in a completely different organism,
Saccharomyces cerevisiae, yielded six related or over-
lapping network motifs [75.38]:
1. Autoregulatory motif: negative feedback in which
a regulator binds to the promotor region of its own
gene
2. Feedforward loop: as described earlier
3. Multicomponent loop: effectively, a closed-loop
with two or more transcription factors
4. Regulator chain: a cascade of serial transcription
factor interactions
5. Single input module: as described earlier (SIM)
6. Multiinput module: a natural extension of the pre-
ceding motif.
In effect, these studies provide strong evidence that,
in both eukaryotic and prokaryotic systems, cell func-
tion is controlled by sophisticated networks of control
loops that are cascading onto and interconnected with
other (transcriptional) control loops. The noteworthy
insight is that the complex networks, which underlie bi-
ological regulation, appear to be made of elementary
systems components like a digital circuit. This lends
credibility to the notion that analysis tools from sys-
tems engineering should find relevance in this problem
domain.
75.3.2 Boolean Networks, Petri Nets,
and Associated Structures
Boolean networks are abstract mathematical models
employed for coarse-grained analysis of biophysical
networks. A Boolean network is represented as a graph
of nodes,with directededges between nodes and a func-
tion for each node (e.g., [75.42]). Boolean networks
are used to model network dynamics; for instance,
a Boolean network can be used to model transcripts and
proteins:
DNA mRNA
t+1
t
DNA
t+1
t
Fig. 75.5 Transcription of transcripts (mRNA)
Part H 75.3

1342 Part H Automation in Medical and Healthcare Systems
mRNA Protein
t+1t
Fig. 75.6 Translation from transcripts to proteins
1. Transcripts and proteins are either on (1) or off (0).
2. The expression of a node at time step t is given by
a logical rule of the expression of its effectors at
time t −1.
3. Transcription depends on transcription factors; re-
pressors are dominant (Fig.75.5).
4. Translation depends on the presence of the tran-
script (Fig.75.6).
5. Transcripts and proteins decay in one step if not
produced.
Place/transition (P/T) nets, or Petri nets, introduced
by Carl Adam Petri in 1962, have been intensively stud-
ied as one of several formal methods used to verify the
correctness of systems, which are described as mathe-
matical objects that can handle characteristics such as
nondeterminism and concurrency (e.g., [75.43]).
There are several additional types of networks
such as Bayesian nets, which combine directed acyclic
graphs with a conditional distribution for each random
variable (vertices in a graph, e.g., [75.44]), and signed
directed graphs, in which a signed directed edge is used
to represent activation versus inhibition (depending on
the sign, e.g., [75.45]). Alternatively, S-systems (bio-
chemical systems theory, BST) provide an approach
wherein polynomial nonlinear dynamic nodes are used
to capture network behavior (e.g., [75.46]).
75.4 Dynamical Models
While the consideration of motifsand network topology
is essential for unraveling design principles in com-
plex biophysical networks, it is necessary to understand
the role of dynamic behavior in ascribing meaning to
the rich hierarchies of regulation. Moreover, because
of the interplay between topology and dynamics, it is
often not enough in systems biology to specify only
the nodes (components) and edges (interactions). The
robust control of biophysical networks requires the an-
swer to many challenging questions such as: (1) What
are the dynamical aspects of the interaction? (2) What is
the characteristic quantity changed by the interaction?
Some of the intrinsically dynamic features of bio-
physical networks have been analyzed in a recent paper
that shows the close relationshipbetween dynamicmea-
sures of robustness and the abundance of particular
network motifs for a wide range of organisms [75.47].
Attempts to detail dynamic behavior in these networks
have fallen into three broad classes of modeling tech-
niques: (1) first-principles approaches, (2) empirical
model identification, and (3) a hybrid approach that
combines minimum biophysical network knowledge
with an objective function to yield a predictive model.
In this section, we outline some key results in the de-
velopment of mechanistic models, and in the following
section, we will address network identification.
Given detailed knowledge of a biological archi-
tecture, mathematical models can be constructed to
describe the behavior of interconnected motifs or tran-
scriptional units (TUs). A number of excellent review
papers have been published in recent years [75.1,36,48,
49]. In the majority of these studies, gene expression
is described as a continuous-time biochemical process,
using combinations of algebraic and ordinary differ-
ential equations (ODEs) [75.12, 36, 50]. In a similar
manner, models at the signal transduction pathway level
have been developed in a continuous-time framework,
yielding ODEs [75.51]. At the TU level, a detailed
mathematical treatment of transcriptional regulation is
described in [75.41]. Mechanistic models for a number
of specific biological systems have been reported, in-
cluding basic operons and regulons in E. coli (trp, lac,
and pho) and bacteriophage systems (T7 and λ) [75.52].
Systems theory has found an enabling role in
the analysis of the complex mathematical structures
that result from the previously described modeling
approaches. The language of systems theory now
dominates the quantitative characterization of biolog-
ical regulation, as robustness, complexity, modularity,
feedback, and fragility are invoked to describe these
systems. Even classical control theoretic results, such
as the Bode sensitivity integral, are being applied to
describe the inherent tradeoffs in sensitivity across fre-
quency [75.53]. Robustness has been introduced as
both a biological system-specific attribute, as well as
a measure of model validity [75.54, 55]. In the next
section, brief accounts of systems-theoretic analysis of
biological regulatory structures are given, emphasizing
where new insights into biological regulation have been
uncovered.
Part H 75.4

Automatic Control in Systems Biology 75.4 Dynamical Models 1343
75.4.1 Stochastic Systems
Discrete stochastic modeling has recently gained
popularity owing to its relevance in biological pro-
cesses [75.8, 56–59] that achieve their functions with
low copy numbers of some key chemical species. Un-
like the solutions to stochastic differential equations,
the states/outputs of discrete stochastic systems evolve
according to discrete jump Markov processes, which
naturally lead to a probabilistic description of the sys-
tem dynamics. A first-order Markov process is a random
process in which the future probabilities are dependent
only on the present value, and not on past values. Such
descriptions canfind relevance in systems biologywhen
the magnitude of the fluctuations in a stochastic system
approaches the levels of the actual variables (e.g., pro-
tein concentrations). In addition, there are qualitative
phenomena that are intrinsic to such descriptions that
arise in biological systems, as will be mentioned later.
The idea that stochastic phenomena are essential for
understanding complex transcriptional processes was
nicely illustrated by Arkin and coworkers in the analy-
sis of the phage λ lysis–lysogenydecision circuit [75.8].
The probabilistic division of the initially homogeneous
cell population into subpopulations corresponding to
the two possible fate outcomes was shown to require
a stochastic description (and could not be described
with a continuous deterministic model). In particular,
the coexistence of the two subpopulations necessi-
tated such a formal characterization, and the relative
sensitivity of the subpopulations to model parameters
including external variables could be analyzed with
the resulting models. In a more recent work, Samilov
and coworkers [75.60] have shown another example
of a biological behavior that is intrinsically stochastic
in nature – namely the dynamic switching behavior in
a class of biochemical reactions (enzymatic futile cy-
cles). In this case, the behavior is more subtle than the
lysis–lysogeny switch, where the existence of a bifurca-
tion was at least evident in the continuous differential
equation model. In the enzymatic futile cycle prob-
lem, the deterministic model gives no indication of
multiplicity, yet the discrete stochastic model generates
behaviors, including switching as well as oscillations,
that indicate characteristics of bifurcation regimes. It is
suggested that such noise-induced mechanisms may be
responsible for control of switch and cycle behavior in
regulatory networks.
In the discrete stochastic setting, the states and out-
puts are random variables governed by a probability
density function, whichfollows a chemicalmaster equa-
tion (CME) [75.61]. The rate of reaction no longer
describes the amount of chemical species being pro-
duced or consumed per unit time in a reaction but
rather the likelihood of a certain reaction to occur
in a time window. Though analytical solution of the
CME is rarely available, the density function can be
constructed using the stochastic simulation algorithm
(SSA) [75.61].
The discrete stochastic system of interest is de-
scribed by a CME [75.62]
d f(x, t|x
0
, t
0
)
dt
=
m
k=1
a
k
(x−v
k
, p) f(x−v
k
, t|x
0
, t
0
)
−a
k
(x, p) f(x, t|x
0
, t
0
) , (75.1)
where f(x, t|x
0
, t
0
) is the conditional probability of the
system to be at state x and time t, given theinitial condi-
tion x
0
at time t
0
. The state vector x gives the molecular
counts of the species in the system. Here, a
k
denotes
the propensity functions, v
k
denotes the stoichiomet-
ric change in x when the k-th reaction occurs and m is
the total number of reactions. The propensity function
a
k
(x, p)dt gives the probability of the k-th reaction to
occur between time t and t + dt, given the parame-
ters p. As the state values are typically unbounded,
the CME essentially consists of an infinite number of
ODEs, whose analytical solution is rarely available ex-
cept for a few simple problems. The SSA provides an
efficient numerical algorithm for constructing the den-
sity function [75.61]. The algorithm follows a Monte
Carlo approach based on the joint probability for the
time to and the index of the next reaction, which is
a function of the propensities. The SSA indirectly sim-
ulates the CME by generating many realizations of the
states (typically of the order of 10
4
) at specified time t,
given the initial condition and model parameters, from
which the distribution f (x, t|x
0
, t
0
) can be constructed.
There has been simultaneous advancement in ex-
perimental methods for quantifying the characteristics
of biological noise [75.63–65] along with advances in
computing and simulation. Anumber of groups have re-
cently useddual reportermethods totrack the activity of
identical genes in the same cell to measure the impact of
noise on expression. In the work of Elowitz and cowork-
ers, the separate effects of stochastic behavior in the
transcriptional and translational processes in prokary-
otes (so-called intrinsic noise) are distinguished from
noise effects arising fromother cellularcomponents that
influence therate of gene expression (so-called extrinsic
noise [75.63,65]). Raser and O’Shea analyze eukaryotic
systems with both cis-acting and trans-acting mutations
Part H 75.4
1344 Part H Automation in Medical and Healthcare Systems
to distinguish between the noise effects that are intrin-
sic to transcription as opposed to upstream processes
that might ultimately influence expression [75.64].
The interface of discrete stochastic systems and
biology has clearly led to new insights into stochas-
tic phenomena in biological systems, and has also
spurred the development of more efficient computa-
tional methods for stochastic simulation, as well as
analysis methods for these models. This interface will
continue to motivate developments in systems engi-
neering, with improved methods for imaging biological
systems that include the ability to resolve spatial
behaviors. Distributed stochastic models will require
more sophisticated algorithmic developments, particu-
larly as one builds models to truly address systems-scale
phenomena.
75.4.2 Constraints and Optimality
in Modeling Metabolism
To understand complex biological systems, instead of
starting from actual implementations and observations,
one can reduce the problem by first separating the
possible from the impossible, such as configurations
and behaviors that would violate constraints. Systems
approaches try to exploit three broad classes of con-
straints:
1. Empirical: large-scale experimental analysis can
provide constraints on possible network structures,
such as the average or maximal number of interac-
tions per component.
2. Physico-chemical: laws qof physics such as conser-
vation of mass and thermodynamics impose con-
straints on cellular and network behaviors. These
are used, in particular, for structural network ana-
lysis (SNA) with roots in the analysis of chemical
reaction networks [75.66].
3. Functional: biological systems perform certain
functions and their building blocks are confined to
a large, yet finite set. Network structures and behav-
iors have to conform with both aspects.
Functional constraints constitute the main differ-
ences betweencomplex physics and biology. In physics,
they do not exist. Biological (as well as engineered)
systems evolve to fulfill functions, and are constantly
evaluated for their performance. Insufficient perfor-
mance will lead to extinction, and better solutions are
likely to survive. Hence,it is reasonable to assume some
kind of optimality in biological systems. The immediate
consequence of a purpose is a considerably smaller de-
sign space, in which effective and reliable network are
rare and presumably highly structured. Understanding
complexity in biology could, thus, employ a calculus of
purpose – by asking teleological questions such as why
cellular networks are organized as observed, given their
known or assumed function [75.67].
Physico-chemical Constraints in Metabolism
Essential constraints for the operation of metabolic net-
works are imposed by (1) reaction stoichiometries, (2)
thermodynamics that restrict flow directions through
enzymatic reactions,and (3)maximal fluxes forindivid-
ual reactions. For instance, metabolism usually involves
fast reactions and high turnover of substances when
compared with regulatory events. Therefore, on longer
time-scales, it can be regarded as being in quasi-steady
state. The metabolite balancing equation (75.2)for
a system of m internal metabolites and q reactions with
the m×q stoichiometric matrix N and the q×1 vector of
reaction rates (fluxes) r formalizes this main constraint
in SNA. As for most real networks q * m, the system
of linear equation (75.2) is underdetermined. However,
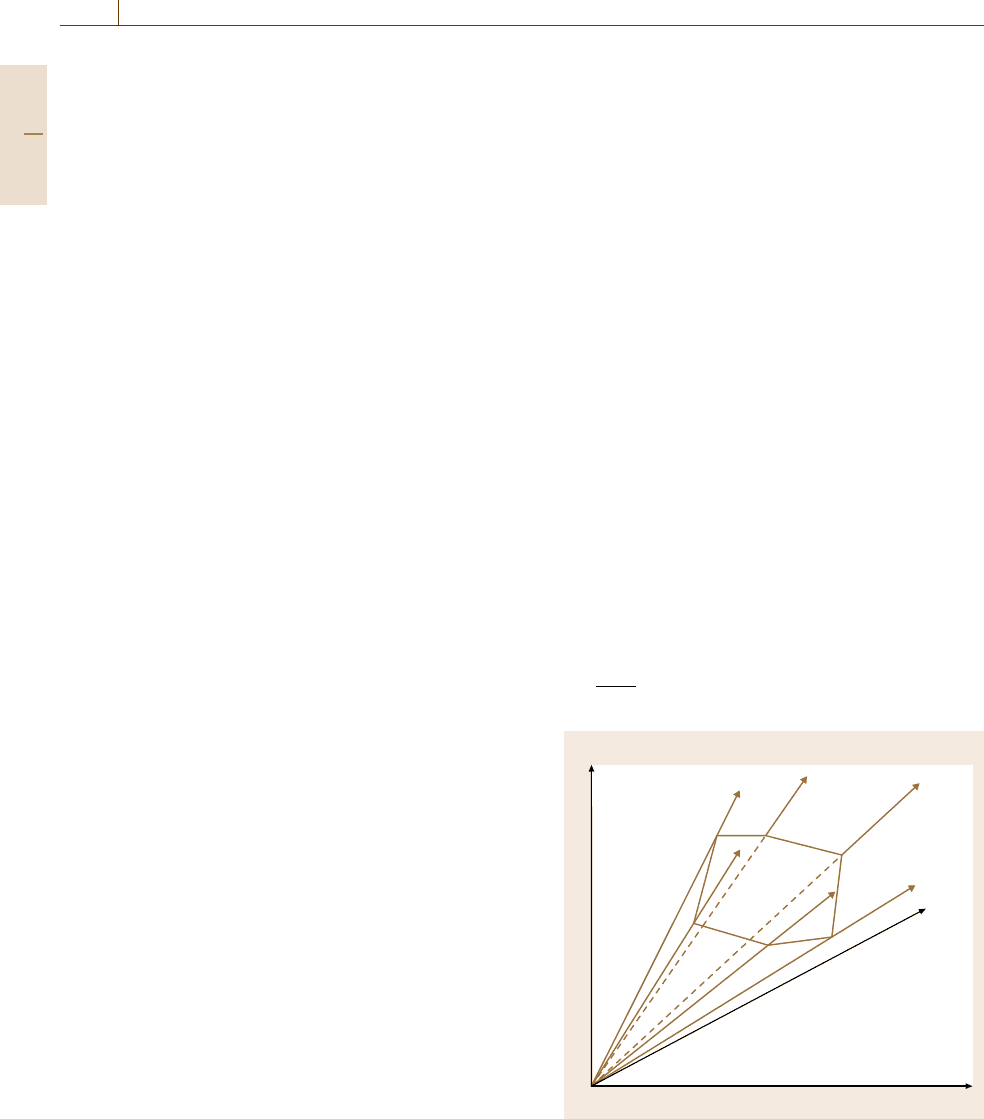
all possible solutions are contained in a convex vector
space, or flux cone (Fig.75.7). Methods from convex
analysis allow to investigate this space [75.68,69]
dx(t)
dt
=N·r =0 .
(75.2)
Rate 2
Rate 1
Rate 3
Fig. 75.7 Linear constraints specify a flux cone, with path-
ways as generating rays, projection on three-dimensional
flux space (after [75.4])
Part H 75.4
