Lennarz W.J., Lane M.D. (eds.) Encyclopedia of Biological Chemistry. Four-Volume Set . V. 4
Подождите немного. Документ загружается.


Secretory Pathway
Karen J. Colley
University of Illinois at Chicago, Chicago, Illinois, USA
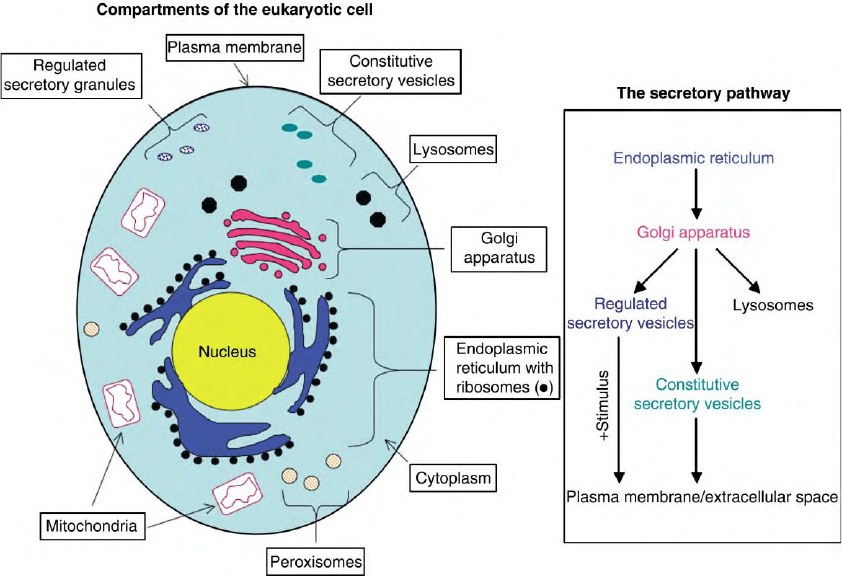
The eukaryotic cell is separated into several functionally
distinct, membrane-enclosed compartments (Figure 1). Each
compartment contains proteins required to accomplish specific
functions. Consequently, each protein must be sorted to its
proper location to ensure cell viability. Proteins possess specific
signals, either encoded in their amino acid sequences or added
as posttranslational modifications, which target them for the
various compartments of the cell. The pioneering work of
Dr. George Palade provided scientists with their first picture of
the functional organization of the mammalian secretory path-
way. Later work showed that the secretory pathway acts as a
folding, modification, and quality control system for proteins
that function in the endoplasmic reticulum (ER) and Golgi
apparatus, and for those that are targeted to the lysosome,
plasma membrane, and extracellular space. This article will
focus on protein targeting to and within the compartments of
the secretory pathway, and how proteins within this pathway
function to ensure that correctly folded and modified proteins
are delivered to the cell surface and secreted from cells.
Targeting of New Proteins to
the Secretory Pathway
WHAT KINDS OF PROTEINS
ARE
TARGETEDTOTHE
SECRETORY PATHWAY?
The proteins that are targeted to the secretory pathway
can be separated into two groups – those that function
in the ER and Golgi to ensure proper protein folding and
modification (i.e., resident proteins), and those that are
processed in the ER and Golgi, and are transported to
later compartments like the lysosome, plasma mem-
brane, and extracellular space (Figure 1). Each of these
proteins not only possesses a signal to enter the secretory
pathway, but also may have a secondary signal to
localize it to a particular organelle within the pathway.
SIGNALS AND MECHANISMS OF
SECRETORY PATHWAY ENTRY
The 1999 Nobel Prize in physiology or medicine was
awarded to Dr. Gu
¨
nter Blobel for his contributions to
our understanding of the mechanism of secretory path-
way entry. Dr. Blobel and his colleagues found that in
order to enter the secretory pathway, proteins are
synthesized with an amino terminal signal peptide that
allows them to cross the membrane of the endoplasmic
reticulum (ER). The signal peptide is recognized by a
complex of proteins and ribonucleic acid called the
signal recognition particle (SRP) (Figure 2). As the signal
peptide emerges from the ribosome during translation,
SRP binds to it and halts translation, and then targets the
new protein–ribosome complex to the cytoplasmic face
of the ER membrane where it binds to the SRP receptor.
Subsequently, the new protein–ribosome complex is
released from SRP and its receptor, and transferred to an
aqueous membrane channel known as the “translocon.”
Translation resumes and the new protein is co-transla-
tionally transferred through the translocon into the
lumen of the ER, where in many cases the signal peptide
is cleaved by a specific signal peptidase (Figure 2).
SOLUBLE AND INTEGRAL
MEMBRANE PROTEINS
Soluble proteins are completely translocated across the
ER membrane into the lumen (Figure 2). These proteins
will either remain in the ER, be targeted to another
organelle, or be secreted from the cell. Integral
membrane proteins that possess one or more hydro-
phobic membrane-spanning regions will use these
sequences to insert into the membrane of the ER and
either stay as ER-resident transmembrane proteins, or be
targeted to another cellular membrane. A type-I
membrane protein has a cleavable signal peptide and a
separate hydrophobic stretch of amino acids that acts as
a membrane-spanning region. This type of protein has
its amino terminus in the lumen of an organelle or the
outside of the cell (which are topologically equivalent),
and its carboxy terminus in the cytoplasm (Figure 2). In
contrast, a type-II membrane protein has an uncleavable
signal peptide, or signal anchor that is not at the
protein’s amino terminus and serves a dual function as
both signal peptide and a membrane-spanning region.
Type-II membrane proteins employ a more elaborate
insertion mechanism than do type-I membrane proteins.
Encyclopedia of Biological Chemistry, Volume 4. q 2004, Elsevier Inc. All Rights Reserved. 11
For this reason, a type-II membrane protein will have its
carboxy terminus in the lumen of an organelle or outside
the cell, and its amino terminus in the cytoplasm
(Figure 2). Other proteins span the membrane several
times and are called type-III membrane proteins. They
can start with either cleavable signal peptides or
uncleavable signal anchors and possess variable num-
bers of hydrophobic membrane-spanning segments.
Protein Folding and Modification
in the ER
THE INITIATION OF PROTEIN N-LINKED
GLYCOSYLATION IN THE ER
As proteins enter the ER lumen, they fold and assemble
with the help of chaperone proteins. Many proteins are
also co-translationally modified by the addition of
carbohydrates to asparagine residues in the process of
N-linked glycosylation (Figure 2). A preformed oligosac-
charide, consisting of three glucoses, nine mannoses, and
two N-acetylglucosamine residues (Glc
3
Man
9
GlcNAc
2
)
is transferred to accessible asparagine residues in the
tripeptide sequence asparagine-X-serine or threonine
(X cannot be proline) by the oligosaccharide protein
transferase complex. Subsequent modification by glyco-
sidases (enzymes that remove monosaccharides) and
glycosyltransferases (enzymes that add monosacchar-
ides) in the ER and Golgi lead to the remodeling of the
N-linked oligosaccharides. These N-linked carbo-
hydrates help proteins fold, protect them from proteo-
lytic degradation and, in some cases, are critical for
modulating and mediating protein and cell interactions
at the cell surface and in the extracellular space.
CHAPERONES AND THE ER QUALITY
CONTROL SYSTEM
An important function of the ER is to serve as a site of
protein folding and quality control. Protein folding in
the ER includes the formation of intra-molecular
disulfide bonds, prolyl isomerization, and the sequestra-
tion of hydrophobic amino acids into the interior of the
protein. Protein disulfide bonds are formed as the
protein exits the translocon and may at first form
incorrectly between cysteine residues close together
in the protein’s linear amino acid sequence. Thiol-
oxidoreductases, such as protein disulfide isomerase
(PDI), help to form and reorganize proteins’ disulfide
bonds into the most energetically favorable configur-
ation. Different types of chaperones monitor a protein’s
FIGURE 1 Compartments of eukaryotic cells and the organization of the secretory pathway. Diagrammatic representation of the compartments
of the eukaryotic cell is shown. The anterograde flow of membrane and protein traffic in the secretory pathway is shown in the box. Anterograde
flow is indicated by arrows. Retrograde flow between the ER and Golgi, endosome/lysosome system and Golgi, and plasma membrane and Golgi
does occur, but is not shown.
12 SECRETORY PATHWAY
folding and prevent exit of unfolded and unassembled
proteins from the ER. The chaperone BiP, originally
identified as an immunoglobulin heavy-chain-binding
protein, interacts with the exposed hydrophobic
sequences of folding intermediates of many proteins
and prevents their aggregation. Two chaperones called
calnexin and calreticulin recognize a monoglucosylated
carbohydrate structure (Glc
1
Man
9
GlcNAc
2
)thatis
formed by a special glucosyltransferase that recognizes
unfolded or misfolded proteins and adds a single glucose
to the Man
9
GlcNAc
2
structure. Proteins that are not
folded properly or are not assembled into oligomers with
partner subunits, are prevented from exiting the ER by
chaperone interactions, and can be targeted back across
the ER membrane through the translocon into the
cytoplasm where they are degraded by the proteosome
complex in a process called ER associated degradation
(ERAD).
Protein Transport through and
Localization in the Secretory
Pathway
VESICULAR TRANSPORT BETWEEN
THE
ER AND GOLGI
Proteins move between the ER and Golgi in vesicular
carriers. These vesicles are coated with specific sets of
cytoplasmic proteins that form the COP-I and COP-II
coats. COP-II-coated vesicles move from the ER to the
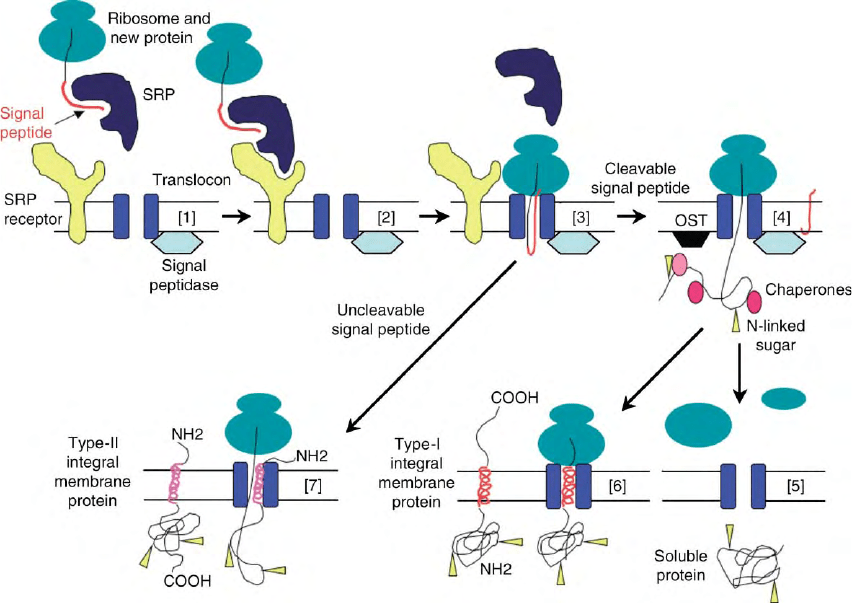
FIGURE 2 Entry into the secretory pathway. Many proteins are targeted for the secretory pathway by an amino terminal hydrophobic signal
peptide that allows their co-translational translocation across the ER membrane. [1] Signal recognition particle (SRP) recognizes the new protein’s
signal peptide. [2] The ribosome–new protein– SRP complex interacts with the SRP receptor on the cytoplasmic face of the ER membrane. [3] The
ribosome–new protein complex is transferred to the translocon channel, protein synthesis continues and the protein moves through the aqueous
channel. [4] As the new protein enters the lumen of the ER, its signal peptide is cleaved by the signal peptidase, chaperone proteins bind to aid in
folding and oligosaccharide protein transferase complex (OST) transfers oligosaccharides (arrowheads) to asparagine residues in the process of
N-linked glycosylation. [5] Soluble proteins lack additional hydrophobic sequences and are translocated through the translocon to complete their
folding and modification in the ER lumen. [6] Type-I integral membrane proteins have a second hydrophobic sequence that partitions into the lipid
bilayer and acts as a membrane-spanning segment. These proteins have their amino termini in the lumen of an organelle or outstide the cell and their
carboxy-termini in the cell cytoplasm. [7] Unlike proteins with cleavable amino-terminal signal peptides, type-II integral membrane proteins have
an uncleavable signal anchor that target the protein to the secretory pathway and then partitions into the lipid bilayer to act as a membrane-
spanning segment. These proteins have their amino termini in the cell cytoplasm and their carboxy-termini in the lumen of an organelle or outside
the cell. Soluble and integral membrane proteins that enter at the level of the ER need not stay there, and can be transported out of the ER to other
locations in the pathway (see Figure 1, Secretory Pathway box).
SECRETORY PATHWAY 13
intermediate compartment (IC)/cis Golgi, while COP-I-
coated vesicles move from the Golgi back to the ER and
may also mediate transport between Golgi cisternae in
both the anterograde (toward the plasma membrane)
and retrograde (toward the ER) directions (Figure 3).
The process of vesicular transport can be separated into
three stages—cargo selection and budding, targeting,
and fusion. In the first stage, the COP coats serve to
select cargo for exit from a compartment and help to
deform the membrane for vesicle budding. They
assemble on the membrane with the help of small
GTPases called ARF (specific for COP I) and Sar1p
(specific for COP II). After vesicle budding, the
hydrolysis of GTP by ARF and Sar1p leads to the
uncoating of the vesicle. This uncoating reveals other
vesicle proteins that are essential for vesicle targeting
and fusion. In the second and third stages, tethering
proteins on the transport vesicle and target membrane
interact weakly bringing the membranes together. This
allows vesicle-associated SNARE proteins and target
membrane-associated SNARE proteins to form com-
plexes. Subsequent conformational changes in the
SNARE protein complex bring the membranes together
for fusion. Another group of small GTPases (Rabs)
control the process of vesicular transport at several
levels by recruiting and activating various proteins in the
pathway.
PROTEIN LOCALIZATION IN THE ER
Proteins involved in protein folding, modification, and
quality control must remain in the ER, while proteins
destined for the Golgi, lysosome, cell surface or those
that are secreted from the cell must exit. Exit from the
ER is a selective process that involves cargo receptors
that interact with COP-II coat components (Figure 3). It
is likely that most resident ER proteins are not selected
to exit the ER. It is clear, however, that some resident
proteins escape from the ER and are retrieved from the
Golgi and intermediate compartment by COP-I vesicles
(Figure 3). These proteins have specific amino acid
signals that allow their incorporation into COP-I
vesicles either by direct interaction with COP-I
components or indirectly by interaction with cargo
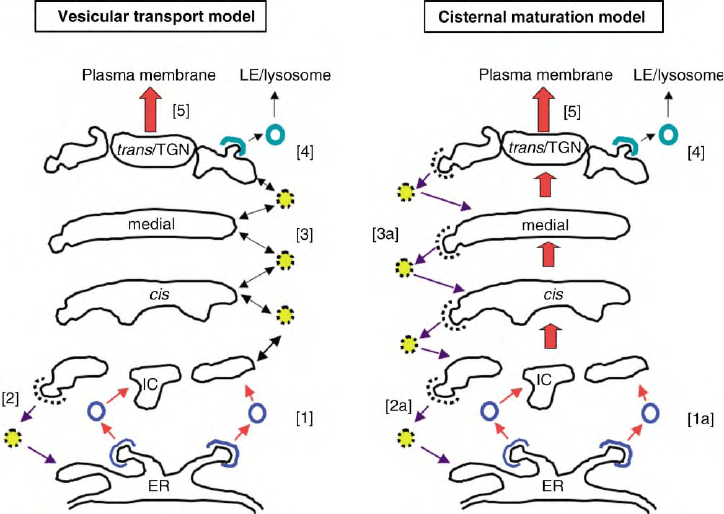
FIGURE 3 Comparison of two models of protein transport through the Golgi apparatus. In the vesicular transport model cargo proteins move
between the cisternae in vesicles, while Golgi enzymes are retained in their resident cisternae. [1] COP-II-coated vesicles transport new proteins
from the ER to the intermediate compartment (IC). [2] Resident ER proteins that escape the ER can be retrieved from the IC in COP-I-coated
vesicles. [3] COP-I-coated vesicles also transport anterograde cargo proteins between the Golgi cisternae in both a retrograde and anterograde
fashion (“percolating vesicles”). In the cisternal maturation model, cargo proteins enter a new cisterna at the cis face of the stack, and are modified
(matured) by “resident” Golgi enzymes that are continuously transported in a retrograde fashion into the sequentially maturing cisternae. [1a]
COP-II-coated vesicles transport new proteins from the ER to the IC where a new cis cisterna forms. [2a] Resident ER proteins that escape the ER
can be retrieved from the IC in COP-I-coated vesicles. [3a] Golgi enzymes are transported in a retrograde fashion in COP-I-coated vesicles to modify
the cargo proteins in earlier cisternae. Mechanisms of protein exit from the TGN are common to both models: [4] clathrin-coated vesicles mediate
late endosome (LE)-lysosome transport, while [5] other proteins are secreted in either a regulated or constitutive fashion to the plasma membrane or
extracellular space.
14 SECRETORY PATHWAY
receptors. For example, mammalian BiP is a soluble ER
protein that has the carboxy-terminal four amino acid
sequence lysine–aspartate–glutamate–leucine (KDEL).
This KDEL sequence is recognized by the KDEL
receptor that mediates their incorporation into COP I
vesicles moving from the intermediate compartment (IC)
back to the ER.
PROTEIN MODIFICATION IN THE GOLGI
The Golgi apparatus consists stacks of flattened cister-
nae that contain enzymes and other proteins involved in
the further modification and processing of newly made
proteins. It is separated into cis, medial, and trans
cisternae, followed by a meshwork of tubules and
vesicles called the trans Golgi network (TGN). The
process of N-linked glycosylation is completed through
the action of glycosidases and glycosyltransferases
localized in specific cisternae. Likewise, the glycosyla-
tion of serine and threonine residues (O-linked glycosy-
lation) is accomplished by other glycosyltransferases.
Additional modifications also occur in the Golgi. For
example, proteins and carbohydrate are sulfated by
sulfotransferases and some proteins are phosphorylated
on serine and threonine residues by Golgi kinases. In
addition, proteins like digestive enzymes (trypsin,
carboxypeptidase) and hormones (insulin) are made as
inactive precursors that must be proteolytically pro-
cessed to their active forms in the late Golgi or post-
Golgi compartments.
TRANSPORT OF PROTEINS
THROUGH THE
GOLGI
Currently there are two different models to explain
protein transport through the Golgi (Figure 3). The
vesicular transport model proposes that proteins move
sequentially between the Golgi cisterna in COP-I-coated
vesicles, while the cisternae themselves are stationary.
Proteins not retained in the cis Golgi, for example, would
be incorporated into coated vesicles and be transported to
the medial Golgi, and then to the trans Golgi. Proteins
destined for post-Golgi compartments move through
successive Golgi cisternae in this fashion, being modified
by the resident enzymes in each compartment (Figure 3).
In the cisternal maturation model a new cisterna is
formed on cis face of the Golgi stack from ER-derived
membrane. This requires both the anterograde transport
of newly synthesized proteins from the ER in COP-II-
coated vesicles and the retrograde transport of cis Golgi
enzymes from the pre-existing cis cisterna in COP I-
coated vesicles. The new cis cisterna and its contents
progressively mature through the stack as resident Golgi
enzymes are successively introduced by COP-I coated
vesicles (Figure 3). In the vesicular transport model, the
resident Golgi enzymes are retained in the cisternae while
the cargo moves in vesicles between the different
cisternae. In contrast, in the cisternal maturation
model, the “resident” enzymes are continuously moving
in a retrograde fashion, while the anterograde cargo
remains in the cisternae. Evidence for both mechanisms is
compelling, suggesting that both mechanisms may work
in parallel.
LOCALIZATION OF RESIDENT
GOLGI ENZYMES
In the context of the vesicular transport model, Golgi
enzymes are retained in specific cisternae. Two mechan-
isms have been suggested for Golgi protein retention.
The “bilayer thickness” mechanism suggests that the
relatively short transmembrane regions of Golgi proteins
prevent their incorporation into the wider, cholesterol-
rich lipid bilayers of the transport vesicles destined for
post-Golgi compartments (such as the plasma mem-
brane), and as a result, these proteins are retained in the
relatively cholesterol-poor Golgi. The “oligomeriza-
tion” mechanism predicts that once an enzyme has
reached its resident cisterna it forms homo- or hetero-
oligomers that prevent its incorporation into transport
vesicles moving to the next compartment. In the context
of the cisternal maturation model, resident Golgi
enzymes are actively incorporated into COP-I vesicles
for retrograde transport to a new cisterna, and one
might predict that the cytoplasmic tails of these proteins
would interact with COP-I-coat components to allow
vesicle incorporation. Interestingly, there are only a few
examples where the cytoplasmic tail of a Golgi enzyme
plays a primary role in its localization, whereas the
membrane-spanning regions of these proteins seem to be
more critical. Again, it is possible that some or all of
these mechanisms work together to maintain the steady-
state localization of the resident Golgi proteins.
Protein Exit from the Golgi and
Targeting to Post-Golgi Locations
PROTEIN EXIT FROM THE GOLGI
Once proteins reach the TGN they are sorted to post-
Golgi compartments that include the lysosome, the
plasma membrane, and the extracellular space
(Figure 3). Trafficking to the lysosome–endosome system
involves clathrin-coated vesicles similar to those that
function in the uptake of proteins in endocytosis. In
contrast, transport to the plasma membrane, or exocy-
tosis, can occur either constitutively in secretory vesicles/
tubules or in a regulated fashion from secretory granules
found in specific cell types.
SECRETORY PATHWAY 15
PROTEIN TARGETING TO THE LYSOSOME
The lysosome is a degradative compartment that
contains numerous acid hydrolases that function to
digest proteins, lipids, and carbohydrates. The traffick-
ing of the majority of lysosomal enzymes to the lysosome
requires mannose 6-phosphate residues on these
enzymes’ N-linked sugars. The mannose 6-phosphate
residues are recognized by receptors in the TGN that
mediate the incorporation of the new lysosomal enzymes
into clathrin-coated vesicles destined for the late
endosome compartment (Figure 3). These clathrin-
coated vesicles move from the TGN and fuse with the
late endosome, where a decrease in lumenal pH causes
the lysosomal enzymes to dissociate from the mannose
6-phosphate receptors. The enzymes are then trans-
ported to the lysosome, while the receptors recycle to the
TGN. Some lysosomal membrane proteins are also
trafficked in clathrin-coated vesicles to the lysosome like
the soluble enzymes but without the use of a mannose
6-phosphate marker, while others are transported to the
cell surface, incorporated into a different set of clathrin-
coated vesicles used in the process of endocytosis, and
then trafficked to the lysosome via the late endosome.
CONSTITUTIVE AND REGULATED
SECRETION
In many cell types, membrane-associated and soluble
proteins move to the plasma membrane constitutively
without a requirement for specific signals. Constitutively
secreted proteins include receptors, channel proteins,
cell adhesion molecules, and soluble extracellular matrix
and serum proteins. Other proteins like hormones and
neurotransmitters are targeted to secretory granules that
are involved in regulated secretion from endocrine and
exocrine cells, some types of immune cells, and neurons.
These granules remain in a secretion-ready state
until extracellular signals that lead to an increase in
intracellular calcium levels trigger the exocytosis of
their contents.
SEE ALSO THE FOLLOWING ARTICLES
Chaperones, Molecular † Endoplasmic Reticulum-
Associated Protein Degradation † Glycoproteins,
N-linked † Golgi Complex † Protein Folding and
Assembly † Protein Glycosylation, Overview
GLOSSARY
chaperone A protein that aids in the folding and assembly of other
proteins, frequently by preventing the aggregation of folding
intermediates.
cisternal maturation/progression One model of protein transport
through the Golgi apparatus that suggests that secretory cargo
enters a new cisternae that forms at the cis face of the Golgi stack,
and that this cisternae and its cargo progresses or matures through
the stack by the sequential introduction of Golgi modification
enzymes.
glycosylation The modification of lipids and proteins with carbo-
hydrates in the endoplasmic reticulum and Golgi apparatus of the
secretory pathway.
secretory pathway An intracellular pathway consisting of the
endoplasmic reticulum, Golgi apparatus, and associated vesicles
that is responsible for the folding, modification, and transport of
proteins to the lysosome, plasma membrane, and extracellular
space.
vesicular transport One model of protein transport through the Golgi
apparatus, which suggests that secretory cargo moves sequentially
between stationary Golgi cisternae in transport vesicles and is
modified by resident Golgi enzymes in the process.
FURTHER READING
Ellgaard, L., Molinari, M., and Helenius, A. (1999). Setting the
standards: Quality control in the secretory pathway. Science 286,
1882–1888.
Farquhar, M. G., and Palade, G. E. 1998. The Golgi apparatus: 100
years of progress and controversy. Trends Cell Biol. 8, 2–10.
Intracellular compartments and protein sorting (chapter 12) and
intracellular vesicular traffic (chapter 13). In The Molecular
Biology of the Cell (B. Alberts, A. Johnson, J. Lewis, M. Raff, K.
Roberts, and P. Walter, eds.), 4th edition, pp. 659–766. Garland
Science, New York.
Kornfeld, S., and Mellman, I. (1989). The biogenesis of lysosomes.
Annu. Rev. Cell Biol. 5, 483–525.
Palade, G. (1975). Intracellular aspects of the process of protein
synthesis. Science 189, 347– 358.
Rapoport, T. A., Jungnickel, B., and Kutay, U. (1996). Protein
transport across the eukaryotic endoplasmic reticulum and
bacterial inner membranes. Annu. Rev. Biochem. 65, 271– 303.
Rockefeller University web site describing Dr. Gu
¨
nter Blobel’s Nobel
Prize research (http://www.rockefeller.edu).
BIOGRAPHY
Karen J. Colley is a Professor in the Department of Biochemistry and
Molecular Genetics at the University of Illinois College of Medicine
in Chicago. She holds a Ph.D. from Washington University in St.
Louis, and received her postdoctoral training at the University of
California, Los Angeles. Her principal research interests are in
protein trafficking and glycosylation. Her recent studies focus on
the elucidation of the signals and mechanisms of Golgi glycosyl-
transferase localization.
16 SECRETORY PATHWAY

Selenoprotein Synthesis
August Bo
¨
ck
University of Munich, Munich, Germany
Selenoproteins contain one or more residues of the nonstan-
dard amino acid selenocysteine, which is an analogue of
cysteine in which a selenol group replaces a thiol. The majority
of these proteins catalyze some oxidation/reduction function
in which the selenol of the selenocysteine that is present in the
active site of the respective enzyme takes part in the reaction.
The advantage of having a selenol instead of a thiol lies in the
fact that it confers to these enzymes a higher kinetic efficiency.
In some biological systems, selenoproteins may also fulfill a
structural role because of their capacity to oligomerize proteins
via the formation of diselenide or mixed disulfide– selenide
bridges. The biosynthesis of selenoproteins is unique since the
incorporation of selenocysteine occurs co-translationally by
the ribosome and not posttranslationally. Selenocysteine
insertion is DNA encoded, requires the function of a cognate
tRNA and of a specific translation elongation factor different
from elongation factor Tu. Selenocysteine, therefore, has been
designated as the 21st amino acid.
Bacterial Selenoprotein Synthesis
The structure and the function of the components
involved in selenocysteine biosynthesis have been charac-
terized to a considerable extent in the case of the bacterial
system. The process can be divided into three functional
steps, namely the biosynthesis of selenocysteine in the
tRNA-bound state, the formation of a complex between
elongation factor SelB, GTP, selenocysteyl-tRNA
Sec
and
the mRNA, and the decoding event at the ribosome. As
far as it is known, though there are some major
differences, similarities also exist between bacterial
selenoprotein synthesis and the process characteristic of
eukarya and archaea.
SELENOCYSTEINE BIOSYNTHESIS
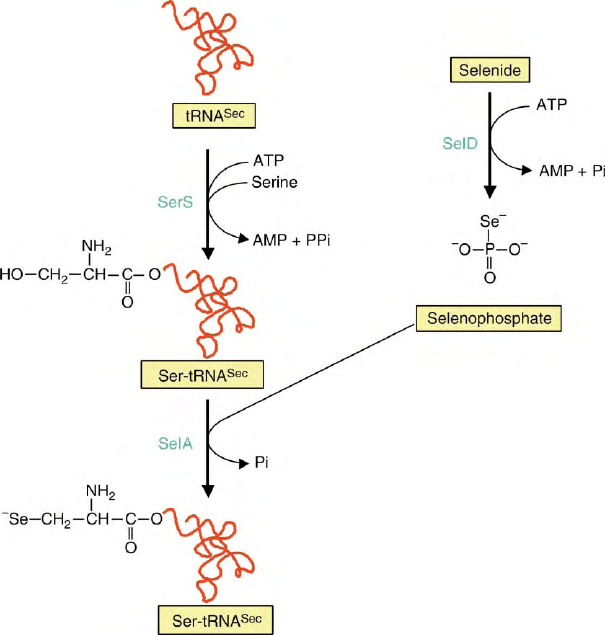
Figure 1 summarizes the process of selenocysteine
biosynthesis as it has been worked out for Escherichia
coli. It requires the activities of three enzymes, namely
seryl-tRNA synthetase (SerS), selenophosphate synthe-
tase (SelD), and selenocysteine synthase (SelA) plus the
specific tRNA (tRNA
Sec
). Seryl-tRNA synthetase
charges tRNA
Sec
with L-serine, selenocysteine synthase
converts the seryl-tRNA
Sec
into selenocysteyl-tRNA
Sec
using selenophosphate as a source for activated sel-
enium. Selenophosphate is provided by selenophosphate
synthetase from selenide in an ATP-dependent reaction.
The genes for these components had been identified with
the aid of E. coli mutants isolated by Mandrand–
Berthelot as being pleiotropically deficient in formate
dehydrogenase activities.
tRNA
Sec
tRNA
Sec
(Figure 2) is the key molecule of selenoprotein
synthesis since it serves both as the adaptor for
selenocysteine biosynthesis and for incorporation of the
amino acid at the ribosome. It deviates in size, secondary
structure, and in normally conserved sequence positions
from canonical elongator tRNA species. Because of the
elongated extra arm and the one base-pair-extended
aminoacyl acceptor arm, tRNA
Sec
species are the largest
members of the elongator tRNA family. All tRNA
Sec
species identified thus far possess a UCA anticodon which
enables them to pair with UGA stop codons (but only if
these are in a special mRNA sequence context). More-
over, tRNA
Sec
species deviate from canonical elongator
tRNA species in sequence positions which are usually
invariant and which are involved in the establishment of
novel tertiary interactions within the molecule.
On the basis of its serine identity elements, tRNA
Sec
is
charged by the cellular seryl-tRNA synthetase which
also aminoacylates serine inserting isoacceptors. How-
ever, both the affinity and the rate of aminacylation are
diminished in comparison to the charging of tRNA
Ser
,
resulting in an overall 100-fold reduced efficiency.
Selenocysteine Synthase
The overall reaction catalyzed by selenocysteine
synthase consists in the exchange of the hydroxylgroup
of the serine moiety of seryl-tRNA
Sec
by a selenol
group (Figure 1). The reaction occurs in two steps; first,
the amino group of serine forms a Schiff base with
the carbonyl of the pyridoxal 5
0
-phosphate cofactor of
selenocysteine synthase leads to the 2,3-elimination of a
water molecule and the formation of dehydroalanyl-
tRNA
Sec
and second, nucleophilic addition of reduced
Encyclopedia of Biological Chemistry, Volume 4. q 2004, Elsevier Inc. All Rights Reserved. 17
selenium to the double bond of dehydroalanyl-tRNA
Sec
from selenophosphate as a donor yields selenocysteyl-
tRNA
Sec
.
Selenocysteine synthase from E. coli is a homodeca-
meric enzyme and low resolution electron microscopy
revealed that it is made up of two pentameric rings
stacked on top of each other. Two subunits each are able
to bind one molecule of seryl-tRNA
Sec
, so the fully
loaded enzyme can complex five charged tRNA mol-
ecules. As serine isoacceptor tRNAs are not recognized,
the tRNA must have determinants for the specific
recognition of seryl-tRNA
Sec
by selenocysteine synthase
and antideterminants for the rejection of seryl-tRNA
Ser
species. The specificity for discrimination of the sel-
enium donor is not as strict since the purified enzyme
accepts thiophosphate instead of selenophosphate as a
substrate. This results in the formation of cysteyl-
tRNA
Sec
. So the discrimination between sulfur and
selenium must take place at some other step of
selenocysteine biosynthesis.
Selenophosphate Synthetase
Purified selenocysteine synthase does not exhibit an
absolute requirement for selenophosphate, as a substrate
to convert seryl-tRNA
Sec
into selenocysteyl-tRNA
Sec
,
since the reaction also occurs in the presence of high
concentrations of selenide. Even sulfide is accepted
although at a very low efficiency. So, the necessity for
selenophosphate as a substrate may reside in one or
more of the following three aspects, i.e., (1) to
discriminate sulfide from selenide, (2) to efficiently use
low concentrations of selenium compounds, and (3) to
accelerate the reaction rate effected by the activation of
the trace element. Indeed, selenophosphate synthetase
efficiently discriminates between sulfide and selenide,
and thus excludes sulfur from intrusion into the
selenium pathway. Selenophosphate synthetase from
E. coli is a monomeric enzyme with a unique reaction
mechanism since formally it transfers the
g
-phosphate of
ATP to selenide with the intermediate formation of
enzyme-bound ADP which is subsequently hydrolysed
into AMP and inorganic phosphate.
FORMATION OF THE SelB 3
GTP 3 S
ELENOCYSTEYL-TRNA 3
SECIS C
OMPLEX
Elongation factor Tu, which forms a complex with all 20
standard aminoacyl-tRNAs and donates them to the
ribosomal A-site, displays only minimal binding affinity
FIGURE 1 Path of selenocysteine biosynthesis. For explanation see text.
18 SELENOPROTEIN SYNTHESIS
for selenocysteyl-tRNA
Sec
. Consequently, insertion of
selenocysteine requires the function of an alternate
elongation factor which is SelB. SelB from E. coli is a
70 kDa protein which contains the sequence elements of
elongation factor Tu in the N-terminal two-third of the
molecule (designated domains I, II, and III) plus a
domain IV of about 25 kDa which can be subdivided
into domains IVa and IVb. Domains I, II and III share
their functions with those of elongation factor Tu,
namely the binding of guanosine nucleotides and of
charged tRNA. An important difference, however, is that
they can discriminate between the serylated and the
selenocysteylated forms of tRNA
Sec
. In this way, the
insertion of serine instead of selenocysteine, which
would lead to an inactive enzyme, is prevented. The
structural basis for this discrimination ability has not yet
been resolved. A second difference is that the overall
affinity for GTP is about 10-fold higher than that for
GDP which obviates the need for the function of a
guanosine nucleotide release factor since GDP is
chemically replaced by GTP. In accordance, the structure
of SelB lacks those subdomains which in elongation
factor Tu are responsible for interaction with the GDP
release factor EF-Ts. The 25 kDa C-terminal extension
of SelB (domain IV) is required for the function in
selenoprotein synthesis since its truncation inactivates
the molecule. The reason is that subdomain IVb binds to
a secondary structure of the mRNA (the SECIS element)
coding for selenoprotein synthesis. SelB, thus, is able to
form a quaternary complex with GTP and two RNA
ligands, namely selenocysteyl-tRNA and the SECIS
element (Figure 3). The isolated domains IV or IVb
retain the binding capacity for the SECIS element.
Formation of the quaternary complex follows random
order kinetics. An important feature also is that the
stability of the complex is increased when both RNA
ligands are bound.
The SECIS element itself is a hairpin structure formed
within a section of 39 bases of the selenoprotein mRNA
which follows the codon specifying selenocysteine
insertion at the 3
0
-side. Binding of SelB takes place to
its apical stem loop minihelix of 17 nucleotides. Genetic
and structural analysis showed that bases in the loop
region plus a bulged-out U in the helix are required for
the interaction with SelB. This apical part of the SECIS
element is separated by a short unpaired region from a
helix at the base of the hairpin. Pairing within this
second helix is not essential but it increases the efficiency
of selenocysteine insertion. An absolute requirement,
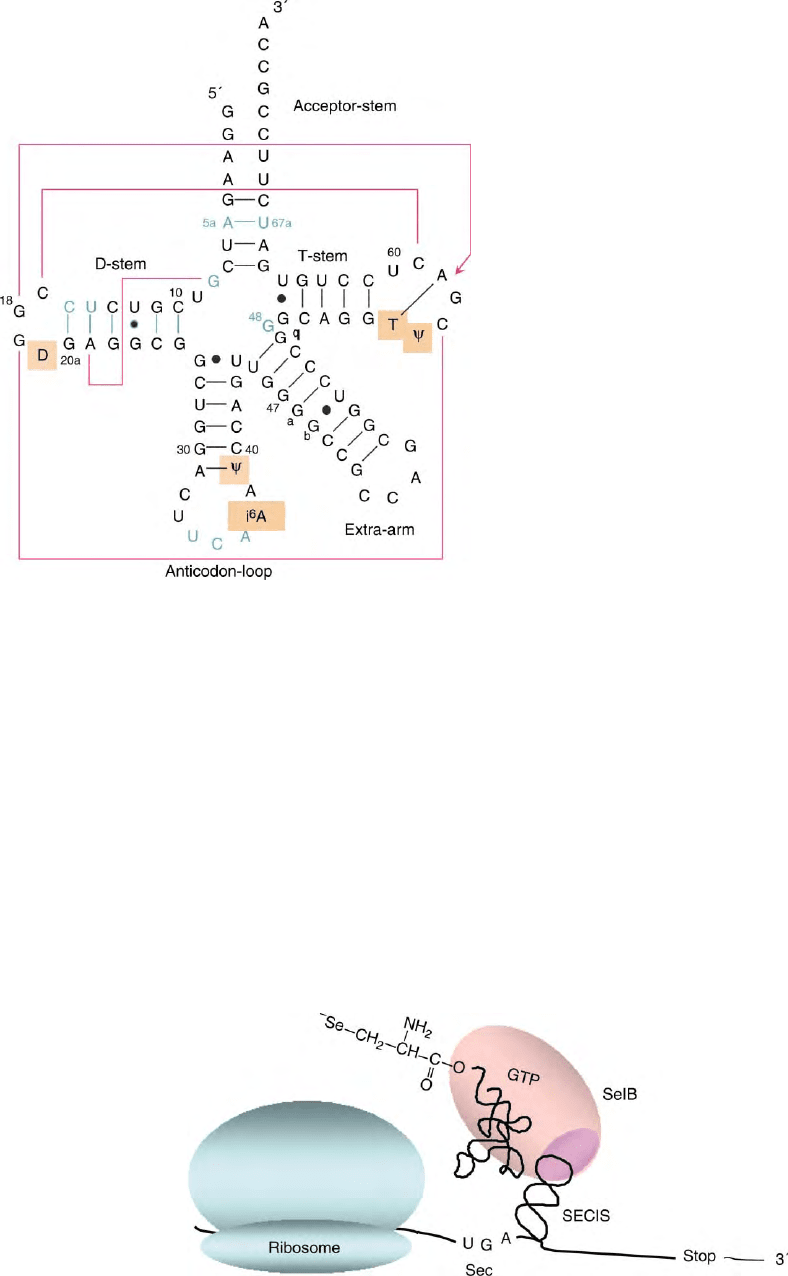
FIGURE 2 Cloverleaf presentation of the structure of tRNA
Sec
from
E. coli. Modified bases are shaded. Tertiary interactions via base
pairing are indicated by connecting red lines, and those involving
intercalation are denoted by arrows. Bases and base pairings deviating
from the consensus are indicated in green.
FIGURE 3 Translation of prokaryotic selenoprotein mRNA. Note that the SECIS element is within the mRNA reading frame and is complexed to
domain IVb of SelB carrying selenocysteyl-tRNA
Sec
and GTP.
SELENOPROTEIN SYNTHESIS 19
however, is that the codon determining selenocysteine
insertion lies within a critical distance relative to the
binding site of SelB.
Bacterial SECIS elements lie within the reading frame
of their selenoprotein mRNAs; they are thus subject to
stringent sequence constraints in order to deliver a
functional gene product. However, they do not need to
be translated since they also function when placed in the
3
0
-untranslated region at the correct distance to the
selenocysteine codon within an upstream reading frame.
The sequence constraint (which depends on the protein
to be formed) and the requirement for binding of SelB
restricts the number of selenoprotein mRNAs to be
expressed in a single organism and explains why the vast
majority of selenoprotein genes cannot be heterolo-
gously expressed unless the cognate SelB gene is
coexpressed. Thus, SelB and their SECIS elements are
subject to coevolution.
DECODING EVENT AT THE RIBOSOME
In all biological systems analyzed thus far, selenocysteine
insertion is directed by the opal stop codon UGA but
only if it is followed by an SECIS element at the correct
distance. This violates the dogma that no codon can
have more than one meaning within a single cell. The
questions to be answered therefore are: (1) what
prevents the UGA to be used as a termination signal,
and (2) which mechanism interferes with insertion of
selenocysteine at ordinary UGA stop codons?
Counteraction of Stop at the UGA?
A convincing answer to the question why the
selenocysteine-specific UGA codon does not function
as an efficient termination signal must await structural
information on the decoding complex. It is, however,
clear that termination always competes with seleno-
cysteine insertion, especially under conditions when
the capacity for decoding the UGA with selenocysteine
is a limiting factor. This can be, for example, a surplus
of selenoprotein mRNA in relation to the amount of
SelB quaternary complex which forces the ribosome
to stall at the UGA. One fact identified to be involved
in the suppression of termination is that the base
following the UGA at the 3
0
-side in selenoprotein
mRNAs is prodominately an A or C, which renders the
UGA a weak termination signal. Also, the two amino
acids preceding selenocysteine in the nascent polypep-
tide chain are predominantly hydrophobic which
counteracts the dissociation of the nascent polypeptide
from the ribosome, when translation pauses at a
“hungry” codon present in the A site. Additional
mechanisms, however, must exist which contribute to
the suppression of termination.
Selenocysteine Specificity of UGA Codons
From the colinearity between the mRNA nucleotide
sequence and the amino acid sequence of the translation
product, it is clear that UGA determines the position
where selenocysteine is to be inserted during translation.
The specificity of the UGA, however, is determined by
the codon context, i.e., by the existence of a SECIS
element at the 3
0
side. The results of extensive
biochemical and biophysical analysis suggest the follow-
ing scenario for the decoding process: (1) SelB forms the
quaternary complex at the mRNA in which the two
RNA ligands display cooperativity in their interaction
with the protein; (2) in this quaternary complex SelB
attains a conformation compatible for interaction with
the ribosome which then results in stimulation of GTP
hydrolysis by SelB which in turn causes the release of the
charged tRNA in the vicinity of the ribosomal A-site; (3)
loss of the tRNA ligand causes the SelB protein to return
to a conformation with about tenfold lower affinity for
the SECIS element. As a consequence, the mRNA is
released from the protein and freed for the translation of
codons downstream of the UGA. The consequence of the
complex cascade of reactions is that the efficiency of the
decoding of UGA with selenocysteine is lower than that
of any of the standard sense codons. It is also reflected by
a considerable pause taking place when the ribosome
encounters the quaternary complex at the mRNA. In the
absence of selenocysteyl-tRNA, binding of SelB alone to
the mRNA does not retard the rate of translation.
Archaeal and Eukaryal
Selenoprotein Synthesis
tRNA
Sec
species from archaea and eukarya share several
structural similarities with the bacterial counterparts but
they are more related to each other than either one is to
bacterial tRNA
Sec
. There is also considerable sequence
similarity between selenophosphate synthetases from all
three lines of descent rendering their annotation in
genome projects easy. On the other hand, homologues
for the bacterial selenocysteine synthase have not been
identified yet in any of the genomic sequences from
organisms known to synthesize selenoproteins.
Whereas UGA directs selenocysteine insertion also in
archaea and eukarya, a fundamental difference is that
the SECIS element is not positioned within the reading
frame but in the 3
0
-nontranslated region of the mRNA.
SECIS elements from organisms of the three lines of
descent are different by sequence and by secondary
structure. They may be positioned at different distances
from the actual termination codon and/or the seleno-
cysteine inserting UGA codon but a critical distance
must not be underpassed. It is thought that the selective
value for having the SECIS element in the nontranslated
20
SELENOPROTEIN SYNTHESIS
