Lennarz W.J., Lane M.D. (eds.) Encyclopedia of Biological Chemistry. Four-Volume Set . V. 4
Подождите немного. Документ загружается.

FURTHER READING
Holmes, F. L. (1991). Hans Krebs, the Formation of a Scientific Life,
1900 to 1933. Oxford University Press, Oxford.
Holmes,F.L.(1993).Hans Krebs, Architect of Intermediary
Metabolism, 1933 to 1937. Oxford University Press, Oxford.
Krebs, H. A. (1954). The citric acid cycle. In Les Prix Nobel en 1953,
pp. 139–150. Boktryckeriet P.A. Norstedt and Soner, Stockholm,
Kungl.
Krebs, H. A. (1981). Hans Krebs: Reminiscences and Reflections.
Clarendon Press, Oxford.
Krebs, H. A., and Henseleit, K. (1932). Untersuchungen u
¨
ber die
Hamstoffbildung in Tierko
¨
rper. Hope-Seyler’s Z. Physiol. Chem.
210, 33–66.
Krebs, H. A., and Johnson, W. A. (1937). The role of citric acid in
intermediate metabolism in animal tissue. Enzymologia 4,
148–156.
Krebs, H. A., and Veech, R. L. (1969). Pyridine nucleotide
interrelations. In The Energy Level and Metabolic Control in
Mitochondria (S. Papa, J. M. Tager, E. Quagliariello and E. C. Slater,
eds.) pp. 329– 382. Adriatica Editrice, Bari.
Morowitz, H. J., Kostelnik, J. D., Yang, J., and Cody, G. D. (2000).
The origin of intermediary metabolism. Proc. Natl. Acad. Sci USA
97, 7704– 7708.
Ogston, A. G. (1948). Mechanism of fixation of carbon dioxide in the
Krebs cycle. Nature 162, 963.
Wood, H. G., Werkman, C. H., Hemingway, A., and Nier, A. O.
(1941). Interpretation of experiments on metabolic processes using
isotopic tracer elements. J. Biol. Chem. 139, 483.
BIOGRAPHY
Richard L. Veech is the Head of the Laboratory of Metabolic Control,
NIAAA, NIH, DHHS, in Rockville, Maryland. He is an M.D. from
Harvard Medical School, and has a D.Phil. in Biochemistry from
Oxford University where his supervisor was Hans Krebs. He works on
problems involving the control of the processes of intermediary
metabolism, particularly in disease states.
262 TRICARBOXYLIC ACID CYCLE

tRNA Synthetases
Karla L. Ewalt and Paul Schimmel
Scripps Research Institute, La Jolla, California, USA
Protein biosynthesis is the culmination of the transfer of genetic
information from DNA to proteins. It is a highly coordinated
process that requires many cellular factors including RNAs,
proteins, nucleotides, and amino acids. An essential component
for the translation of genetic information from genes to
proteins, is the group of enzymes known as aminoacyl-tRNA
synthetases. These enzymes covalently link amino acids to
transfer RNAs (tRNAs) and thereby establish the rules of the
genetic code that pair amino acids to codons. The translation of
codons on the messenger RNA into amino acids occurs at the
ribosome – the center of protein biosynthesis.
Enzymatic Reaction
Accurate protein biosynthesis requires a collection of
amino acids attached to tRNAs (aminoacyl-tRNAs),
which are typically generated by 20 amino acid-specific
enzymes called aminoacyl-tRNA synthetases. The ami-
noacylation reaction is carried out at the enzymes active
sites in two-steps. In the first step, an amino acid (AA) is
condensed with ATP yielding an aminoacyl-adenylate
(AA-AMP) and pyrophosphate (PPi) (Scheme 1). In the
second step, the aminoacyl group is transferred to either
the 2
0
or 3
0
-ribose hydroxyl on the terminal adenosine of
tRNA (AA-tRNA). Certain tRNA synthetases contain a
second active site (editing active site) for the hydrolysis
of incorrectly paired amino acids and tRNAs. At this
site, the tRNA synthetase corrects its errors.
Translational Accuracy
Aminoacyl-tRNA synthetases establish and determine
the accuracy of the genetic code and of protein
biosynthesis, as they pair amino acids with codons
through selection of amino acids and tRNAs. Accuracy
is achieved during selection of amino acids and tRNAs,
as well as through editing reactions that correct errors.
AMINO ACIDS
Each tRNA synthetase is specific for one of the 20 amino
acids. A selective binding site for the amino acid
accomplishes discrimination among the various amino
acid substrates. However, amino acid substrates for a
few enzymes are closely similar in size and functional
groups to other amino acids. For example, isoleucyl-
tRNA synthetase selects between isoleucine and valine,
which are both
b
-branched aliphatic amino acids
differing in length by only one methylene. The
misactivation of valine to valyl-adenylate by isoleucyl-
tRNA synthetase occurs at a low frequency. Threonyl-
tRNA synthetase must select between threonine and
serine, which differ by only a single methylene. The
active site is a “course sieve” that allows the correct
amino acid and occasionally similar, but slightly smaller
amino acids to be transferred to tRNA.
tRNAs
tRNAs are adapters that covalently connect anticodons
with amino acids. Sequences of tRNAs are 70–90
nucleotides (76 is most common) that form a common
cloverleaf secondary structure with four major arms.
This secondary structure in turn folds into an L-shaped
tertiary structure made up of two domains (Figure 1).
Amino acids are attached to tRNAs at the 3
0
-terminal
adenosine at the end of a CCA trinucleotide sequence
that is common to all tRNAs. In the second domain is a
trinucleotide sequence known as the anticodon, located
, 75A
˚
from the amino acid attachment site. During
protein synthesis, the anticodon triplet binds through
complementary base pairing to a codon on mRNA
and thereby delivers an amino acid to the growing
polypeptide chain.
Since the genetic code is degenerate, more than one
tRNA may exist for a single amino acid. The 61 sense
codons of the code are recognized by a combination of
distinct tRNAs (one tRNA specific for each codon) and
tRNAs that employ wobble-base pairing (one tRNA that
recognizes sets of related codons). (A notably reduced
set of tRNAs is found in the mitochondria of eukaryotes,
where 22 tRNAs read the codons.) Although, trypto-
phan and methionine each have a single tRNA, there are
as many as five tRNAs specific for the six leucine codons.
The multiple tRNAs specifying a single amino acid are
termed isoacceptors, and are substrates for a single
Encyclopedia of Biological Chemistry, Volume 4. q 2004, Elsevier Inc. All Rights Reserved. 263

aminoacyl-tRNA synthetase. Specific recognition of
tRNA is primarily through a nucleotide determinant
located in the acceptor stem. Nucleotides at other
positions including the anticodon stem are also import-
ant in many instances. The specific recognition of tRNAs
by synthetases based on the nucleotides present in the
acceptor stem, has led to the idea that these features
constitute a second genetic code that may be left over
from a smaller, historical RNA, which predated the
current tRNA structure.
EDITING
Several tRNA synthetases have an editing activity to
ensure correct pairing of amino acids with tRNAs
(Scheme 2). This editing, or proofreading, function
prevents misincorporation of amino acids into proteins
by hydrolyzing incorrectly aminoacylated tRNAs (post-
transfer editing). The tRNA synthetases for isoleucine,
leucine, valine, threonine, and alanine have been shown
to have specific domains with editing active sites that are
separate from the active site for aminoacylation. The
editing site operates as a “fine sieve” to exclude the
correct aminoacyl-tRNA while binding the smaller and
incorrect aminoacyl-tRNA, which is then hydrolyzed. It
is currently thought that tRNA synthetases can also
hydrolyze misactivated amino acids prior to attachment
to the tRNA (pre-transfer editing). Through the combi-
nation of proper substrate selection and editing, the
general error rate for protein synthesis is estimated to
be # 1/3000.
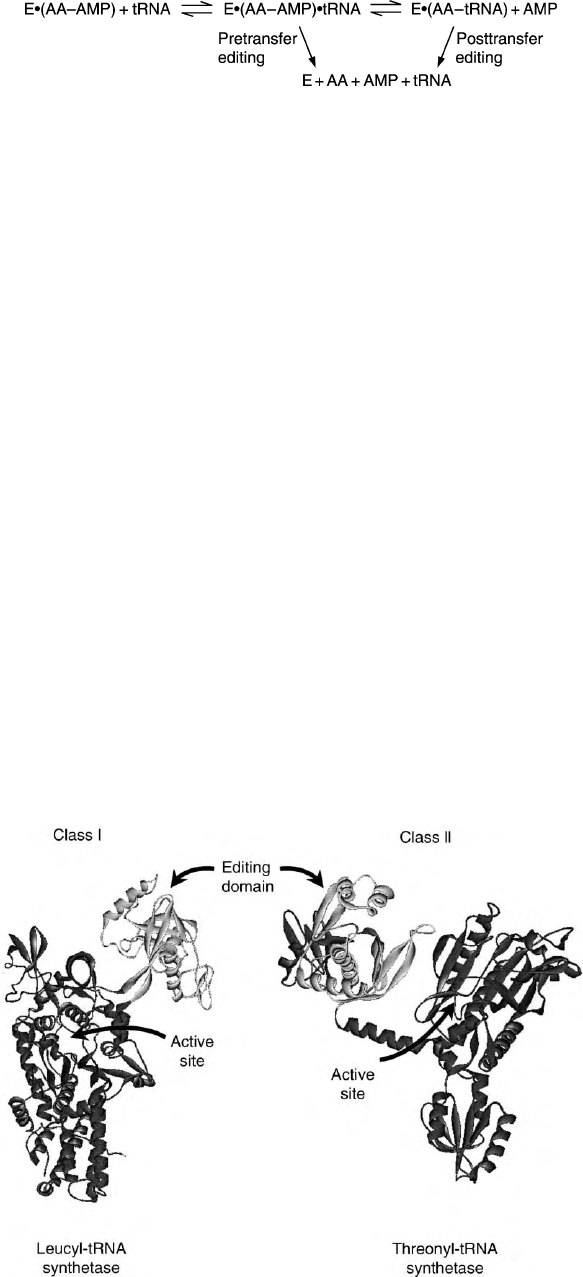
Two Classes of Enzymes
STRUCTURES
Aminoacyl-tRNA synthetases are divided into two
groups of ten enzymes – class I and class II – based
upon the sequences and structures of the active site
domains. Ten different class I enzymes activate and
transfer the amino acids Arg, Cys, Glu, Gln, Ile, Leu,
Met, Tyr, Trp, and Val to tRNAs. All class I enzymes
contain a Rossmann nucleotide-binding domain and an
active site with two signature sequences, an 11 amino
acid element that ends in HIGH and a KMSKS
pentapeptide. The active site is interrupted by a
polypeptide insertion called CP1 that encodes an editing
domain. (Figure 2). Similarly, there are ten class II
enzymes that activate and transfer the amino acids Ala,
Asn, Asp, Gly, His, Lys, Phe, Pro, Ser, and Thr to tRNAs.
These ten enzymes share an active site architecture
formed by three conserved motifs (motif 1, 2, and 3) in
an overall structure having a seven-stranded antiparallel
b
-sheet flanked by
a
-helices. Enzymes in both classes
have additional motifs or domains that are idiosyncratic
to each enzyme. Some of these domains provide RNA
binding elements, while others have functions such as
signaling in cytokine pathways.
MECHANISMS
The enzymes in the two classes are distinguished by the
orientation with which they bind tRNA. Class I enzymes
generally bind tRNA on the minor groove side of the
RNA helix and attach the amino acid to the 2
0
-ribose
hydroxyl of the terminal adenosine. Most class II
enzymes bind tRNA on the major-groove side and
attach the amino acid to the 3
0
-ribose hydroxyl. While
aminoacylation is specific to a particular hydroxyl, the
amino acid rapidly migrates between the 2
0
-and
3
0
-ribose hydroxyl groups.
Alternative Pathways and Enzymes
In a few cases, amino acids are modified while attached
to specific tRNAs in order to generate different
aminoacyl-tRNA species. This type of mechanism has
evolved as a biosynthetic pathway for glutamine,
asparagine, and selenocysteine.
tRNA DEPENDENT AMIDATION
Bacteria and archea have developed an alternate two-
step route for producing aminoacylated tRNAs for
glutamine and asparagine. In these organisms, glutamic
acid is attached to the glutamine specific tRNA
SCHEME 1 Two-step aminoacylation reaction catalyzed by amino-
acyl tRNA synthetases (E). Amino acid (AA) and ATP react to form an
activated aminoacyl-adenylate (AA-AMP) and pyrophosphate (PPi).
The aminoacyl group is transferred to either the 2
0
or 3
0
ribose
hydroxyl of the terminal adenine on tRNA.
FIGURE 1 The cloverleaf secondary and L-shape tertiary structure
of tRNA.
264 tRNA SYNTHETASES
(tRNA
Gln
) by glutamyl-tRNA synthetase. This system
requires that the glutamyl-tRNA synthetase have a
relaxed specificity that permits the acylation of glutamic
acid onto both tRNA
Glu
and tRNA
Gln
. In the second
step, glutamine is produced as Gln-tRNA
Gln
from Glu-
tRNA
Gln
, by the enzyme glutamyl-tRNA
Gln
amidotrans-
ferase. In these two kingdoms, this system is the
predominant mechanism for producing Gln-tRNA
Gln
.
Although some bacteria and archea have the gene
encoding asparaginyl-tRNA synthetase, an analogous
system also exists for the amidation of Asp-tRNA
Asn
to
produce Asn-tRNA
Asn
.
SELENOCYSTEINE
Sometimes called the 21st amino acid, selenocysteine is
incorporated into proteins at the UGA codon that
normally is a stop codon. However, a special tRNA
(tRNA
Sec
) exists for the incorporation of selenocysteine
at this position. Selenocysteyl-tRNA
Sec
is produced by
modification of Ser-tRNA
Sec
. Serine is attached to the
tRNA
Sec
by seryl-tRNA synthetase, and is then modified
by selenocysteine synthase. The enzyme incorporates
selenide through a pyridoxal 5-phosphate-mediated
reaction. Incorporation of selenocysteine into proteins
at UGA is dependent on the presence of a selenocysteine-
insertion-sequence in the mRNA that is immediately
downstream of the codon.
Novel Functions of
tRNA Synthetases
In addition to their essential role in protein biosynthesis,
several tRNA synthetases are involved in other cellular
processes. The novel functions that appear in tRNA
synthetases are not common features to all of the
enzymes for a particular amino acid, but rather are
specialized features of the enzymes in selected organ-
isms. For examples, mitochondrial leucyl- and tyrosyl-
tRNA synthetase promote splicing in certain fungi.
Bacterial threonyl-tRNA synthetase regulates its own
translation by binding to an upstream region of the
SCHEME 2 Pretransfer and post-transfer editing reactions catalyzed by aminoacyl-tRNA synthetases (E). Pretransfer editing occurs prior to
the aminoacyl group transfer to tRNA. Posttransfer editing occurs after the aminoacyl group has been transferred to tRNA. The net result of
both pathways is the enzymatic hydrolysis of the aminoacyl-group (AA-AMP or AA-tRNA) to the free amino acid (AA) with release of AMP
and tRNA.
FIGURE 2 The crystallographic structure of a class I enzyme, leucyl-tRNA synthetase, and a class II enzyme, threonyl-tRNA synthetase,
indicating the active sites and editing domains.
tRNA SYNTHETASES 265
mRNA, and a histidyl-tRNA synthetase related protein
called GCN2 regulates translation in eukaryotes.
Increasingly, roles for tRNA synthetases as cell signaling
factors are being discovered in higher eukaryotes.
Fragments of tyrosyl- and tryptophanyl-tRNA synthe-
tase are mediators of angiogenesis, while histidyl- and
asparaginyl-tRNA synthetase specifically stimulate
immune cells. These extra functions demonstrate how
tRNA synthetases have been adapted and utilized for a
wide variety of functions in addition to their enzymatic
role in protein biosynthesis.
SEE ALSO THE FOLLOWING ARTICLES
Amino Acid Metabolism † Pre-tRNA and Pre-rRNA
Processing in Bacteria † Pre-tRNA and Pre-rRNA
Processing in Eukaryotes † RNA Editing
GLOSSARY
aminoacyl-tRNA synthetases A family of enzymes that attach each
amino acid to its appropriate tRNA.
anticodon The three nucleotide sequence on tRNA that binds to the
codon through complementary base pairing.
codon A three nucleotide RNA sequence that specifies a particular
amino acid.
translation The process of protein biosynthesis from messenger RNA.
tRNA A small RNA molecule that carries an amino acid to the
ribosome during protein biosynthesis, recognizes the codon
specifying the amino acid and delivers it to the growing
polypeptide chain.
FURTHER READING
Alberts, B., Bray, D., Lewis, J., Raff, M., Roberts, K., and Watson, J. D.
(2002). Molecular Biology of the Cell. Garland Publishing,
New York.
Schimmel, P., and Ribas de Pouplana, L. (2000). Footprints of
aminoacyl-tRNA synthetases are everywhere. Trends Biochem.
Sci. 25(5), 207–209.
Soll, D., and RajBhandary, U. L. (1995). tRNA Structure, Biosynthesis,
and Function. ASM Press, Washington, DC.
Voet, D., and Voet, J. G. (1995). Biochemistry. Wiley, New York.
BIOGRAPHY
Paul Schimmel is the Ernest and Jean Hahn Professor and member
at the Skaggs Institute for Chemical Biology at the Scripps
Research Institute. He holds a Ph.D. from the Massachusetts Institute
of Technology, where he went on to become the John D. and Catherine
T. MacArthur Professor of Biochemistry and Biophysics in the
Department of Biology prior to joining the Scripps Research Institute.
His major research activities have concentrated on the decoding of
genetic information, with an emphasis on the rules of the universal
genetic code established by aminoacyl-tRNA synthetases.
Karla L. Ewalt is a Staff Scientist in the Department of Molecular
Biology at the Scripps Research Institute. She holds a Ph.D. from the
University of California, San Diego. Her research focus is on the
functional specializations found in components of eukaryotic protein
translation machinery.
266 tRNA SYNTHETASES

trp Operon and Attenuation
Paul Gollnick
State University of New York, Buffalo, New York, USA
Transcription attenuation can be defined as any mechanism
that utilizes transcription pausing or termination to control
expression of downstream genes. This mechanism of regu-
lation is used to control expression of many genes in bacteria in
responses to changes in their environment. Moreover, bacteria
have evolved several elaborate and elegant variations on the
attenuation theme, mainly based on the mechanism used to
control formation the RNA structures that signal RNA
polymerase to pause or terminate.
Attenuation
The first attenuation mechanism to be described
was for the Escherichia coli tryptophan biosynthetic
(trpEDCBA) operon. When Charles Yanofsky and his
co-workers elucidated this mechanism, it was the first
demonstration that organisms can utilize changes in
RNA structure to regulate gene expression. Sub-
sequently many other examples of transcription attenu-
ation have been discovered. In each case the cis-acting
genetic information is contained within a 150–300 bp
region located after the start of transcription and prior
to the start of the coding sequence for the first struc-
tural gene of the operon. This region is called the
leader region. In this article, the features of transcrip-
tion attenuation control of the E. coli trp operon are
discussed, followed by the modifications in this classic
system that allow it to be adapted to control other amino
acid biosynthetic operons. Thereafter attenuation con-
trol of the trp operon in the gram-positive bacterium
Bacillus subtilis, which involves a larger variation on the
attenuation theme, is described. There are several recent
reviews that cover the attenuation in more depth as well
as describe attenuation control of other bacterial genes.
The E. coli trp Operon: Attenuation
Based on Translation of a
Leader Peptide
The E. coli trpEDCBA operon encodes the enzymes
required to synthesize L-tryptophan from chorismic
acid. Transcription of the trp operon is regulated in
response to changes in intracellular tryptophan
levels. When the cells contain adequate amounts of
tryptophan, for example when it is present in the
growth medium, transcription of the operon is down-
regulated. In contrast, when tryptophan is limiting,
the trp operon is actively transcribed in order to
express the enzymes required for its synthesis.
Initiation of transcription is regulated by the trp
repressor, a DNA-binding protein encoded by the
trpR gene. In addition, after transcription has
initiated, the elongating transcription complex is subject
to regulation by attenuation. Together, repression (80-
fold) and attenuation (8-fold) serve to allow , 600-
fold overall control of transcription of the trp operon
in response to various levels of tryptophan
availability.
ATTENUATION CONTROL OF THE
E. COLI TRP OPERON
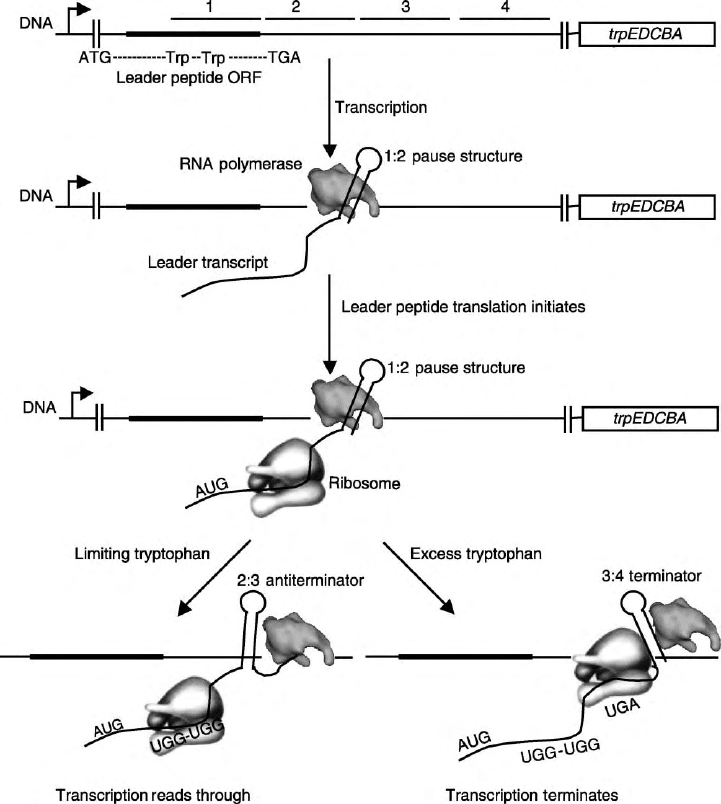
The E. coli trp operon contains a 162 bp leader region
prior to the start of the trpE coding sequence. The trp
leader transcript contains several inverted repeats,
composed of the segments labeled 1–4 in Figure 1,
that can form three different overlapping base-paired
RNA secondary structures. These structures include an
intrinsic transcription terminator (3:4), an overlapping
antiterminator (2:3), and a pause structure (1:2).
In addition, the leader transcription contains a small
open reading frame (ORF) that encodes a 14-amino
acid leader peptide, which contains two critical tandem
UGG Trp codons. The cell’s ability to efficiently
translate these two Trp codons determines which RNA
structure forms in the nascent leader transcript, which
in turn controls whether transcription halts in the
leader region or continues into the structure genes of
the operon.
Shortly after transcription initiates from the trp
promoter, the 1:2 pause hairpin forms (Figure 1).
This structure signals RNA polymerase to pause
transcription after nucleotide 92. This pausing of RNA
polymerase is a critical feature of the attenuation
mechanism because it allows time for a ribosome to
Encyclopedia of Biological Chemistry, Volume 4. q 2004, Elsevier Inc. All Rights Reserved. 267
initiate translation of the leader peptide. When the
ribosome begins translating the leader peptide this
releases the paused RNA polymerase to resume tran-
scription. Transcription and translation are now
coupled, with the ribosome closely following RNA
polymerase. This situation is essential to allow events
involving the ribosome to affect transcription by the
associated RNA polymerase.
At this point there are two possible pathways for
the attenuation mechanism to follow depending on
the level of tryptophan in the cell. The choice depends
on how efficiently the tandem Trp codons in the
leader peptide are translated. This efficiency reflects
the availability of aminoacylated tRNA
Trp
in the cell.
Under conditions where tryptophan is limiting, the
amount of charged tRNA
Trp
is low. As a result of this
low concentration of tryptophanyl-tRNA
Trp
trans-
lation of the tandem Trp codons is inefficient and
the ribosome stalls at one of these two codons. The
associated RNA polymerase continues to transcribe
through the trp leader region and transcription and
translation become uncoupled. As RNA polymerase
proceeds through segments 2 and 3 of the leader
region, the antiterminator structure (2:3) forms,
which prevents formation of the overlapping intrinsic
terminator (3:4) structure. Hence transcription con-
tinues through the leader region and into the trp
structural genes.
FIGURE 1 Model of transcription attenuation of the E. coli trp operon. RNA polymerase pauses following formation of the pause
structure. This provides time for a ribosome to initiate translation of the leader peptide. Under tryptophan-limiting conditions the ribosome
stalls at the tandem Trp codons, resulting in transcription read through. Under conditions of tryptophan excess the ribosome reaches
the leader peptide stop codon. This ribosome position blocks formation of the antiterminator leading to terminator formation and
transcription termination.
268 trp OPERON AND ATTENUATION
When tryptophan is plentiful, the level of
charged tRNA
Trp
in the cell is high. This allows effi-
cient translation of the tandem Trp codons and hence
the ribosome proceeds rapidly to the end of the
leader peptide. When the ribosome reaches the leader
peptide stop codon, it covers part of RNA segment 2
and thus prevents formation of the 2:3 anti-
terminator structure as transcription proceeds. This
frees RNA segment 3 to base pair with segment 4
and form the terminator. Under these conditions
transcription terminates in the leader region prior
to the trp structural genes, which are therefore
not expressed.
In this attenuation mechanism the regulatory signal
is the level of charged tRNA
Trp
and the sensory event is
the efficiency with which the ribosome can translate
the tandem Trp codons in the leader peptide. This
system can easily be adapted to regulate other bacterial
amino acid biosynthetic operons. The only needed
modification is to change the identity of the critical
codons in the leader peptide, which controls the amino
acid the system will respond to. There are numerous
examples of such adaptations of this attenuation
mechanism, particularly in enteric bacteria, including
the his, phe, and leu operons, which contain seven His,
seven Phe and four Leu codons in their respective
leader peptides.
The Bacillus subtilis trp
Operon: Attenuation Mediated
by an RNA-Binding Protein
The transcription attenuation mechanism that regulates
the trp operon in the gram-positive bacterium Bacillus
subtilis differs more dramatically from that the E. coli
trp operon than those described above for other amino
acid biosynthetic operons in gram-negative bacteria.
Most notably there is no leader peptide, and ribosomes
are not involved in this attenuation mechanism. Instead
an RNA-binding protein senses the level of tryptophan
in the cell and determines whether transcription
terminates in the leader region or continues into the
structural genes.
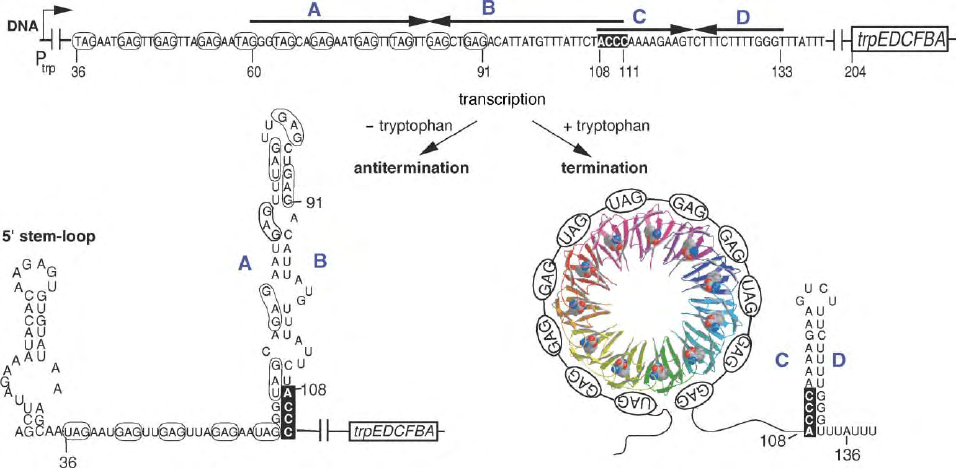
THE B. SUBTILIS TRP LEADER
Expression of the trpEDCFBA operon in B. subtilis and
several related bacilli is regulated by the trp RNA-
binding Attenuation Protein (TRAP). Transcription of
the operon initiates from the trp promoter 203
nucleotides upstream of the start codon of the first
structural gene, trpE (Figure 2). There is no evidence
for any regulation of initiation of transcription from
this promoter. Hence there is no homologue, either
FIGURE 2 Model of transcription attenuation of the B. subtilis trpEDCFBA operon. The large blue letters indicate the complementary strands of
the terminator and antiterminator RNA structures. The TRAP protein is shown as a ribbon diagram with each of the 11 subunits as a different color
and the bound RNA is shown forming a matching circle upon binding to TRAP. The bound tryptophans are shown as space filling models. The GAG
and UAG repeats involved in TRAP binding are shown in ovals and are also outlined in the sequence of the antiterminator structure. Numbers
indicate the residue positions relative to the start of transcription. Nucleotides 108– 111 overlap between the antiterminator and terminator
structures and are shown as darkly outlined letters.
trp OPERON AND ATTENUATION 269
functional or based on amino acid sequence, of the
E. coli DNA binding trp repressor in B. subtilis.
The B. subtilis trp leader transcript also contains
inverted repeats that can form an intrinsic transcription
terminator (C:D) and an antiterminator (A:B) RNA
secondary structure (Figure 2). These two structures
overlap by four nucleotides and hence their formation is
mutually exclusive.
In the presence of excess tryptophan, TRAP binds to
a series of 11 trinucleotide repeats in the trp leader
transcript consisting of seven GAGs and four UAGs
(Figure 2). The (G/U)AG repeats are separated from
each other by two or three “spacer” nucleotides, whose
sequence is not conserved. These triplet repeats in part
overlap the 5
0
portion of the antiterminator structure
and hence TRAP-binding interferes with form-
ation of the antiterminator (A:B) thus favoring form-
ation of the terminator (C:D) hairpin. Therefore under
these conditions transcription halts in the
leader region prior to the trp structural genes, which
are not expressed.
When tryptophan is limiting, TRAP does not bind to
the trp leader RNA transcript and formation of the A:B
antiterminator is favored. This situation allows tran-
scription to proceed into the trp operon structural genes,
which can now be expressed to produce the enzymes to
synthesize tryptophan.
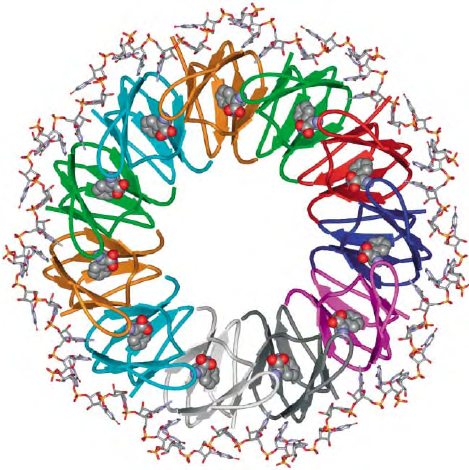
THE TRAP PROTEIN
In this system the ability of TRAP to sense the levels of
intracellular tryptophan and to then influence the
structure of the nascent trp leader mRNA transcript
is the basis for attenuation control of the trp operon.
TRAP is a ring-shaped protein composed of 11
identical subunits (Figure 3). TRAP is activated to
bind its RNA targets by binding 11 molecules of
L-tryptophan in clefts between adjacent subunits.
The detailed mechanism by which tryptophan binding
activates TRAP to bind RNA is not known, however,
recent studies suggest that tryptophan binding induces
a conformational change in TRAP as well as reduces
the flexibility of the protein. Upon activation by
tryptophan, TRAP binds to its RNA target consisting
of the 11 (G/U)AG repeats by wrapping the single-
stranded RNA around the outer perimeter of the
protein ring (Figure 3). The phosphodiester backbone
is on the outside of the RNA ring and the bases point
in toward the protein. The specificity of this interaction
derives mainly from hydrogen bonds between several
amino acids of each TRAP subunit including Glu36,
Lys37, Lys56, and Arg58, and the bases on the
(G/U)AG repeats. The spacer nucleotides do not
contact the protein, and their identity is not crucial
for TRAP recognition and binding. This mechanism of
wrapping the RNA around the protein ring explains
both the specificity of the TRAP–RNA interaction as
well as how TRAP binding alters the structure of the
trp leader RNA to regulate attenuation.
THE ROLE OF PAUSING IN TRAP
M
EDIATED ATTENUATION
In addition to the terminator and antiterminator
structures, there is an RNA hairpin at the 5
0
end of
the trp leader mRNA (Figure 2;5
0
stem-loop).
Although this structure has been shown to be import-
ant for proper attenuation control of the trp operon, it
is not a transcriptional pause signal analogous to the 1:2
structure of the E. coli trp attenuation system. The role
of this 5
0
stem-loop may be to enhance the rate of
TRAP binding to the trp leader RNA. This is an
important consideration because TRAP must bind to its
target before RNA polymerase transcribes beyond
the terminator structure, otherwise TRAP binding
would not be able to influence attenuation. Recent
studies published by Yakhnin and Babitzke indicate
that RNA polymerase pauses at U107 (Figure 2)
during transcription of the trp leader region, and that
this pausing is dependent on the NusA transcription
factor. This pause would provide additional time for
TRAP to bind to the nascent trp leader RNA and
promoter termination.
FIGURE 3 Ribbon diagram of TRAP complexed with an RNA
containing 11 GAG repeats separated by AU spacers. The TRAP
protein is shown in ribbon diagrams with each subunit depicted
in a different color. The RNA is shown in stick models and
the 11 molecules of bound L-tryptophan are shown in space-
filling models.
270 trp OPERON AND ATTENUATION
ANTI-TRAP AND THE ROLE
OF T
RNA
T
RP
IN REGULATING
THE
B. SUBTILIS TRP GENES
In this attenuation system the regulatory signal is the
level of free tryptophan available to activate TRAP to
bind RNA. In the previously described E. coli trp
attenuation system, free tryptophan is sensed by the
DNA binding trp repressor protein and the regulatory
signal for attenuation is the availability of charged
tRNA
Trp
. The level of charged tRNA
Trp
also influences
attenuation control of the B. subtilis trp operon,
although by a very different mechanism than in the
E. coli system. In B. subtilis the rtpA gene encodes a
protein called anti-TRAP (AT). AT influences expression
of the trp genes by binding to tryptophan-activated
TRAP and preventing it from binding RNA, thus
elevating expression of trp genes. Expression of rtpA,
which is in a two-gene operon together with ycbK
(unknown function), is regulated in response to changes
in the level of uncharged tRNA
Trp
by a mechanism
known as T-box antitermination. High levels of
uncharged tRNA
Trp
induce expression of AT, which
prevents TRAP from down-regulating expression of the
trp genes. Hence even if free tryptophan levels are high
enough to activate TRAP, if the level of charging of
tRNA
Trp
is low, then the trp genes will be expressed.
Thus while both E. coli and B. subtilis regulate
expression of the trp genes in response to changes in
levels of both free tryptophan and aminoacylated
tRNA
Trp
, they have evolved very different mechanisms
to do so.
OCCURRENCES OF TRAP MEDIATED
ATTENUATION IN BACTERIA
TRAP mediated attenuation control of the trp operon
has till date been only observed in B. subtilis and several
related bacilli including B. pumilus, B. stearothermo-
philus, B. caldotenax,andB. halodurans, as well as in
Clostridium thermocellum. In contrast to the leader-
peptide-dependent mechanism described previously,
the RNA-binding protein-dependent attenuation mecha-
nism has never been characterized for any other amino
acid biosynthetic operons beside tryptophan even
though it would seem to be easily adaptable by simply
changing the amino acid that activates the protein to
bind its RNA target. Perhaps with the explosion of
bacterial genomic information currently becoming
available we might soon discover such a system.
SEE ALSO THE FOLLOWING ARTICLES
DNA Polymerase I, Bacterial † DNA Polymerase II,
Bacterial † DNA Polymerase III, Bacterial † Ribosome
Structure † RNA Polymerase Reaction in Bacteria †
RNA Polymerase Structure, Bacterial † T7 RNA
Polymerase
GLOSSARY
intrinsic terminator A signal in the nascent RNA transcript that
signals RNA polymerase to halt transcription and dissociate
from the DNA template. Intrinsic terminators consist of a short
base-paired stem-loop structure followed by a short stretch of U
residues in the RNA. Also called factor-independent terminator or
Rho-independent terminator.
ribosome The large RNA–protein complex that translated mRNA
into protein. It consists of two subunits, termed small and large.
RNA polymerase The enzyme that transcribes DNA into RNA.
In bacteria there is only one version of this enzyme.
transcriptional pausing In response to signals in the RNA, RNA
polymerase will pause and discontinue transcription but not
dissociate from the DNA template. Either after some period of
time, or in response to a signal, a paused RNA polymerase will
resume transcription.
tRNA The adaptor RNA molecule that reads the genetic code in the
mRNA by base-pairing with the codon triplets. An amino acid is
attached to the 3
0
end of the last base in the tRNA by an enzyme
called an aminoacyl tRNA synthetase in a process called
aminoacylation or charging. There are tRNAs corresponding
each of the 20 amino acids.
FURTHER READING
Antson, A. A., Otridge, J. B., Brzozowski, A. M., Dodson, E. J.,
Dodson, G. G., Wilson, K. S., Smith, T. M., Yang, M., Kurecki, T.,
and Gollnick, P. (1995). The three dimensional structure of trp
RNA-binding attenuation protein. Nature 374, 693–700.
Antson, A. A., Dodson, E. J., Dodson, G. G., Greaves, R. B., Chen,
X.-P., and Gollnick, P. (1999). Structure of the trp RNA-binding
attenuation protein, TRAP, bound to RNA. Nature 401, 235–242.
Babitzke, P., and Gollnick, P. (2001). Posttranscriptional initiation
control of tryptophan metabolism in Bacillus subtilis by the trp
RNA-binding attenuation protein (TRAP), anti-TRAP, and RNA
structure. J. Bacteriol. 183, 5795–5802.
Valbuzzi, A., and Yanofsky, C. (2001). Inhibition of the B-subtilis
regulatory protein TRAP by the TRAP-inhibitory protein, AT.
Science 293, 2057 –2059.
Yakhnin, A. V., and Babitzke, P. (2002). NasA-stimulated RNA
polymerase pausing and termination participates in the Bacillus
subtilis trp operon attenuation mechanism in vitro. Proc. Natl.
Acad. Sci. USA 99, 11067–11072.
Yanofsky, C. (2000). Transcription attenuation: Once viewed as a
novel regulatory strategy. J. Bacteriol. 182,1–8.
Yanofsky, C. (2001). Advancing our knowledge in biochemistry,
genetics, and microbiology through studies on tryptophan metab-
olism. Annu. Rev. Biochem. 70, 1–37.
BIOGRAPHY
Paul Gollnick is a Professor in the Department of Biological Sciences at
the University of Buffalo, the State University of New York. His
principal research interests are in RNA–protein interactions and
regulation of gene expression. He holds a Ph.D. from Iowa State
University and received postdoctoral training at Stanford University.
He and his collaborator Dr. Alfred Antson at York University
in England have determined the structure of TRAP and the
TRAP:RNA complex.
trp OPERON AND ATTENUATION 271
