Lennarz W.J., Lane M.D. (eds.) Encyclopedia of Biological Chemistry. Four-Volume Set . V. 4
Подождите немного. Документ загружается.


Translation Termination
and Ribosome Recycling
Nadja Koloteva-Levin and Mick F. Tuite
University of Kent, Canterbury, UK
The decoding of genetic information stored in an mRNA
molecule is initiated by a ribosome locating the translation
start codon AUG. The codons that define the encoded
polypeptide chain are then read sequentially by the ribosome
with the amino acids being delivered to the ribosome by
tRNAs. The chain of amino acids is therefore assembled as
directed by the order of codons in the mRNA which in turn are
recognized by the anticodon sequence of the tRNA. Trans-
lation termination is the last stage in this process and results in
the release of the completed polypeptide chain from the
ribosome once the end of the coding sequence has been
reached. The trigger for the termination process is the
appearance of one of the three possible stop codons at
the decoding center of the ribosome. Following termination
the ribosome dissociates from the mRNA and is recycled to
take part in a new protein synthesis cycle.
Translation Termination Apparatus
in Prokaryotes and Eukaryotes
The facilitators of the final stage of the translation cycle
are the protein release factors (RFs). Translation
termination occurs when one or other of the three stop
codons—UAA, UAG, or UGA—at the end of the
mRNA’s coding sequence are positioned at the riboso-
mal “acceptor” (A) site, i.e., at the decoding site within
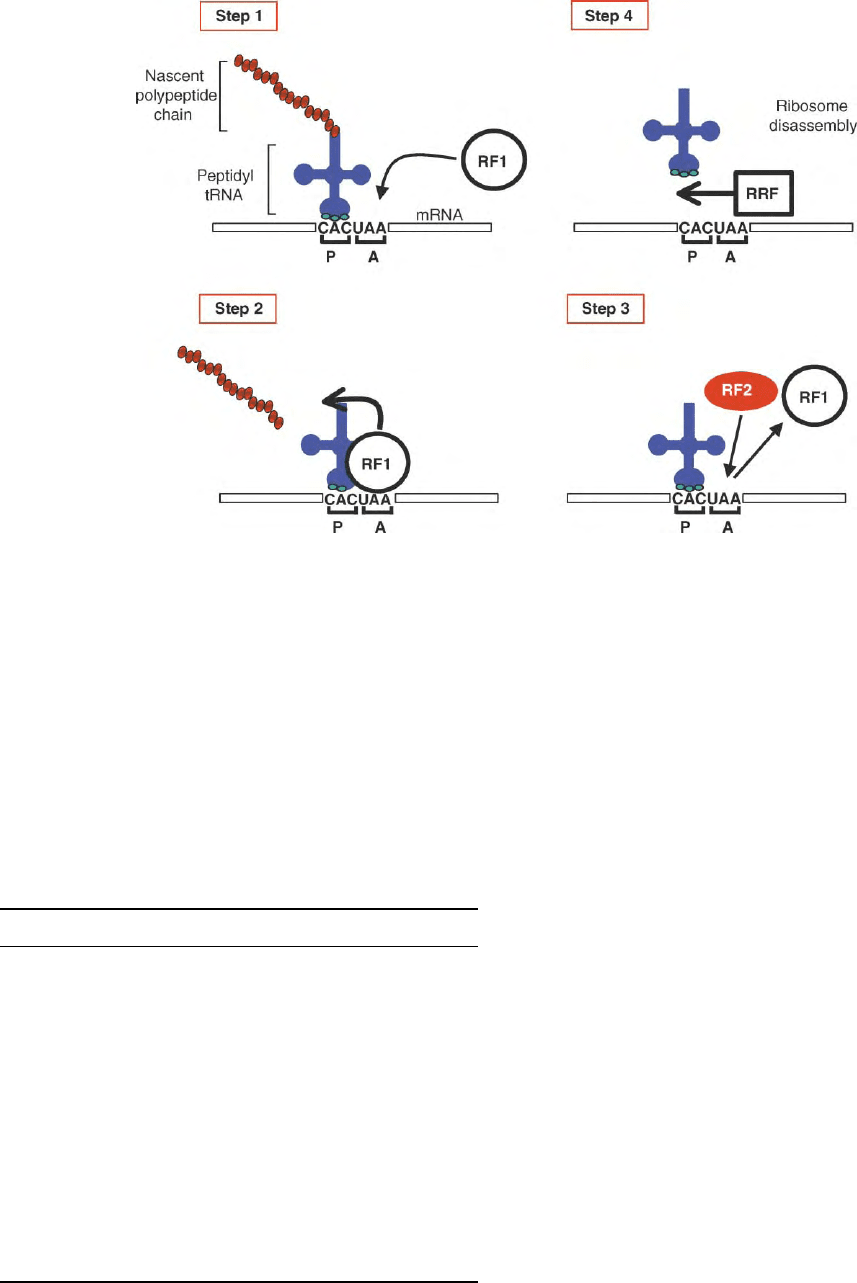
the small ribosomal subunit (Figure 1, step 1). The stop
codon is recognized by an individual RF or by an RF
complex that then triggers hydrolysis of peptidyl-tRNA
from the nascent polypeptide chain positioned at the
adjacent ribosomal “peptidyl” (P) site (Figure 1, step 2).
This is mediated by the peptidyl-transferase center
(PTC) of the large ribosomal subunit transferring the
peptidyl group to water rather than to an amino-
acyl tRNA.
RFs fall into one of two classes: class-I RFs recognize
and bind to the stop codon while class-II RFs facilitate
the release of the class-I factor after peptidyl-tRNA
hydrolysis (Figure 1, step 3). In the final stage, a
ribosome recycling factor (RRF) triggers the disassembly
of the terminated ribosome from the mRNA and the
release of the deacylated tRNA from the P ribosome
(Figure 1, step 4). Table I summarizes the known protein
factors involved in translation termination.
The ribosome also plays an active role in the process
of termination: interaction of the small ribosomal
subunit RNAs and ribosomal proteins with the stop
codon and a class-I RF ensures that the stop codon is
accurately decoded at the A site, whereas the interaction
of large ribosomal subunit RNAs with the RF or the RF
complex facilitates the catalysis of peptidyl-tRNA
hydrolysis at the P site via the ribosomal PTC.
Release Factors
PROKARYOTIC POLYPEPTIDE CHAIN
RELEASE FACTORS
In bacteria, translation termination is controlled by
three different RFs (Table I). Two class-I protein release
factors, RF1 and RF2, each decode two of three stop
codons, UAA or UAG (RF1) and UAA or UGA (RF2).
Recognition of the stop codon by the RFs is mediated via
a conserved tripeptide motif: Pro-Ala-Thr (PAT) in RF1
and Ser-Pro-Phe (SPF) in RF2. These tripeptides are
referred to as peptide anticodons. The other important
functional domain of class-I RFs contains the highly
conserved amino acid motif Gly-Gly-Gln (GGQ) and it
is this domain that triggers hydrolysis of the protein-
tRNA bond. An understanding of how RFs trigger the
release of the completed polypeptide from the tRNA at
the P site has come from a study of RF2 and its
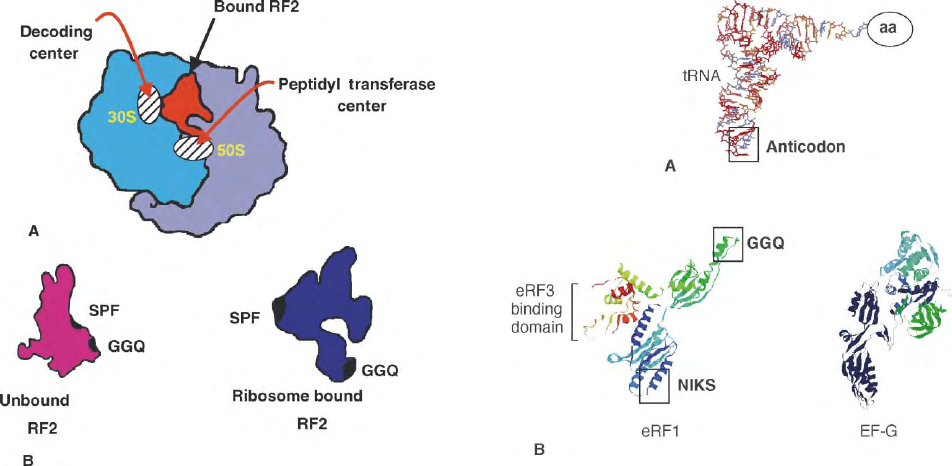
interaction with the terminating ribosome. When RF2
binds to the ribosome with a stop codon positioned at
the A site, RF2 changes its three-dimensional confor-
mation such that the domain with the conserved
“peptide anticodon” (SPF) interacts with the mRNA at
the decoding center, and the GGQ-containing
domain comes in the contact with the ribosomal PTC
to trigger hydrolysis of the peptidyl-tRNA linkage
(Figure 2).
Encyclopedia of Biological Chemistry, Volume 4. q 2004, Elsevier Inc. All Rights Reserved. 242
The single class-II RF in bacteria, namely RF3, is a
GTP-binding protein that accelerates dissociation of
either RF1 or RF2 from the ribosomal A-site after
release of the completed polypeptide chain from the
ribosome. RF3 bound to GDP accesses the ribosome
which, in complex with RF1 or RF2, acts as guanine
nucleotide exchange factors (GEFs) and triggers dis-
sociation of GDP from RF3. This leads to the formation
of a stable ribosome-RF1 (or RF2-) RF3 complex.
Hydrolysis of the peptidyl-tRNA linkage triggered by
RF1 or RF2 allows GTP binding to RF3. This induces an
altered RF3 conformation with a high affinity for the
ribosome and leads to rapid dissociation of RF1 or RF2
from the termination complex. To leave the ribosome,
RF3 requires GTP hydrolysis, which converts it to the
GDP-bound form of RF3 which has a lower affinity for
the ribosome. Once RF3 leaves the ribosome, it is ready
to enter the next translation cycle.
EUKARYOTIC CHAIN RELEASE FACTORS
In contrast to bacteria, in eukaryotic cells translation is
terminated by a single heterodimer consisting of two
different RFs, eRF1 and eRF3. eRF1 is a class-I RF that
decodes all three stop codons and triggers peptidyl-
tRNA hydrolysis by the ribosome to release the nascent
polypeptide. In eRF1, the stop codon recognition site is
located close to the amino terminus of the protein
molecule in a region that contains an evolutionarily
conserved tetrapeptide sequence, Asn-Ile-Lys-Ser
(NIKS). This sequence may be functionally equivalent
to the bacterial RF peptide anticodon. The GGQ
motif found in bacterial RF1 and RF2 is located in a
FIGURE 1 An overview of the mechanism of translation termination. The four key steps of the process triggered by the arrival of a stop codon at
the ribosomal A site are shown. The peptidyl tRNA is positioned at the ribosomal peptidyl P site. The major protein factors associated with each
step are indicated: RF1, class-I release factor; RF2, class-II release factor; RRF, ribosome recycling factor.
TABLE I
Protein Factors Involved in Translation Termination
Organisms RF Function
Archaebacteria
aRF1 Class-I RF, recognizes UAA/UAG/UGA
Prokaryotes
RF1 Class-I RF, recognizes UAA/UAG in
mRNA
RF2 Class-I RF, recognizes UAA/UGA in
mRNA
RF3 Class-II RF, GTPase, accelerates dissociation
of termination complex from the ribosome
RRF Ribosome recycling factor
Eukaryotes
a
eRF1 Class-I RF, recognizes UAA/UAG/UGA
eRF3 Class-II RF, GTPase, increases termination
efficiency, precise function unknown
a
No RRF homologue has yet been described for eukaryotic cells
other than a mitochondrially encoded homologue.
TRANSLATION TERMINATION AND RIBOSOME RECYCLING 243
separate domain of the protein to the NIKS sequence.
The carboxy-terminal part of the eRF1 binds eRF3.
The crystal structure of human eRF1 shows that it is a
Y-shaped molecule that resembles the structure of a
tRNA (Figure 3). Since both bacterial and eukaryotic
class-I RFs carry out essentially the same function, one
would expect them to interact similarly with ribosomal
A site. As with RF2 (see Figure 2), eRF1 must also
undergo a conformational change after binding to the
ribosome in order that it can interact with both the
ribosomal decoding site and the ribosomal PTC.
The eukaryotic class-II RF, eRF3, forms a complex
with eRF1, and via this interaction enhances the
efficiency of the translation termination although its
function remains to be fully defined. The GTPase
activity of eRF3, which is triggered by stop codons, is
both eRF1- and ribosome dependent. The carboxy-
terminal half of the eRF3 molecule shows significant
amino acid identity to the translation elongation factor
eEF1A that is responsible for bringing the aminoacyl
tRNAs to the ribosome during polypeptide chain
elongation. This suggests that eRF3 may act in a manner
analogous to eEF1A, a protein factor bringing the RF
complex to the ribosome. A number of functions have
been attributed to eRF3, e.g., it may displace eRF1 from
the ribosome (i.e., the function assigned to prokaryotic
RF3), or proofread the eRF1:stop codon interaction, or
stimulate ribosome disassembly (i.e., the function
assigned to prokaryotic RRF; see below), although
direct experimental proof for any one specific function
is lacking. In mammalian cells, eRF3 binds to the
poly(A)-binding protein (PABP), a protein associated
with the poly(A) tails located at the 3
0
end of the
majority of eukaryotic mRNAs. This interaction might
be important for the regulation of both mRNA stability
and translation initiation, since disruption of the
eRF3::PABP interaction suppresses the recycling of the
ribosome on the same mRNA.
In Bakers’ yeast (Saccharomyces cerevisiae), the eRF3
protein (also known as Sup35p) has an additional
remarkable property; it is a prion protein that acts as
the protein determinant of a non-Mendelian genetic
element called [PSI
þ
]. Like the mammalian prion
protein PrP, eRF3 can exist in one of two different
conformations with the aggregation-prone prion con-
former catalyzing the conversion of the normal soluble
form, to the prion form. In [PSI
þ
] cells the majority of
eRF3 is found as an inactive, high molecular weight
complex, and thus [PSI
þ
] cells have a defect in
translation termination albeit without detriment to
yeast cell growth. This mechanism may represent a
novel means of regulating the efficiency of translation
termination in yeast. There is no evidence that mamma-
lian eRF3 is a prion.
FIGURE 2 The three-dimensional conformation of a release factor
changes once it is bound to the ribosome. (A) The RF2:bacterial
ribosome complex indicating the points of contact between RF2 and the
decoding center and the PTC of the ribosome. (B) The three-
dimensional structures of the unbound and bound forms of bacterial
RF2 indicating the location of the “peptide anticodon” sequence
Ser-Pro-Phe (SPF) and the conserved Gly-Gly-Gln (GGQ) amino acid
motif.
FIGURE 3 Many of the protein factors involved in translation
elongation and termination have three-dimensional shapes that mimic
a tRNA molecule. (A) Three-dimensional structure of a tRNA showing
the location of the anticodon and the site to which the amino acid is
covalently attached. (B) The three-dimensional structures of the
mammalian RF eRF1 (left) and the bacterial translation elongation
factor EF-G (right). The position of the conserved NIKS and GGQ
amino acid motifs are indicated on eRF1, together with the domain to
which the class-II factor eRF3 binds.
244 TRANSLATION TERMINATION AND RIBOSOME RECYCLING
In Archaebacteria, the single class-I RF found (aRF1)
shares significant amino acid sequence and structural
homology with eRF1, but not with either RF1 or RF2.
Like eRF1, aRF1 is able to decode all three stop codons.
No eRF3 homologue has been identified in any
Archaebacterial genome.
Molecular Mimicry
RFs decode stop codons at the ribosomal A site and in
that respect can functionally be compared to aminoacyl
tRNAs which decode sense codons of the mRNA when
positioned at the ribosomal A site during translation
elongation. In so doing, RF1 and RF2 physically interact
with the stop codons in the mRNA. That a common
ribosomal binding site for tRNAs, translation
elongation factors, and RFs exists has led to the
suggestion that these protein and RNA molecules may
also have similar three-dimensional structures, the so-
called “molecular mimicry” model. The structural
similarity—discovered by Clark, Nyborg, and col-
leagues in 1995—between bacterial elongation factor
EF-G and the tRNA-EF-Tu-GTP complex provided
direct evidence for this model. One of the structural
domains of the EF-G protein strongly mimics the
structure of the T-stem and anticodon loop of a tRNA
(Figure 3). The “molecular mimicry” model can also be
extended to include the RFs and confirmation of this
came from the solution of the crystal structure of human
eRF1, whose overall shape and dimensions resemble a
tRNA molecule with the three clearly identifiable
structural domains corresponding to the anticodon
loop, the aminoacyl acceptor stem, and T-stem of a
tRNA molecule (Figure 3). There is also significant
amino acid sequence similarity between several RF
domains and the carboxy-terminal portion of elongation
factor EF-G, the region of the molecule that interacts
with the ribosome.
Posttermination Events
Once the translation of an mRNA has been terminated
and the completed polypeptide chain released, the
ribosome must dissociate from the mRNA in order to
be able to participate in a new round of protein
synthesis. In bacteria, another protein factor, the RRF,
is involved in this posttermination step together with the
elongation factor that mediates ribosomal translocation,
i.e., EF-G. The three-dimensional structure of RRF, like
eRF1, also mimics a tRNA molecule which, in turn, is
important for its activity in the dissociation of the
posttermination ribosome. After RF3 catalyzes the
release of RF1/RF2 from the ribosome, RRF enters
the A-site of the ribosome, and it is then translocated by
EF-G to the P site, similarly to the translocation of the
peptidyl-tRNA during translation elongation (Figure 1,
step 4). This releases the deacylated tRNA from the P
site which then moves to the ribosomal exit (E) site from
where it rapidly dissociates from the ribosome. The
release of the mRNA from the ribosome is accompanied
by the release of RRF and EF-G. The final step before the
ribosome can re-enter the translation cycle is dis-
sociation of the ribosome into its two subunits: an
event that requires an additional protein factor, the
translation initiation factor 3 (IF3).
In eukaryotes, the posttermination step still remains
to be characterized. A gene encoding a eukaryotic RRF-
like protein is found in most eukaryotic genomes but
encodes an RRF that only participates in translation
in the mitochondria. It is conceivable that eRF3 fulfills
this function.
Bypassing Stop Codons: The Causes
and Consequences
The accurate recognition of a stop codon as a
termination signal is important for cell viability,
although cells can tolerate a certain level of stop codon
translation by nonsense suppressor tRNAs. However,
the efficiency with which RFs mediate translation
termination at a stop codon can be affected by several
factors. These include the nucleotide context of the stop
codons in the mRNA (i.e., the identity of the bases
directly preceding or following the termination codon),
the cellular concentration of RFs, and the presence of
naturally occurring, nonmutant tRNAs, which are able
to recognize a stop codon as well as their usual normal
sense codon(s) albeit at much lower efficiency. For
example, in yeast the tRNA that decodes the CAA codon
as a glutamine residue can also recognize the related stop
codon UAA at low efficiency.
A number of nonstandard translational events are
known to compete with the termination machinery in
order to facilitate the translational bypassing of specific
stop codons. Such a bypass can extend the decoding
properties of a single gene and is an event that can be
regulated. For example, certain animal viruses can cause
ribosomes to shift into a different reading frame just
before a stop codon is reached thereby generating a
second extended form of the translation product – which
in this case is the viral gag-pol protein. In certain plant
viruses, e.g., Tobacco Mosaic Virus (TMV), host-
encoded tRNAs can decode a viral stop codon as a
sense codon if that stop codon is present in the
appropriate nucleotide context.
A further example where stop codons are treated as
sense codons is in the translation of genes encoding
certain selenoproteins, i.e., contains one or more
TRANSLATION TERMINATION AND RIBOSOME RECYCLING 245
selenocysteine residues. In these mRNAs the UGA codon
is used both as a signal for termination and as a signal
for selenocysteine incorporation. How the UGA codon
is read is defined by both a specific translation factor
(e.g., the SelB protein in bacteria) and a structural
element within the mRNA molecule that is being
translated (e.g., the SECIS element in eukaryotic
selenoprotein-encoding mRNAs).
SEE ALSO THE FOLLOWING ARTICLES
Prions and Epigenetic Inheritance † Prions, Overview †
Ribosome Structure
GLOSSARY
aminoacyl tRNA A transfer RNA molecule that has an appropriate
amino acid residue esterified to its 3
0
-terminal adenosine.
anticodon A contiguous sequence of three nucleotides in transfer
RNA that are complementary to a specific codon in mRNA.
codon A contiguous sequence of three nucleotides in mRNA that are
used to specify a particular amino acid, e.g., CAG is the codon
specifying the amino acid glutamine.
nonsense suppressor tRNAs tRNAs that contain a mutation in their
anticodon sequence such that they can recognize an mRNA stop
codon positioned at the ribosomal A site by standard base
pair interactions.
peptidyl-transferase center The region of the ribosome that is
responsible for two reactions important for protein synthesis:
peptide bond formation during translation elongation and nascent
peptide chain release during termination.
prion A protein that can exist in a self-perpetuating conformationally
altered isoform. Prions have been identified as infectious agents
responsible for certain neurodegenerative diseases of animals and
as epigenetic determinants in yeast.
reading frame Any one of three ways in which an mRNA sequence
can be translated one codon at a time. The “open” reading frame
encoding a gene product is set by the initiation codon AUG.
FURTHER READING
Bertram, G., Innes, S., Minella, O., Richardson, J., and Stansfield, I.
(2001). Endless possibilities: Translation termination and stop
codon recognition. Microbiology UK 147, 255–269.
Ehrenberg, M., and Tenson, T. (2002). A new beginning of the end of
translation. Nat. Struct. Biol. 9, 85–87.
Kisselev, L., Ehrenberg, M., and Frolova, L. (2003). Termination of
translation: Interplay of mRNA, rRNAs, and release factors? Eur.
Molecul. Biol. Orgn. J. 22, 175– 182.
Klaholz, B. P., Pape, T., Zavialov, A. V., Myasnikov, A. G., Orlova,
E. V., Vestergaard, B., Ehrenberg, M., and van Heel, M. (2003).
Structure of the Escherichia coli ribosomal termination complex
with release factor 2. Nature 421, 90–94.
Nakamura, Y., and Ito, K. (2003). Making sense of mimic in
translation termination. Trends Biochem. Sci. 28, 99– 105.
Nissen, P., Kjeldgaard, M., Thirup, S., Polekhina, G., Reshetnikova, L.,
Clark, B. F., and Nyborg, J. (1995). Crystal structure of the ternary
complex of Phe-tRNAPhe, EF-Tu, and a GTP analog. Science 270,
1464–1472.
Rawat, U. B., Zavialov, A. V., Sengupta, J., Valle, M., Grassucci, R. A.,
Linde, J., Vestergaard, B., Ehrenberg, M., and Frank, J. (2003). A
cryo-electron microscopic study of ribosome-bound termination
factor RF2. Nature 421, 87–90.
BIOGRAPHY
Mick Tuite is a Professor of Molecular Biology at the University of
Kent and the former Head of the Department of Biosciences at this
institution. He has had a long-standing interest in the mechanism and
regulation of translation termination using the yeast S. cerevisiae as a
model system. This has recently led to a study of the prion-like
properties of one of the release factors eRF3 (Sup35p). He has a D.Phil.
from the University of Oxford and undertook postdoctoral research
at both Oxford and the University of California, Irvine before joining
the faculty at Kent. He has published over 150 research papers
and reviews.
Nadja Koloteva-Levin is a Senior Postdoctoral Researcher in Mick
Tuite’s laboratory working on the prion behavior of eRF3 (Sup35p).
She received her Ph.D. from University of Manchester Institute of
Science and Technology (UMIST) in the United Kingdom and followed
this with 4 years of postdoctoral research on posttranscriptional gene
regulation at Tel Aviv University in Israel.
246 TRANSLATION TERMINATION AND RIBOSOME RECYCLING

Translesion DNA
Polymerases, Eukaryotic
Alexandra Vaisman and Roger Woodgate
National Institutes of Health, Bethesda, Maryland, USA
Translesion DNA polymerases are enzymes capable of copying
templates containing DNA-distorting lesions. Unlike the highly
processive and accurate replicative polymerases, translesion
polymerases are relatively distributive and lack intrinsic 3
0
–5
0
-
exonuclease activity and are, therefore, unable to proofread
any errors made during replication. As a consequence, they
have low base substitution and frameshift fidelity on unda-
maged DNA templates. Translesion DNA polymerases are also
characterized by an ability to extend mismatched and
misaligned primer termini.
Lesion bypass is believed to occur in two mechanistically
discrete steps, both of which can be either relatively accurate or
highly error-prone. The first step is nucleotide incorporation
opposite the damaged base(s) itself, and the second step is
extension of the nascent chain beyond the damage. For some
lesions, translesion replication is facilitated by a single enzyme,
but for certain lesions two polymerases are required; one
enzyme for incorporation opposite the lesion and another for
extension. The polymerase that gains access to the lesion
terminus largely determines the fidelity of translesion synthesis,
i.e., whether it is accurate or mutagenic. After lesion bypass is
completed, the inherently distributive translesion polymerases
dissociate from the DNA template so as to allow replicative
polymerases to resume chromosomal duplication.
Y-Family DNA Polymerases: pol
h
;
pol
i
; pol
k
and Rev1
Most of the enzymes implicated in replication of
imperfect DNA belong to the Y-family of DNA
polymerases. Phylogenetic analysis of this family
suggests that it can be subdivided into several discrete
branches consisting of UmuC-, DinB-, Rev1-, and
Rad30-like proteins. The DinB subfamily is the most
diverse and is found in prokaryotes, archaea, and
eukaryotes. Members of the UmuC subfamily are
found exclusively in bacteria, and proteins from the
Rad30 and Rev1 branches are found only in eukaryotes.
Multiple Y-family orthologues are often present in one
organism. Humans, for example, have four Y-family
DNA polymerases, two RAD30 paralogues, pol
h
and
pol
i
, a DinB homologue pol
k
, and Rev1.
BIOCHEMICAL PROPERTIES
Enzymatic characterization of the Y-family DNA poly-
merases in vitro reveals that despite numerous simi-
larities, each member of the family exhibits distinctive
properties during DNA synthesis. For example, pol
h
is
truly unique among eukaryotic DNA polymerases in its
efficiency and accuracy of replication past UV-induced
cyclobutane pyrimidine dimers (CPDs). In contrast, Pol
i
;
a homologue of Rad30, differs from other Y-family DNA
polymerases in that it has a 5
0
-deoxyribose phosphate
lyase activity, similar to X-family pols
b
and
l
: A most
unusual biochemical feature of pol
i
is, however, its
sequence context fidelity when replicating undamaged
DNA templates. Remarkably, Pol
i
prefers making
dGMP·T and dGMP·U wobble mispairs – three- to
tenfold greater than the correct dAMP·T and dAMP·U
base pairs. Yet at the same time, pol
i
discriminates
against certain other mismatches quite well. Thus, errors
made by pol
i
at template A occur up to 100-fold less
frequently than that made by human pol
h
or
k
: Pol
k
on
the other hand, readily catalyzes extension of misaligned
primer termini resulting in -1 frameshift mutations
and is considerably more proficient at extending
terminal mismatches compared to pol
h
and pol
i
.
Finally, unlike other Y-family DNA polymerases that
utilize all four deoxynucleotide triphosphates (dNTPs) in
a template-dependent manner, Saccharomyces cerevisiae
and human Rev1 proteins appear to be specialized
deoxycytidyl transferases, since they utilize the three
other dNTPs only very sparingly.
STRUCTURAL FEATURES
Sequence alignment of the Y-family DNA polymerases
shows a high degree of conservation in the N-terminal
region containing the catalytic domain, while the
Encyclopedia of Biological Chemistry, Volume 4. q 2004, Elsevier Inc. All Rights Reserved. 247
C-terminal part is unique for each family member.
Even though the amino acid sequences of these
polymerases show no similarity to classical DNA
polymerases, the general structure of polymerases
from all families appears to be remarkably conserved.
Crystallographic data have shown that like poly-
merases from the A-, B-, and X-families, the Y-family
polymerases adopt a right-hand structure with charac-
teristic finger, thumb, and palm domains. The Y-family
polymerases differ, however, from classical poly-
merases in having a fourth small domain that has
been termed the “little finger.” DNA is held in a cleft
between the thumb and the little finger domains
(Figure 1). The active site of Y-family polymerases is
more solvent accessible and, due to shorter fingers and
thumb domains, forms a more open structure than
that of replicative polymerases. Therefore, if the
geometric constraints on the active site of Y-family
polymerases are less stringent, DNA lesions and
mispairs can be more readily accommodated within
their active site. Such observations thereby help
explain their proficiency at lesion bypass and a
concomitant decrease in fidelity on undamaged DNA.
B-Family DNA Polymerase: pol
z
The ability to accommodate distorted DNA is not
just limited to Y-family polymerases. Pol
z
; a eukaryotic
B-family polymerase, is another important enzyme
involved in the replication of damaged DNA. Catalyti-
cally active pol
z
comprises a heterodimer made from the
REV3 and REV7 proteins. The fidelity of pol
z
on
undamaged DNA is significantly higher than that of
Y-family polymerases (Table I). However, pol
z
is
extremely efficient at the extension of aberrant DNA
primer ends, including lesion-containing sites and
misaligned primer-template termini.
X-Family DNA Polymerases: pol
b
;
pol
l
; and pol
m
Although much attention in the last few years has
focused on the Y-family polymerases, earlier studies
raised the possibility that DNA pol
b
may play a role in
translesion synthesis, even though its major function is
in base excision repair. Pol
b
is a distributive enzyme in
its mode of action, however, it is more processive than
most Y-family DNA polymerases and the inherent
ability of pol
b
to select correct rather than incorrect
dNTPs for incorporation is considerably higher than
that of the Y-family polymerases (Table I).
Lesion bypass with varying efficiencies can also be
achieved in vitro by other X-family polymerases, such as
the recently discovered pols
l
and
m
: Pol
m
is closely
related to terminal deoxynucleotidyl transferase (TdT),
but in contrast to TdT, it is a partially processive
template-directed DNA polymerase. In vitro, pol
m
is
highly error-prone and makes 2 1 frameshift errors in
repetitive sequences. Pol
l
is a close homologue of pol
b
and like pol
b
is more accurate than pol
m
and possesses
an intrinsic deoxyribophosphate lyase activity (Table I).
Lesion Bypass Specificity
DNA molecules are intrinsically unstable and are
constantly subjected to the damaging effects of endogen-
ous or exogenous agents. DNA synthesis past a wide
variety of DNA lesions has been studied using both
biological and biochemical approaches. Among the
most common lesions that have been investigated are
UV-induced cis-syn CPDs and the 6-4 pyrimidine-
pyrimidone photoproducts (6-4PPs); abasic sites occur-
ring spontaneously or from the action of DNA
glycosylases; cisplatin adducts (CP) formed at adjacent
guanines by anticancer drugs; and benzo[a]pyrene diol
epoxide (BaP DE) adducts, a naturally occurring meta-
bolite of carcinogens commonly found in cigarette
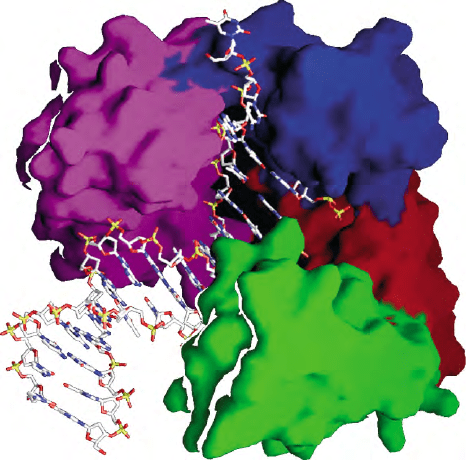
FIGURE 1 Crystal structure of Dpo4, an archaeal Y-family
polymerase, in a ternary complex with a DNA template and an
incoming nucleoside triphosphate. The surface of the enzyme is color-
coded to indicate the four subdomains found in all Y-family
polymerases. These consist of the palm (red), thumb (green), finger
(blue), and little finger (purple) domains. The DNA molecule is drawn
as sticks. Unlike replicative DNA polymerases, the catalytic cleft in
Dpo4 is cavernous and solvent exposed, which explains in part why
Y-family polymerases exhibit low-fidelity synthesis on undamaged
DNA and can accommodate bulky DNA adducts/mismatched bases
in their active sites.
248 TRANSLESION DNA POLYMERASES, EUKARYOTIC

smoke. The accuracy and efficiency of translesion
replication varies not only on the chemical nature of
the lesion and specific distortion that it imposes upon
DNA, but also on the local sequence context within
which the lesion is located and on the specific
polymerase or combination of polymerases involved in
a given translesion pathway. For example, in both
S. cerevisiae and humans, pol
h
is the most proficient and
accurate enzyme at replicating past thymine –thymine
CPDs. Pols
i
; pol
m
and to a lesser extent pol
z
and pol
b
are also able to bypass the lesion but with a much lower
efficiency and fidelity. By comparison, both pol
i
and
pol
h
are proficient at incorporating a nucleotide
opposite the 3
0
base of a thymine-thymine 6-4PP, and
pol
z
can readily extend the resulting primer termini.
Similarly, translesion replication past an abasic site
might involve pols
h
;
i
;
k
;
b
;
l
; or Rev1 to incorporate a
base opposite the lesion and pol
z
for its subsequent
extension. Pol
h
and pol
b
are both very efficient and
accurate when replicating past a CP lesion, whereas
bypass by pol
m
is highly error-prone since it generally
results in 2, 3, and 4 nucleotide deletions, pol
k
; pol
i
,
and pol
l
are completely blocked by CP adducts in
template DNA.
The ability of the polymerase to facilitate lesion
bypass also depends on the nucleotide at which a
specific adduct is formed. For example, in human cells,
efficient and accurate bypass of BaP DE adducts
formed at deoxyadenosine (dA) can be achieved
through the sequential action of pol
i
and pol
k
; but
not by pol
h
or pol
b
: In contrast, the ability of
translesion polymerases to bypass the same BaP DE
adduct formed at deoxyguanosine (dG) base is quite
different. Whereas human pol
h
and pol
i
bypass BaP
DE dG adducts inefficiently and inaccurately, pol
k
bypasses the lesion with high efficiency and predomi-
nantly incorporates the correct base, dCMP, opposite
the adducted dG.
Protein Partners
Despite extensive biochemical characterization of the
polymerases involved in translesion synthesis, it is still
unclear how the process physically occurs in living cells.
It has been shown that at least three of the translesion
polymerases, pol
h
; pol
i
; and Rev1 are associated with
the replication machinery in an undamaged cell and that
upon DNA damage the polymerase accumulates at sites
of stalled replication forks.
The catalytic activity of pols
h
;
i
; and
k
on
undamaged and lesion-containing DNA is substantially
increased through protein– protein interactions with
accessory proteins normally utilized by replicative
polymerases. These include proliferating cell nuclear
antigen (PCNA), which serves as a sliding clamp that
helps recruit polymerases to their site of action, and a
clamp loader, replication factor C (RFC). Given the
sixfold symmetry of the PCNA complex, it is possible
that multiple DNA polymerases connect simultaneously
to a replicative single clamp. However, how a cell
chooses between the various translesion polymerases it
has at its disposal and how the switch from the
replicative polymerase to a translesion polymerase and
then back to the replicase occurs in vivo still remains to
be determined.
Cellular Role(s)
Pol
h
is clearly responsible for the accurate and robust
replication of CPD-containing DNA in vivo. Although
the lack of pol
h
in human cells does not confer a
substantial hypersensitivity to killing by UV light,
humans with a defective pol
h
do exhibit the Xeroderma
Pigmentosum Variant (XP-V) syndrome and are
predisposed to the manifestation of skin cancers.
TABLE I
Eukaryotic DNA Polymerases Known to Participate in Translesion Replication
DNA
polymerase Gene or synonym Family
Error rate on
undamaged DNA Additional proposed functions
b
; beta POLB X10
24
–10
26
Base excision repair, double-strand break repair, synapsis and
recombination, meiosis, neurogenesis
z
; zeta POLZ (REV3L/REV7L) B10
24
–10
25
Somatic hypermutation, mismatch extension, developmental
cell proliferation
h
; eta POLH (RAD30A, XPV) Y10
22
–10
23
Somatic hypermutation
i
; iota POLI (RAD30B) Y10
1
–10
24
Base excision repair, somatic hypermutation
k
; kappa POLK (DINB1) Y10
23
–10
24
Spontaneous mutagenesis
l
; lamda POLL (POL4) X10
23
Base excision repair, repair during meiosis
m
; mu POLM X ? Nonhomologous end-joining pathway, lymphoid formation
Rev1 REV1L Y
TRANSLESION DNA POLYMERASES, EUKARYOTIC 249
To date, XP-V is the only known example of a human
disorder caused by a defect in translesion replication.
Apart from a clear role for pol
h
in the bypass of CPDs,
there is indirect evidence suggesting the participation of
other enzymes in translesion replication in vivo. For
example, cells with a reduced level of the deoxycytidyl
transferase Rev1 show lower levels of UV-induced
mutagenesis. In vitro assays suggest that pol
k
may
play an important role in accurate bypass of certain
lesions generated by polycyclic aromatic hydrocarbons
and recent studies with transgenic mice devoid of pol
k
support such hypotheses.
Just as translesion replication often requires multiple
DNA polymerases, so a particular translesion poly-
merase might have evolved to participate in one or
more functional tasks in a cell. Indeed, various lines of
evidence support a role for translesion polymerases
h
and
i
in the somatic hypermutation of rearranged
immunoglobulin genes. While S. cerevisiae strains with
deletions in their REV3 or REV7 genes (encoding pol
z
)
are viable, disruption of murine REV3 unexpectedly
leads to embryonic lethality in mice, suggesting that, in
addition to lesion bypass, mammalian pol
z
may play
additional role(s) in cellular development. Genetic and
biochemical studies have shown that S. cerevisiae
Rev1, in addition to participating in the bypass of
UV-induced lesions and abasic sites, also has a second
function that is independent of its dCMP transferase
activity. Pol
b
whose primary function is in base
excision repair, has also been implicated in double-
strand-break repair, meiotic events associated with
synapsis and recombination and in neurogenesis. On
the other hand, owing to an associated 5
0
-deoxyribose
phosphate lyase activity, it has been hypothesized that
pol
i
and pol
l
might possibly substitute for pol
b
in
certain forms of specialized base excision repair.
Finally, pol
m
’s main cellular role may actually be to
participate in a non-homologous end-joining pathway
that repairs DNA double-strand breaks and its ability
to traverse certain DNA lesions may, in fact, be rarely
utilized in vivo.
SEE ALSO THE FOLLOWING ARTICLES
DNA polymerase
a
, Eukaryotic † DNA polymerase
b
,
Eukaryotic † DNA polymerase
d
, Eukaryotic † DNA
polymerase 1, Eukaryotic † DNA Polymerases: Kinetics
and Mechanisms
GLOSSARY
DNA polymerase families DNA polymerases have historically been
classified into “families” based upon the phylogenetic relations of
their primary amino acid sequence. A-family polymerases are
related to Escherichia coli pol I; B-family polymerases to E. coli
pol II; C-family polymerases to E. coli pol III; X-family polymerases
to mammalian pol
b
; and Y-family polymerases to UmuC, DinB,
Rad30, and Rev1 proteins.
fidelity The accuracy with which a polymerase replicates a DNA
template.
polymerase processivity The number of nucleotides incorporated into
a nascent DNA strand per polymerase– template binding event.
proofreading The removal of misincorporated nucleotides at a
growing 3
0
end by a 3
0
exonuclease often associated with the
polymerase.
replicative polymerases Enzymes involved in the accurate copying of
genetic material. The active sites of these enzymes are usually much
smaller and constrained than the translesion polymerases, so as to
ensure that only the complementary “Watson and Crick” bases are
incorporated into nascent DNA.
translesion replication Also referred to as lesion bypass or translesion
synthesis (TLS), it is an inherently error-prone process that permits
cells to tolerate the presence of persistent DNA damage and
involves the direct replication through and beyond the DNA
damaged site by DNA polymerases.
FURTHER READING
Friedberg, E. C., Wagner, R., and Radman, M. (2002). Specialized
DNA polymerases, cellular survival, and the genesis of mutations.
Science 296, 1627 –1630.
Goodman, M. F. (2002). Error-prone repair DNA polymerases in
prokaryotes and eukaryotes. Annu. Rev. Biochem. 71, 17– 50.
Ohmori, H., Friedberg, E. C., Fuchs, R. P. P., Goodman, M. F.,
Hanaoka, F., Hinkle, D., Kunkel, T. A., Lawrence, C. W., Livneh,
Z., Nohmi, T., Prakash, L., Prakash, S., Todo, T., Walker, G. C.,
Wang, Z., and Woodgate, R. (2001). The Y-family of DNA
polymerases. Molecul. Cell 8,7–8.
BIOGRAPHY
Alexandra Vaisman is a Senior Research Fellow in the Laboratory of
Genetic Integrity at the National Institute of Child Health and Human
Development. Her principal research interests are in the field of
mammalian DNA repair and replication. She holds a Ph.D. from the
Moscow Institute of Fine Chemical Technology and received her
postdoctoral training at the Russian Academy of Sciences and the
University of North Carolina at Chapel Hill.
Roger Woodgate has devoted his scientific career to understanding the
molecular mechanisms of damage-induced mutagenesis in prokaryotes
and eukaryotes. He received his Ph.D. from the University of Sussex in
Brighton, United Kingdom and for the past 15 years has worked at the
National Institutes of Health in Bethesda, Maryland. At present, Dr.
Woodgate is Chief of the Laboratory of Genomic Integrity in the
National Institute of Child Health and Human Development.
250 TRANSLESION DNA POLYMERASES, EUKARYOTIC

Trehalose Metabolism
Alan D. Elbein
University of Arkansas for Medical Sciences, Little Rock, Arkansas, USA
Trehalose is a nonreducing disaccharide of glucose in which
the two glucoses are linked in an
a
,
a
-1,1-linkage. This rather
simple sugar is widespread throughout the biological king-
dom, being found in bacteria, yeast, fungi, protozoa,
nematodes, insects, plants, and so on. In these various
organisms, this sugar can serve a number of key functions:
as a storehouse of energy and carbon, as a protectant and
stabilizer of cell membranes and proteins during stress, as a
signaling molecule, and as a structural component of various
glycolipids in bacterial cell walls. There are at least three
different pathways for the synthesis of trehalose in various
organisms, and several or all of these pathways may be
present in the same organism. The fact that an organism may
have multiple routes for the synthesis of trehalose strongly
suggests that this sugar must be essential for the survival of
the organism.
Pathways of Biosynthesis
of Trehalose
The best-known and most-widely distributed pathway
for synthesis of trehalose (Figure 1)involvestwo
enzymes known as trehalose-phosphate synthase (TPS)
and trehalose-phosphate phosphatase (TPP). The reac-
tions catalyzed by these two enzymes are shown in
Figure 2. TPS catalyzes the transfer of glucose from the
sugar nucleotide, uridine diphosphate glucose (UDP-
glucose), to glucose-6-phosphate to produce trehalose-
6-phosphate and UDP. TPP then cleaves the phosphate
from trehalose-6-P to give free trehalose and inorganic
phosphate. These two enzymes have been isolated and
characterized from the cytoplasm of many cells includ-
ing bacteria, yeast, fungi, insects, and plants. The genes
for these enzymes have been cloned from several
different organisms and expressed in Escherichia coli.
In several cases, the purified recombinant enzymes have
been well characterized.
A more recently described pathway of trehalose
formation, so far demonstrated in only a few bacteria,
including Mycobacterium tuberculosis, involves the
intramolecular rearrangement of maltose to convert
the
a
1,4-glycosidic linkage of this disaccharide to the
a
,
a
1,1-glycosidic linkage of trehalose. This reaction is
catalyzed by the enzyme trehalose synthase (TS) as
shown in Figure 2. The gene for TS from Pimelobacter
species was cloned to produce a 573 amino acid protein
that had considerable homology to the glycosidase,
maltase. The mechanism of action of TS is not known,
but this enzyme can convert trehalose to maltose, or
maltose to trehalose. Its physiological function in these
cells is also not known.
A third pathway also only identified in a few bacteria
involves several different enzymes, the first of which
converts the glucose linked in an
a
1,4-bond at the
reducing end of a malto-oligosaccharide to the
a
,
a
1,1-
bond of trehalose. This enzyme is called malto-
oligosyltrehalose synthase (TreY). The second enzyme
then releases the trehalose to leave a malto-oligosac-
charide i.e., two glucoses shorter (at the reducing end)
than the starting oligosaccharide. This second enzyme
is called malto-oligosyltrehalose trehalohydrolase
(TreZ) (see Figure 2). These latter two pathways give
rise to free trehalose without the involvement of the
phosphorylated sugar, whereas the first pathway
produces trehalose phosphate as the initial product. In
some bacteria, at least two and possibly all three of
these pathways may be present at the same time. The
presence of these multiple pathways would suggest that
trehalose is an essential component for these cells, and
therefore alternate pathways have evolved to ensure
that levels of trehalose are maintained.
Occurrence of Trehalose in
the Biological World
The first report on the presence of trehalose in living cells
was in 1832 when trehalose was tentatively identified in
ergot of rye by Wiggers. Since that report, this sugar has
been isolated from many different organisms, including
yeast and fungi where it occurs in spores, fruiting bodies,
and vegatative cells. In some of these organisms, such as
the fungus, Neurospora tetrasperma, or the yeast,
Saccharomyces cerevesiae, the amount of trehalose
may be as high as 10% of the dry weight of the cells.
Encyclopedia of Biological Chemistry, Volume 4. q 2004, Elsevier Inc. All Rights Reserved. 251
