Lennarz W.J., Lane M.D. (eds.) Encyclopedia of Biological Chemistry. Four-Volume Set . V. 4
Подождите немного. Документ загружается.

Structure and Function
of the Initiation Factors
Overall, the three initiation factors ensure speed and
accuracy in the initiation phase of protein synthesis.
The properties and specific roles of these proteins are
outlined below.
IF1
Structure
This protein (encoded by infA) consists of , 70 amino
acids (71 in Escherichia coli) and is characterized by a
rigid five-stranded
b
-barrel structure from which pro-
trude the short disordered and highly flexible N- and
C-terminal tails (Figure 3A). This structural motif,
known as the “OB fold,” is characteristic of a class of
proteins that interact with oligonucleotides and RNA
(like IF1) or with oligosaccharides.
Topographical Localization and Function
IF1 binds to the 30S ribosomal subunit mainly through
electrostatic interactions involving the positively
charged surface of the protein and the phosphate
backbone of specific regions of 16S rRNA. Earlier
topographical studies and more recent chemical probing
and crystallographic data indicate that IF1 binds in the
A-site of the 30S ribosomal subunit. More precisely, IF1
fits in the cleft between ribosomal protein S12, the 530
loop and helix 44 of 16S ribosomal RNA (rRNA)
establishing contacts with two functionally important
bases (A1492 and A1493) which belong to this rRNA
helix. This ribosomal localization could cause IF1 to
block the premature access of aminoacyl-tRNA to the
A-site during 30S initiation complex formation and
supports the premise that IF1 contributes to the fidelity
of translation initiation. The existence of a mutual
influence of IF1 and IF2 on their respective interactions
with the 30S ribosomal subunit is well established so
that IF1 is regarded as being a modulator of IF2
recycling on and off the ribosomes. IF1 also increases
the rates of association/dissociation of ribosomal sub-
units and this activity is probably a decisive factor in
favoring the otherwise inefficient ribosome dissociation
activity of IF3. Finally, IF1 can also increase the rate
of 30S initiation complex formation, probably through
a conformational change of the small ribosomal
subunit. Indeed, there is compelling evidence from the
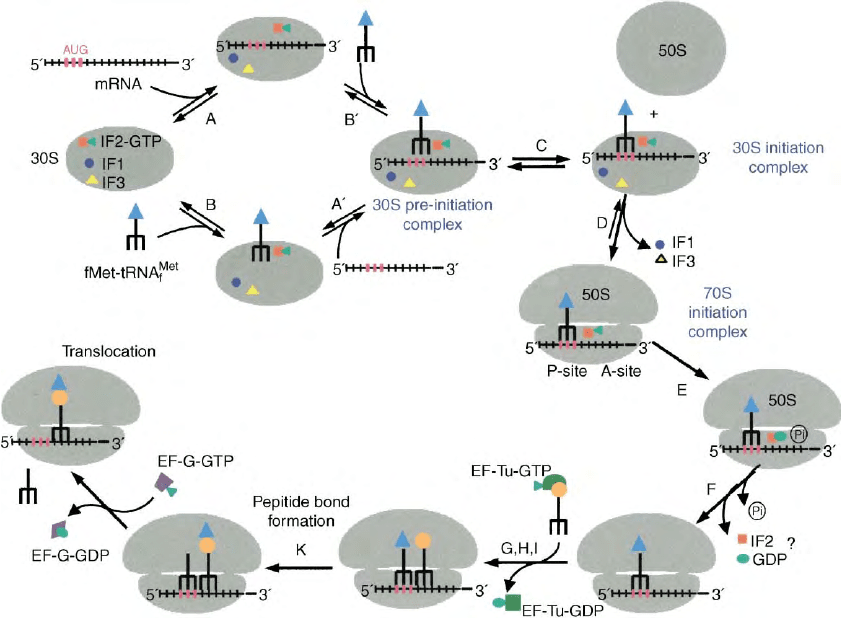
FIGURE 2 Initiation pathway. Scheme depicting the translation initiation pathway in bacteria as determined over the past two decades, mainly
through traditional and fast kinetics analyses. Further details can be found in the text. The symbols and the denominations of the intermediate
complexes are as indicated in the figure.
232 TRANSLATION INITIATION IN BACTERIA: FACTORS AND MECHANISMS
crystallographic data that IF1 binding induces several
localized and long-range changes of the 30S structure.
Overall, IF1 induces a rotation of head, platform and
shoulder of the 30S subunit towards the A-site.
IF2
Structure
Bacterial IF2 (encoded by infB) is the largest initiation
factor (890 residues in E. coli) and consists of three
major parts: (i) a variable N-terminal region, (ii) a highly
conserved 40 kDa region containing two domains, GI
and GII, and (iii) the C-terminal region (25 kDa) which
also consists of two domains, C1 and C2.
So far no three-dimensional (3D) structure has been
directly determined for the whole bacterial IF2 mol-
ecule, and structural information concerning this
factor relies on the NMR structure of the C2 domain
of Bacillus stearothermophilus IF2 and on the crystal
structure of Methanobacterium thermoautotrophicum
eIF5B (Figure 3B), the archaeal homologue of IF2.
However, although it is likely that the structures of the
archaeal and bacterial factors are similar, it should be
noted that the functions of these two proteins have very
little in common. Overall, IF2 is an extended molecule
containing four domains arranged in a unique architec-
tural motif resembling a chalice whose cup is constituted
by GI, GII, and N-terminal part of C1, the stem by the
C-terminal part of the same domain and the foot by the
C2 domain. No structural information is available for
the hydrophilic, positively charged and likely flexible
N-domain which is not present in the archaeal protein
as well as in some bacterial IF2 molecules. The likely
function of this domain, which is dispensable for
translation both in vitro and in vivo, is that of anchoring
the factor on the 30S ribosomal subunit in a more or less
specific way until the acceptor end of an fMet-tRNA
molecule is “captured” by the C2 domain of the factor.
The GI-domain binds GTP/GDP and is highly homolo-
gous to small GTPases and to equivalent domains of
other GTP/GDP binding proteins such as elongation
factors EF-Tu and EF-G. The GI domain is also
responsible for the interaction of IF2 with the 50S
ribosomal subunit and probably contains the GTPase
center of the protein which is naturally activated by the
interaction with the ribosomes.
The structure of this domain consists of an eight-
stranded
b
-sheet flanked by six
a
-helices and a 3
10
helix.
As mentioned above, this domain contains the four
conserved sequence elements characteristic of GTP-
binding proteins (G1/P loop, which participates in
phosphate binding, the G3 and G4 loops, forming the
walls of a hydrophobic pocket where the guanine moiety
of GTP or GDP is bound and G4). The Switch 2 region
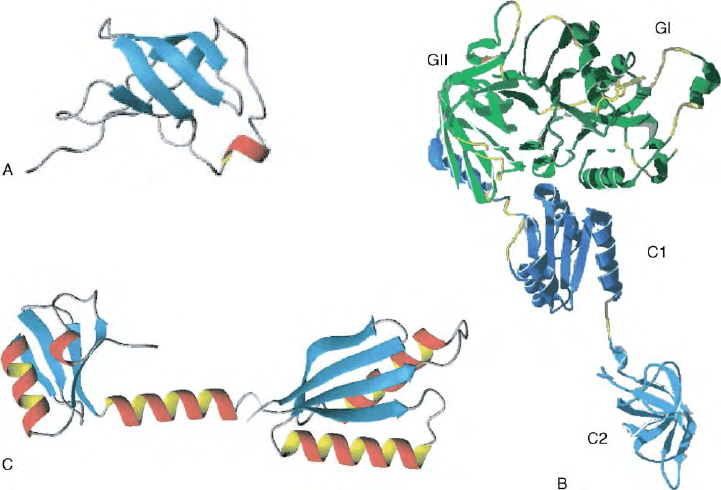
FIGURE 3 Three-dimensional structures of the three translation initiation factors. (A)
b
-Barrel structure of E. coli IF1 in solution as determined
by NMR spectroscopy, (B) predicted structure of bacterial IF2 based on the crystal structure of archaeal eIF5b and on the solution structure of
Bacillus stearothermophilus IF2C2 determined by NMR spectroscopy. The N-terminal domain is missing while the other domains are indicated
with the following color patterns. Dark green, GI, light green GII, dark blue, C1, light blue, C2. (C) Crystallographic structure of
B. stearothermophilus IF3 displaying the N domain (left), linker (center), and the multi-functional C-domain (right).
TRANSLATION INITIATION IN BACTERIA: FACTORS AND MECHANISMS 233
of the GI domain is centrally located in the cup of the
chalice where it makes extensive contacts to domains
GII and C1 and can therefore cause conformational
changes resulting in profound global rearrangements
of the molecule that can reach as far as 90A
˚
to the
C-terminal region of IF2. The GII domain, which
together with C1 is responsible for the interaction of
IF2 with the 30S ribosomal subunit, has a
b
-barrel
structure very similar to that of the C2 domain. Three
b
-strands of this domain interact closely with the
GI-domain in the vicinity of the Switch 2 region. The
GII domain is connected by a 17 residue
a
-helix to
the C1-domain which is characterized by a unique
a
–
b
–
a
sandwich fold consisting of a four-stranded
parallel
b
-sheet flanked on both sides by two
a
-helices.
As mentioned above, the last 40A
˚
long helix (H12) of the
C1 domain extends from the cup to the C2 domain thus
forming the “stem” of the chalice. The C2 domain of
IF2, which is responsible for the specific recognition and
binding of the acceptor end of fMet-tRNA is endowed
with a structure which also consists of an eight-stranded
b
-barrel fold similar to that of GII and to domain II
of EF-Tu and EF-G.
Topographical Localization and Function
IF2 is the only one of the three factors displaying a
specific and fairly high affinity for both ribosomal
subunits, and its interaction with the isolated 50S
subunit is sufficient to elicit its GTPase activity. Recent
experiments localize IF2 in a region of the 30S subunit
topographically adjacent to the A-site, on a surface of
the subunit’s body facing the factor-binding region of the
50S subunit. With respect to the 50S subunit, IF2 was
found to influence the chemical reactivity and/or the
accessibility to nucleolytic cleavage of bases belonging
to helix 89, to the sarcin-ricin domain (SRD) and to the
L11/Thiostrepton-binding region of 23S rRNA leading
to the conclusion that its topographical localization is on
the right edge of the subunit interface site of the particle
and at least partly overlaps that of elongation factors
EF-G and EF-Tu.
The main function of IF2 is that of recognizing and
binding (K
d
in the mM range) the initiator fMet-tRNA
and to stimulate (through an increase of the on-rate) its
binding to the ribosomal P-site. Both specificity and
thermodynamic stability of the IF2-fMet-tRNA inter-
action are properties of the C2 domain (, 11 kDa).
Additional IF2 functions include the stimulation of
subunit association and the positioning of fMet-tRNA in
the ribosomal P-site of the 70S initiation complex which
favors the first transpeptidation. Furthermore, IF2 is a
GTP/GDP-binding protein and a ribosome-dependent
GTPase like EF-Tu and EF-G but, unlike these
elongation factors, the function of the IF2-dependent
GTP binding and hydrolysis is difficult to pin down.
Thus, since neither GTP/GDP-binding nor GTPase
activity seems to be mandatory for any translational
function of IF2 and since the “metabolic alarmone”
ppGpp can bind in place of GTP and inhibit the IF2-
dependent 30S initiation complex formation and
initiation dipeptide synthesis, it has been postulated
that IF2 uses its GDP/GTP-binding site as a receptor for
GTP (under optimal growth conditions) or for ppGpp
(during nutritional stress) and accordingly behaves like a
sensor of the metabolic state of the cell. This raises the
interesting possibility that, in addition to and because of
its roles in translation initiation, IF2 might function as a
global regulator linking translational activity to the
transcriptional control of stable RNA synthesis by
adjusting the translational rate of the cell as a function
of the allowable growth rate.
IF3
Structure
The structure of this medium-sized protein encoded by
infC (180 amino acids in E. coli) is characterized by the
presence of two domains of approximately equal mass
connected by a long (, 45A
˚
) lysine-rich linker
(Figure 3C). Whereas considerable controversy exists
between crystallographic and NMR data as to whether
this linker is a long and rigid
a
-helix or unstructured and
flexible, the 3D structures of both N-terminal (IF3N)
and C-terminal (IF3C) domains seem to be well
established. IF3N contains a globular
a
=
b
fold consisting
of a single
a
-helix, packed against a mixed four-stranded
b
-sheet. IF3C possesses a two-layered
a
=
b
sandwich
fold, comprising a four-stranded
b
-sheet which is packed
against two parallel
a
-helices in a
bababb
topology.
The fold of IF3C is similar to that found in many
eukaryotic RNA-binding proteins (such as U1A) and
indeed IF3C interacts with the 30S subunit via a
protein–RNA interaction involving primarily structural
elements of this domain like strands
b
-7 and
b
-9 that
contain consensus RNP motifs and two loops (L7 and
L8). Regardless of the actual structure of the linker,
several lines of evidence indicate that the two domains
of IF3 do not interact with one another in the free or in
the ribosome-bound protein and that they interact,
independently of each other, with different sites of the
30S subunit.
Topographical Localization and Function
The main interaction of IF3 with the 30S subunit occurs
via IF3C, the domain which encompasses all IF3
activities. Whereas there is good agreement that IF3C
is localized on the platform of the 30S ribosomal
subunit, the ribosomal localization of IF3N is more
controversial, although it seems likely that its binding
site is located somewhere on the head of the particle.
234
TRANSLATION INITIATION IN BACTERIA: FACTORS AND MECHANISMS
Binding of IF3 to the 30S ribosomal subunit interferes
with subunit association thereby shifting to the left of
the equilibrium 30S þ 50S O 70S. In turn, this increases
the pool of free 30S which are amenable to initiate a new
round of translation. Furthermore, the presence of IF3
on the 30S increases the on-rate of “pre-ternary
complex” isomerization which leads to 30S initiation
complex formation (Figure 2) and thereby stimulates
overall mRNA translation. However, since IF3 increases
also the off-rate of the isomerization, in this context it
can promote initiation fidelity favoring the dissociation
from the 30S subunits of both tRNA and template.
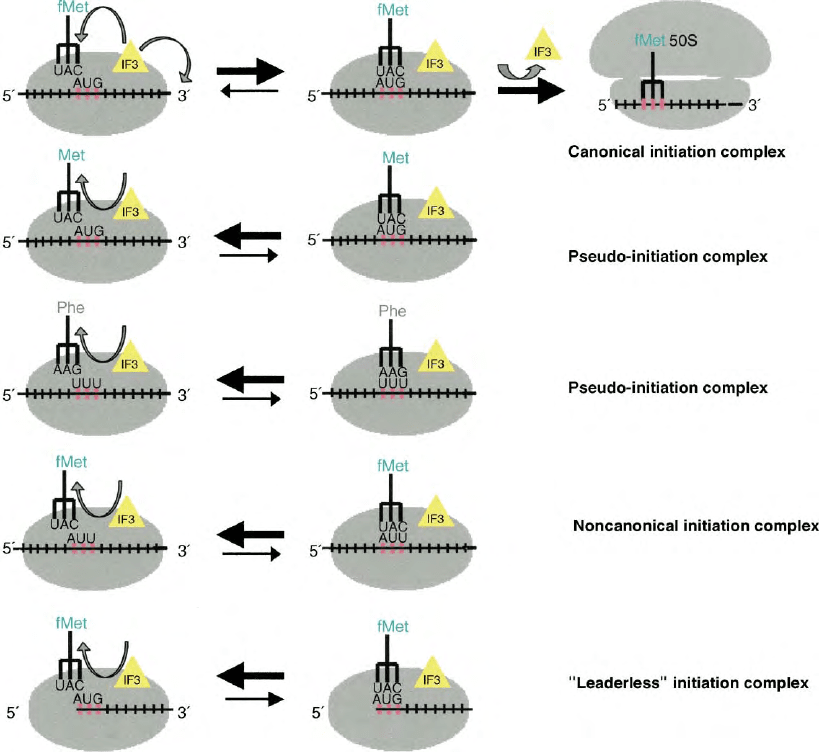
Indeed, the dissociation occurs with different rates
depending on whether the 30S complex is a “canonical,”
a “noncanonical,” a “leaderless,” or a “pseudo”
initiation complex and is in kinetic competition with
the association of the same 30S initiation complex with
the 50S ribosomal subunit which yields a “70S initiation
complex” (Figure 2). The potential 30S initiation
complexes recognized and accepted as “correct” by
IF3 and those kinetically discriminated against as being
“incorrect” are schematically represented in Figure 4.
Contrary to earlier interpretations, IF3 is not required
for mRNA binding to the ribosome but can promote a
re-positioning of 30S-bound mRNA shifting it from the
“stand by site” to the “P-decoding site” of the subunit. It
is likely that this activity may be correlated to the kinetic
selection of the correct initiation triplet by IF3. It is
noteworthy that isolated IF3C not only binds to 30S
subunit but is also capable of performing all the other
known functions of the intact molecule while isolated
IF3N has no autonomous function. Since the affinity of
IF3C for the 30S subunits is approximately two orders
of magnitude lower than that of the intact molecule
and since secondary contacts established by IF3N
stabilize the interaction, it has been suggested that the
two-domain structure is required to modulate binding
and release of IF3 to and from the 30S subunit.
FIGURE 4 Initiation complexes recognized as “correct” and “incorrect” by IF3. The figure presents a scheme of the nature of the various types of
complexes containing 30S ribosomal subunit, template and aminoacyl-tRNAs which are subjected to kinetic positive or negative discrimination by
IF3. Further details may be found in the text.
TRANSLATION INITIATION IN BACTERIA: FACTORS AND MECHANISMS 235
The conformational change of the ribosome induced by
subunit association would push platform and head of
the 30S subunit away from each other thereby
widening the gap between the IF3N- and the IF3C-
binding sites. The loss of the stabilizing interaction
established by IF3N would then facilitate the dis-
sociation of IF3 from the 30S subunit.
SEE ALSO THE FOLLOWING ARTICLES
EF-G and EF-Tu Structures and Translation Elongation
in Bacteria † Ribosome Assembly † Ribosome
Structure † Translation Elongation in Bacteria †
Translation Termination and Ribosome Recycling
GLOSSARY
A-site Aminoacyl-tRNA site, the binding site on the ribosome
occupied by the tRNA carrying the next amino acid to be added
to a growing polypeptide chain.
P-site Peptidyl-tRNA site, the binding site on a ribosome occupied by
the tRNA carrying a growing peptide chain.
Watson–Crick base-pairing Association of two complementary
nucleotides in a DNA or RNA molecule stabilized by hydrogen
bonding between their base components.
wobbling interaction Ability of a tRNA to recognize more than one
codon by unusual (non-G-C, non-A-U) pairing with the third base
of a codon.
FURTHER READING
Boelens, R., and Gualerzi, C. O. (2002). Structure and function of
bacterial initiation factors. Curr. Protein Pep. Sci. 3, 107– 119.
Gualerzi, C. O., Brandi, L., Caserta, E., Garofalo, C., Lammi, M.,
La Teana, A., Petrelli, D., Spurio, R., Tomsic, J., and Pon, C. L.
(2001). Initiation factors in the early events of mRNA trans-
lation in bacteria. Cold Spring Harbor Symp. Quant. Biol. 66,
363–376.
Ramakrishnan, V. (2002). Ribosome structure and the mechanism of
translation. Cell 108, 557–572.
BIOGRAPHY
Cynthia L. Pon is a Professor of Molecular Genetics at the University of
Camerino, Italy. She holds a Ph.D. from Rutgers University and her
current research interest concerns the regulation of gene expression
during cold stress.
Claudio O. Gualerzi is a Professor of Molecular Biology in the
Department of Biology at the University of Camerino, Italy. He holds a
laurea degree from the University of Rome and his current research is
directed toward the search for new antibiotics.
236 TRANSLATION INITIATION IN BACTERIA: FACTORS AND MECHANISMS

Translation Initiation in Eukaryotes:
Factors and Mechanisms
Tatyana V. Pestova
Moscow State University, Moscow, Russia
Christopher U.T. Hellen
State University of New York Downstate Medical Center, Brooklyn, New York, USA
Translation initiation, the first stage in protein synthesis, is the
process of assembly of large (60S) and small (40S) ribosomal
subunits to form an 80S ribosome containing initiator tRNA
(Met-tRNA
Met
i
) that is base paired to the initiation codon of a
mRNA in the ribosomal peptidyl (P) site. This process is
mediated by at least 11 eukaryotic initiation factors (eIFs) and
proceeds via the sequential formation of intermediate com-
plexes. Initiation is the rate-limiting step of translation, and is a
major focus for pathways that regulate gene expression.
The Structure of
Eukaryotic mRNAs
Nearly all eukaryotic mRNAs have a 5
0
-terminal
7-methylguanosine (m
7
G) “cap” and a 3
0
-terminal
poly(A) tail that synergistically enhance the efficiency
of translation initiation. Most eukaryotic mRNAs
contain a single major open reading frame (ORF) that
is translated into protein, and initiation usually begins at
the first AUG triplet from the 5
0
-end of a mRNA, which
follows a 5
0
leader that is , 100 nucleotide long. An AUG
triplet can be bypassed if its context deviates from
the optimal sequence GCC(A/G)CC
AUGG (in which the
initiation codon is underlined and the nucleotides in bold
have the greatest influence), if it occurs very close to the
5
0
-end of an mRNA or if the 5
0
leader has little secondary
structure. Stable structures in the 5
0
leader reduce
initiation efficiency whereas stable structures down-
stream of an AUG codon can enhance initiation at it.
The Mechanism of
Translation Initiation
Translation initiation on most eukaryotic mRNAs
begins with binding of Met-tRNA
Met
i
to a 40S subunit,
followed by ribosomal attachment at the 5
0
-end of a
mRNA, scanning to the initiation codon and joining
with a 60S subunit to form an 80S ribosome. Initiation is
mediated by at least 11 eIFs (Table I), many of which act
at multiple stages in this process (Figure 1). Initiation on
a few mRNAs occurs by noncanonical mechanisms, of
which the most common is 5
0
-end independent internal
ribosomal entry.
DISSOCIATION OF RIBOSOMES INTO
FREE 40S AND 60S SUBUNITS
Initiator tRNA and mRNA initially bind to the 40S
subunit rather than to the 80S ribosome. However,
association of 40S and 60S subunits to form empty 80S
ribosomes is favored under ionic conditions in the
cytoplasm and a mechanism to maintain a pool of free
subunits is therefore a prerequisite for initiation. eIF1A
and eIF3 shift the equilibrium between ribosomes and
their subunits towards dissociation. eIF3 dissociates 80S
ribosomes and, with eIF1A, prevents subunit reassoci-
ation by binding directly to free 40S subunits.
RECRUITMENT OF INITIATOR TRNA
TO THE 40S RIBOSOMAL SUBUNIT
The initiation codon is decoded by a unique initiator
methionyl-tRNA. Its sequence and structural features
distinguish it from the methionyl-tRNAs that decode
AUG triplets during translation elongation. These
features enable eIF2 to select Met-tRNA
Met
i
from the
cytoplasmic pool of aminoacylated and deacylated
initiator and elongator tRNAs, and also exclude Met-
tRNA
Met
i
from translation elongation. eIF2 is a stable
heterotrimeric protein consisting of
a
-,
b
-and
g
-subunits that binds GTP and Met-tRNA
Met
i
to form
a ternary complex. Binding of the ternary complex to a
40S subunit is strongly stabilized by eIF1A and eIF3 and
yields a 43S preinitiation complex. eIF2
0
s activity in
binding Met-tRNA
Met
i
is regulated; its affinity for
Met-tRNA
Met
i
is enhanced by prior binding of GTP
Encyclopedia of Biological Chemistry, Volume 4. q 2004, Elsevier Inc. All Rights Reserved. 237
whereas hydrolysis of eIF2-bound GTP towards the end
of each initiation cycle yields eIF2
.
GDP which dis-
sociates from Met-tRNA
Met
i
, leaving it in the peptidyl (P)
site of the 40S subunit. Biochemical and genetic data
suggest that eIF2
0
s
g
-subunit binds Met-tRNA
Met
i
and
GTP, but despite eIF2
g
having the sequence motifs and
structure characteristic of a conventional GTP-binding
protein, eIF2 does not have an intrinsic GTPase activity.
Hydrolysis of eIF2-bound GTP is induced by eIF5, a
GTPase activating protein specific for eIF2.
ATTACHMENT OF 43S PREINITIATION
COMPLEXES TO MRNA
Binding of initiator tRNA to form a 43S complex is an
obligatory first step before a 40S subunit can bind
mRNA, which for most mRNAs is a 5
0
end-dependent
process. Attachment of 43S complexes is enhanced
synergistically by the 5
0
-terminal cap, the 3
0
poly(A) tail
and the factors that associate with these structures. The
m
7
G “cap” is bound by eIF4E, the cap-binding subunit
of the heterotrimeric eIF4F, which also contains eIF4A
and eIF4G subunits. eIF4A is an ATP-dependent RNA
helicase that cycles in and out of the eIF4F complex.
eIF4G is a large polypeptide that binds mRNA
and eIF4E, eIF4A, and eIF3 and the cytoplasmic
poly(A)-binding protein PABP, thereby coordinating
and in some instances enhancing their activities. eIF4B
and the less abundant eIF4H enhance the helicase
activities of eIF4A and of eIF4F; eIF4B enhances but is
not essential for ribosomal attachment to mRNA. The
3
0
poly(A) tail’s influence is dependent on PABP which
binds to it, and synergism between the cap and the
3
0
poly(A) tail depends on the bridging interaction of
eIF4G with the m
7
G cap/eIF4E and 3
0
poly(A)
PABP
complexes.
The eIF4F complex is required on most mRNAs to
unwind the cap-proximal region to prepare it for
attachment of 43S complexes. Recent studies have
shown that 43S complexes can bind directly to a
mRNA in the absence of eIF4F if the 5
0
-terminal region
is completely unstructured. Ribosomal attachment likely
involves protein–protein interactions between the eIF3
component of 43S complexes and both eIF4G and eIF4B
as well as direct binding of eIF3, eIF4G, and the 40S
subunit to the mRNA. ATP-dependent restructuring of
mRNA and ribosomal attachment to the unwound
region of the 5
0
leader are probably coordinated by
interaction of the eIF4G component of the
eIF4F/PABP/mRNA complex with eIF3.
TABLE I
Mammalian Initiation Factors
Factor Subunits Mass (kDa)
a
Functions
eIF1 13 Promotes scanning and the fidelity of initiation codon recognition
eIF1A 17 Ribosome antiassociation; stabilizes Met-tRNA
i
binding to
40S subunit; promotes scanning
eIF2
a
,
b
,
g
36, 39, 52 Binds GTP and Met-tRNA
i
to 40S subunit; GTPase
eIF2B
a
,
b
,
g
,
d
, 1 34, 39, 50, 58, 80 Guanine-nucleotide exchange factor for eIF2
eIF3 a–l 167, 105, 99, 64, 52, 38,
35, 40, 37, 29, 25, 67
Ribosome dissociation and ribosome subunit antiassociation;
stabilizes binding of the eIF2/GTP/Met-tRNA
i
complex to
40S subunit; required for ribosomal binding to mRNA and
scanning on the 5
0
leader
eIF4A 44 ATP-dependent RNA helicase/RNA-dependent ATPase
eIF4B 69 mRNA-binding cofactor for eIF4A
eIF4E 25 m
7
G “cap”-binding protein
eIF4F, eIF4E,
eIF4A, eIF4G
25, 44, 176 Cap-binding complex comprising eIFs 4A, 4E, and 4G
eIF4G 176 Binds and coordinates the functions of mRNA, PABP, and
eIFs 3, 4A, 4E
eIF4H 25 mRNA-binding cofactor for eIF4A
eIF5 49 GTPase-activating protein specific for eIF2
eIF5A 17 May enhance first cycle of translation elongation
eIF5B 139 GTPase; ribosome subunit joining
PABP 70 Binds the 3
0
poly(A) tail and promotes ribosomal binding
to mRNA
a
Masses (kDa) correspond to those of human proteins, and where appropriate, to the largest isoform.
238 TRANSLATION INITIATION IN EUKARYOTES: FACTORS AND MECHANISMS
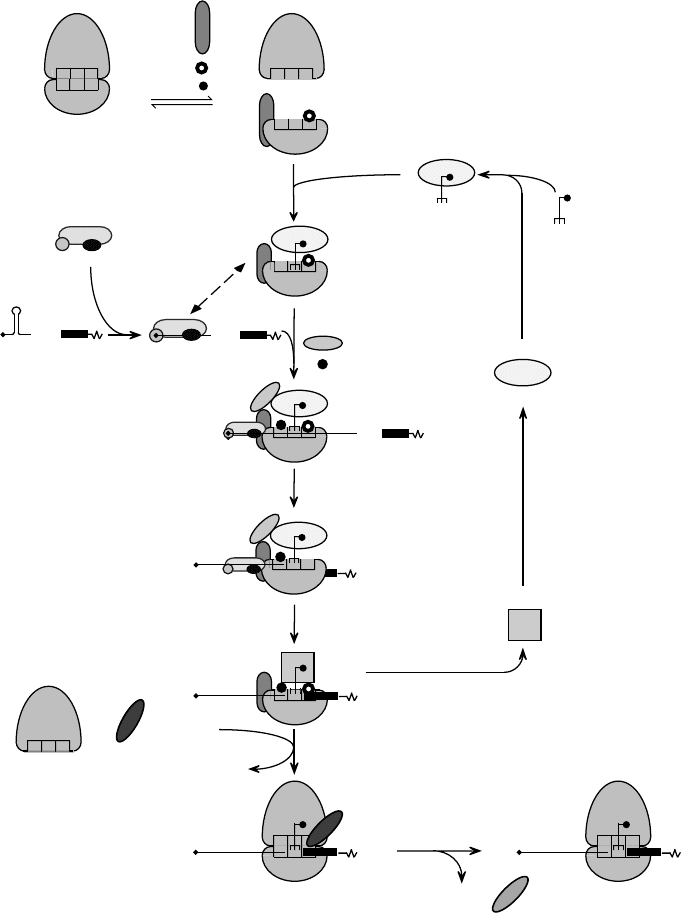
(5) Scanning
60S
Ribosome
dissociation
(1)
eIF3
+
eIF1A
(+ eIF1?)
80S
(9)
Recycling
of eIF2
eIF2B
eIF2 ternary
complex formation
(2)
40S
EAP
43S complex
formation
(3)
43S
EAP
GTP
eIF3
GTP
EAP
AUG
5
(4) Attachment
AUG
m
7
G
ATP
4E 4A
4G
eIF4F
AUG
4E 4A
4G
5
eIF1
Met-tRNA
i
Met
eIF2-GTP
eIF2-GDP
(6)
eIF5
Hydrolyis of
eIF2-bound GTP
48S
GDP
EAP
AUG
eIF5B-GDP
(8)
Displacement
of eIF5B
60S
eIF5B-GTP
eIF3, 1, 1A ?
(7) Subunit joining
+
EAP
AUG
80S
EAP
AUG
GTP
GDP
GTP
eIF2-GTP/
Met-tRNA
i
Met
GTP
E
5
AUG
AP
FIGURE 1 Schematic model of the pathway of 80S initiation complex formation on a model capped eukaryotic mRNA. eIF1A and eIF3 promote
dissociation of the 80S ribosome into 40S and 60S subunits (Step 1). eIF2 binds aminoacylated initiator tRNA (Met-tRNA
Met
i
) and GTP to form a
ternary complex (Step 2). Binding of the ternary complex to a 40S subunit is stabilized by eIF1, eIF1A, and eIF3 to form a 43S complex (Step 3). The
eIF4F complex binds to the 5
0
-terminal m
7
G “cap” of an mRNA and with associated cofactors creates an unstructured cap-proximal site to which
the 43S complex binds (Step 4). This step may be enhanced by the poly(A)-binding protein associated with the mRNA’s 3
0
poly(A) tail (not shown).
The dashed, double-headed arrow indicates interactions that promote attachment of the 43S complex to mRNA, such as those of the eIF3
component of the 43S complex with the eIF4G subunit of cap-bound eIF4F and probably with the mRNA. The 43S complex scans
downstream on the 5
0
-leader until it reaches the initiation codon (Step 5), which is base paired to the anticodon of Met-tRNA
Met
i
in the resulting 48S
complex. Hydrolysis of eIF2-bound GTP is triggered by eIF5, and probably releases of eIF2-GDP (Step 6). The stages at which eIF5 joins and eIF1,
eIF1A, and eIF3 are released from the 40S subunit are not known. The GTP-bound form of eIF5B mediates joining of a 60S subunit to the
resulting complex (Step 7). eIF5B-GDP is released after hydrolysis of eIF5B-bound GTP, which is induced by both ribosomal subunits. The resulting
80S ribosome is then able to begin protein synthesis (Step 8). eIF2B recycles inactive eIF2-GDP to active eIF2-GTP, which can then bind
Met-tRNA
Met
i
again (Step 9).
TRANSLATION INITIATION IN EUKARYOTES: FACTORS AND MECHANISMS 239
RIBOSOMAL SCANNING
ON THE
5
0
-LEADER
Following attachment to the 5
0
-terminal region of a
mRNA, a 43S complex scans downstream until it locates
the initiation codon, which is usually the first AUG
triplet from the 5
0
-end. Scanning consists of ribosomal
movement on the 5
0
-leader and inspection of it to
identify the initiation codon. Recent studies have shown
that ribosomal complexes containing only eIF1, eIF2,
and eIF3 are intrinsically capable of ATP-independent
movement on a 5
0
-leader if it is unstructured; eIFs 1, 1A,
4A, 4B, and 4F all contribute to the processivity of
scanning. eIFs 4A, 4B and 4F, and ATP are involved in
restructuring mRNA to permit scanning and are
essential for scanning if the 5
0
-leader contains even
weak-secondary structure. Many details of the mechan-
ism of scanning remain poorly understood, including the
identity of the set of factors associated with the scanning
complex, the mechanism by which helicase-mediated
unwinding of structured 5
0
-leaders is coupled to
ribosomal movement, the stage at which eIF4F
dissociates from the cap and from the 43S complex
and the mechanism of action of eIF1 and eIF1A.
SELECTION OF THE INITIATION CODON
The scanning 43S complex stops when it encounters an
AUG triplet that it recognizes as an initiation codon.
This is primarily determined by base pairing between it
and the anticodon of initiator tRNA, but in higher
eukaryotes the sequence flanking an AUG triplet also
influences its selection. Genetic analyses in yeast of
mutants that permit initiation at an UUG triplet have
indicated that eIF1, eIF2, and eIF5 all influence start site
selection. Recent studies have shown that eIF1 plays the
principal role in maintaining the fidelity of initiation
codon selection, for example, by destabilizing com-
plexes aberrantly arrested at non-AUG triplets or
assembled on AUG triplets that have a poor context.
DISPLACEMENT OF EIF2
FROM THE 48S COMPLEX
Establishment of base pairing between the initiation
codon and the anticodon of Met-tRNA
Met
i
stimulates
eIF5-induced hydrolysis of eIF2-bound GTP in
ribosomal complexes. eIF2
.
GDP does not bind Met-
tRNA
Met
i
or the 40S subunit and is released from the 48S
complex, leaving Met-tRNA
Met
i
base paired to the
initiation codon. eIF5-induced hydrolysis of eIF2-
bound GTP is involved in conversion of the scanning
ribosomal complex to a complex that is arrested at the
initiation codon. Mutations in eIF5 or eIF2 that alter
the rate of this reaction therefore influence selection of
the initiation codon. eIF2 has a much higher affinity for
GDP than GTP, and GDP has a slow off-rate from eIF2,
so eIF2B, a guanine nucleotide exchange factor specific
for eIF2 is required to regenerate active eIF2
.
GTP from
inactive eIF2
.
GDP.
60S SUBUNIT JOINING TO FORM
AN
80S RIBOSOME
The release of eIF2
.
GDP from 48S complexes that is
induced by eIF5 is not sufficient to permit 60S subunits
to bind to the 40S subunit
Met-tRNA
Met
i
factor com-
plex assembled at the initiation codon. Subunit joining
also requires eIF5B, which is a homologue of the
prokaryotic initiation factor 2, and like it has a GTPase
activity that is maximally stimulated by large and small
ribosomal subunits. Subunit joining leads to the release
of all initiation factors from the 40S subunit and leaves
Met-tRNA
Met
i
in the ribosomal P site. The GTP-bound
form of eIF5B is active in promoting subunit joining but
does not dissociate from the ribosome and instead
blocks the ribosomal A site. GTP hydrolysis leads to the
release of eIF5B
.
GDP from the 80S ribosome, so that it is
able to begin polypeptide synthesis. Translation
initiation therefore requires hydrolysis of two GTP
molecules in reactions catalyzed by eIF2
g
/eIF5 and
eIF5B, respectively. The initial round of elongation may
be enhanced by eIF5A.
Regulation of Translation Initiation
The intrinsic efficiency of translation initiation on a
mRNA is determined by properties such as the degree of
secondary structure in the 5
0
-leader and the sequence
context of the initiation codon. Differences in these
properties account for many of the differences in the
relative levels of translation of different mRNAs.
Translation initiation is also regulated either selectively
on a single species or a small subset of mRNAs, or
globally on all mRNAs, in order to integrate protein
synthesis with physiological demands. Translation of a
single species of mRNA or of a related group of mRNAs
can be repressed by binding of a protein to a specific site
on the mRNA in a manner that interferes with initiation.
For example, the iron regulatory protein that binds to
the 5
0
leader of ferritin mRNA sterically prevents it from
binding to the 43S complex. More commonly, trans-
lation is regulated at a more global level by alteration of
the activities of initiation factors, either as a result of
phosphorylation or by binding to regulatory proteins.
Phosphorylation of eIF2
a
causes eIF2 to bind to and
inhibit eIF2B, ultimately reducing formation of the
eIF2/GTP/Met-tRNA
Met
i
complex and thus down-
regulating initiation globally. Initiation on most
mRNAs is cap-mediated and is down-regulated by
240
TRANSLATION INITIATION IN EUKARYOTES: FACTORS AND MECHANISMS
a reduction in the level of active eIF4F, which can occur
either by proteolysis of eIF4G (for example, during some
viral infections) or by disruption of eIF4E’s interaction
with eIF4G by eIF4E-binding proteins that compete with
eIF4G for binding to eIF4E.
Initiation of Translation by Internal
Ribosomal Entry
Translation on most eukaryotic mRNAs is initiated by
end-dependent ribosomal scanning, but initiation on a
few viral mRNAs is end-independent and is instead
mediated by an internal ribosomal entry site (IRES) that
promotes binding of the 40S subunit to an internal site in
the mRNA without scanning from the 5
0
-end. IRESs are
in general large and contain significant secondary
structure. IRESs from a single virus family are similar,
but the size and structure of IRESs from unrelated virus
families differ greatly from each other. Three groups of
IRESs have been characterized in detail; each mediates
initiation by a different mechanism but all involve direct,
noncanonical interactions of the IRES with canonical
components of the translation apparatus. They all have
simpler initiation factor requirements than cap-mediated
initiation, and therefore escape some mechanisms that
regulate that process. IRESs have been identified in
several cellular mRNAs, but little is known of the
mechanisms by which they promote initiation.
SEE ALSO THE FOLLOWING ARTICLES
mRNA Polyadenylation in Eukaryotes † Pre-tRNA and
Pre-rRNA Processing in Eukaryotes † Ribosome
Assembly † Ribosome Structure
GLOSSARY
eukaryotic initiation factor A protein that acts in one or more steps in
the process of translation initiation.
initiator tRNA The anticodon of initiator transfer RNA is comp-
lementary to the AUG initiation codon and its structural properties
differentiate it from tRNAs that decode AUG triplets during
elongation. Initiator tRNA is activated by covalent linkage to
methionine to form methionyl-tRNA, which is the substrate used
by ribosomes to initiate protein synthesis.
mRNA The RNA template that is translated by ribosomes to
synthesize a protein. Its coding sequence comprises consecutive
triplet codons that are decoded by base pairing with the anticodon
of aminoacylated tRNAs, and that therefore determine the amino
acid sequence of the resulting protein.
ribosome The complex macromolecule that catalyzes mRNA tem-
plate-directed protein synthesis. Its two subunits both consist of
ribosomal RNA and proteins. The 40S subunit binds mRNA and
the anticodon end of tRNA; the 60S subunit aligns the aminoacyl
ends of tRNAs and catalyzes peptide bond formation.
FURTHER READING
Dever, T. E. (2002). Gene-specific regulation by general translation
factors. Cell 108, 545– 556.
Gingras, A.-C., Raught, B., and Sonenberg, N. (1999). eIF4 initiation
factors: Effectors of mRNA recruitment to ribosomes and
regulators of translation. Annu. Rev. Biochem. 68, 913– 963.
Hellen, C. U. T., and Sarnow, P. (2001). Internal ribosomal entry sites
in eukaryotic mRNA molecules. Genes Develop. 15, 1593– 1612.
Kozak, M. (1991). Structural features in eukaryotic mRNAs that
modulate the initiation of translation. J. Biol. Chem. 266,
19867–19870.
Pestova, T. V., and Kolupaeva, V. G. (2002). The roles of individual
eukaryotic translation initiation factors in ribosomal scanning and
initiation codon selection. Genes Develop. 16, 2906– 2922.
Sonenberg, N., Hershey, J. W. B., and Mathews, M. B. (eds.) (2000).
Translational Control of Gene Expression. Cold Spring Harbor
Laboratory Press, Cold Spring Harbor, New York.
BIOGRAPHY
Christopher U. T. Hellen is an Associate Professor in the Department
of Microbiology and Immunology at the State University of New
York Downstate Medical Center in New York. His principal research
interests are in mechanisms of translation initiation by internal
ribosomal entry. He holds a D.Phil. from Oxford University.
Tatyana V. Pestova is an Assistant Professor in the Department of
Microbiology and Immunology at the State University of New York
Downstate Medical Center in New York, and a senior research scientist
in the A. N. Belozersky Laboratory of Physico-chemical Biology,
Moscow State University, Moscow, Russia. Her principal research
interests are in molecular mechanisms of translation initiation in
eukaryotes. She holds a Ph.D. and a D.Sc. from Moscow State
University.
TRANSLATION INITIATION IN EUKARYOTES: FACTORS AND MECHANISMS 241
