Iozzo Renato V. Proteoglycan Protocols
Подождите немного. Документ загружается.

204 McQuillan et al.
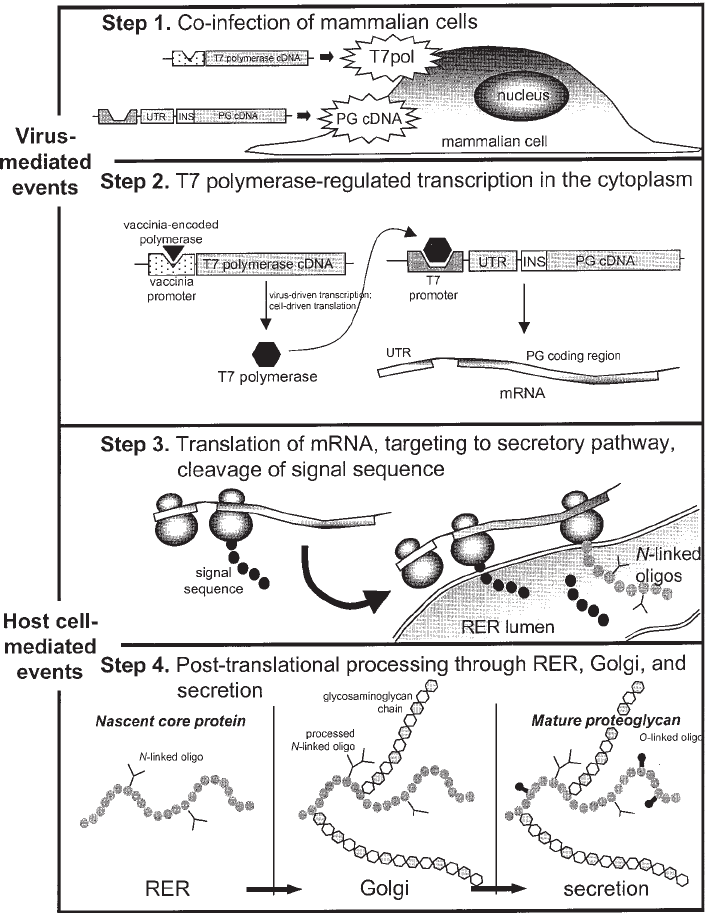
Fig. 1. Schematic representation of vaccinia virus/T7 phage expression system. The major
steps of this powerful eukaryotic recombinant protein expression system are shown. Vaccinia
virus mediates the initial steps 1 and 2. Step 1, co-infection of mammalian cells by a recombi-
nant virus containing the gene encoding phage T7 RNA ploymerase under control of the early/
late vaccinia virus promotor; and a recombinant virus containing the gene encoding the target
protein under control of the phage T7 promotor. Step 2, cytoplasmic expression of T7 RNA

Recombinant Proteoglycan Expression 205
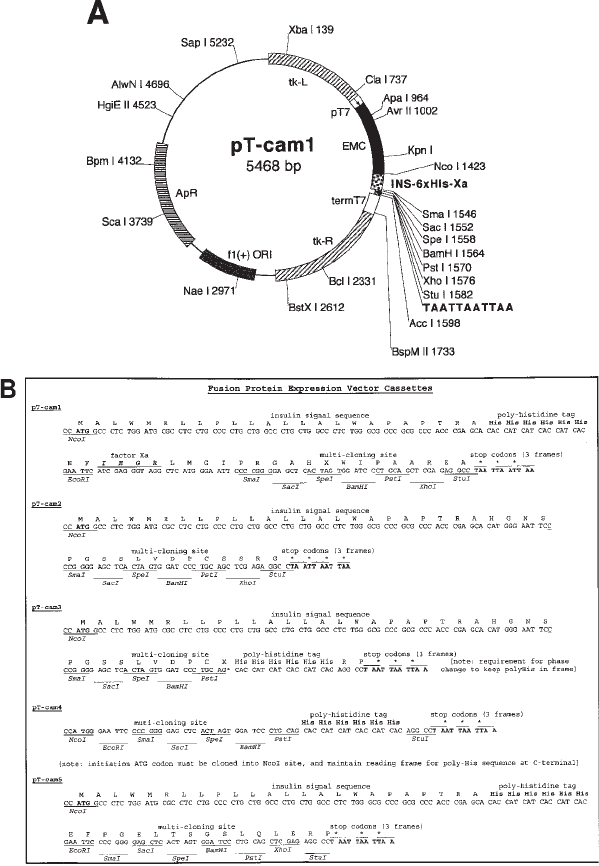
The important domains of these vectors are shown in Fig. 2. The most versatile of the series
is pT-cam1 (see Fig. 2a, b), and comprises (1) a bacteriophage T7 promoter; (2) an ATG start
codon at a precise distance downstream of an encephalomyocarditis virus ribosome-binding
site (EMC UTR); (3) a canine insulin signal sequence, for targeting to the secretory pathway;
(4) a hexa-histidine (6 × His) sequence, for nondenaturing purification by metal-chelating
affinity chromatography; (5) a factor Xa recognition cleavage sequence, for removal of the
histidine tag after purification; (6) a versatile multicloning site, for in-frame insertion of the
cDNA of interest (preferably devoid of endogenous signal sequence); and (7) stop codons in
three reading frames upstream of a polyadenylation signal.
The other vectors shown are essentially modifications of pT-cam1, for situations where
all the elements of the parent vector are not required. If one has an alternative method of
purification (e.g., antibody affinity column or interaction with hyaluronan), pT-cam2 does
not have the hexa-histidine tag nor the factor Xa cleavage sequence. If potential bioactive
domains are predicted to be located in the vicinity of the N-terminus, pT-cam3 has the
hexa-histidine tag located at the C-terminus of the secreted protein. Our experience indi-
cates that the insulin signal sequence can enhance expression of some proteins compared
to the endogenous sequence, but pT-cam4 is designed for insertion of the cDNA inclusive
of endogenous signal sequence with the hexa-histidine tag at the C-terminus. In many
instances, removal of the small hexa-histidine sequence following purification is likely to
be no advantage and potentially cumbersome, and in these circumstances pT-cam5, which
is devoid of the factor Xa cleavage sequence, may be the vector of choice. All vectors and
complete sequences are available upon request from the authors.
2. Materials
1. Cell lines, shown in Table 1.
2. Wild-type vaccinia virus, WR (ATCC # VR-119), titer should be about 5 × 10
9
pfu/mL.
3. DMEM alone (DMEM SF), containing 2.5% FCS (DMEM 2.5%), containing 10% FCS
(DMEM 10%).
4. Lipofectin™ transfection kit (GIBCO-BRL #18292-011) or other efficient transfection
reagents.
5. Plasmid containing cDNA (pT-cam series) at 1 µg/µL.
6. Sterile polystyrene tubes.
7. Bromodeoxyuridine (BrdU) 200-fold concentrated stock solution, 5 mg/mL in water, filter
sterilized.
8. 143B (TK–) cells in log phase, passaged at least once in the presence of BrdU.
9. All culture media for the plaque assays should be supplemented with 25 µg/mL BrdU.
10. Neutral red solution, 3.33 mg/mL, tissue culture grade (Gibco-BRL #15330-079).
11. Low-melting-point agarose (Gibco-BRL #15517-014), 2% solution in PBS, autoclave
and mix well. Can be stored at 45°C for 1–2 wk.
polymerase and transcription of mRNA encoding the target protein. The host mammalian cell
machinery is utilized for steps 3 and 4. Step 3, ribosome directed translation of the target pro-
tein mRNA that is enhanced by inclusion of a ribosome “landing pad” (UTR); targeting to the rough
endoplasmic reticulum by specific signal sequence; signal sequence cleavage; and addition of
N-linked oligosaccharides to the nascent chain. Step 4, posttranslational modifications as appropri-
ate for the target protein, including processing of N-linked oligosaccharides, addition of O-linked
oligosaccharides, synthesis of glycosaminoglycan chains, and finally secretion of a proteoglycan.
206 McQuillan et al.
Fig. 2. pT-cam series of expression vectors. (A) Main structural features of the pT-cam
series of vectors. tk-L and tk-R, thymidine kinase flanking sequences to facilitate insertion of
the expression cassette via homologous recombination into wild-type vaccinia virus genome;
EMC, encephalomyocarditis untranslated region (UTR) that allows for cap-independent ribo-
some binding; INS, insulin signal sequence; 6xHis, hexa-histidine sequence; Xa, factor Xa
cleavage site. (B) cDNA and protein sequences of pT-cam series expression cassettes, showing
unique restriction sites available in the multicloning site (MCS), and positioning of other struc-
tural features in relation to the MCS.
Recombinant Proteoglycan Expression 207
12. Proteinase K stock solution (20 mg/mL, Gibco-BRL #25530-031).
13. TE-saturated phenol.
14. Reagents or kit for standard PCR reaction.
15. Thermal cycler.
16. Crystal violet, 0.1% in 20% ethanol.
17. vTF7-3, recombinant vaccinia virus encoding phage T7 RNA polymerase.
18. Methionine and cysteine-free DMEM.
19. Trans-
35
S-label, a mixture of
35
S-methionine and
35
S-cysteine (ICN Biochemicals,
#51006).
20. Sephadex G-50 (available from Pharmacia-Amersham Biotech).
21. Sepharose 6B conjugated to iminodiacetic acid functional group (slurry or prepacked 1-mL
columns available from Pharmacia-Amersham Biotech).
22. Column buffers: For optimal recovery, column solvents (with the exception of the “charge
buffer”) should contain a detergent (either 0.1% or greater Triton X-100, or 0.2% or
greater CHAPS).
a. Charge buffer: 100 mM NiCl
2
.6H
2
O.
b. Sample and loading buffer: 5 mM imidazole, 0.5 M NaCl, 20 mM Tris-HCl, pH 8.0.
Table 1
Cell Lines Used for Homologous Recombination, Plaque Assays, Amplification,
Titration, and Protein Expression
Cell line (origin, morphology) Culture medium
a
ATTC catalog number
CV-1 (monkey kidney cell, DMEM + 2.5% FCS
b
fibroblastic) 5.0% FCS CCL-70
10.0% FCS
143B (TK–)
(human osteosarcoma, MEM + 5% FCS CRL-8303
fibroblastic) 10% FCS
HeLa
(human adeno-carcinoma, DMEM + 10% FCS CCL-2
epithelial)
UMR-106
(rat osteosarcoma, DMEM + 10% FCS CRL-1661
fibroblastic)
HT-1080
c
(human fibrosarcoma, DMEM + 10% FCS CCL-121
epithelial)
a
Inclusion of antibiotics is optional, except in the case of the initial transfection for the homologous
recombination. If using Lipofectin, then antibiotics should be omitted, as it can inhibit transfection
efficiency.
b
Fetal calf serum, or serum substitutes.
c
Any other cell line that is likely to process proteoglycans appropriately can be substituted here,
with the exception of CHO cells.
208 McQuillan et al.
c. Wash buffer: 20 mM imidazole, 0.5 M NaCl, 20 mM Tris-HCl, pH 8.0.
d. Elution #1: 60 mM imidazole, 0.5 M NaCl, 20 mM Tris-HCl, pH 8.0.
e. Elution #2: 250 mM imidazole, 0.5 M NaCl, 20 mM Tris-HCl, pH 8.0.
f. Strip buffer: 100 mM EDTA, 0.5 M NaCl, 20 mM Tris-HCl, pH 8.0.
23. Sonicator, either probe or cup (e.g., Fisher Sonic Dismembranator 550).
24. Phase-contrast light microscope.
25. Biohazard containment.
3. Methods
3.1. In Vitro Transcription and Translation (
see
Note 1)
We routinely use the TNT
®
coupled reticulocyte lysate system available from
Promega (#L4610), and essentially follow the manufacturer’s instructions. In a
typical cell-free transcription/translation reaction, we would use the following
protocol.
1. Mix in an eppendorf tube for a final reaction volume of 50 µL: 25 µL rabbit reticulocyte
lysate, 2 µL T
NT reaction buffer, 1 µL T7 RNA polymerase, 1 µL methionine-free amino
acid mixture, 4 µL Trans
35
S-label, a mixture of
35
S-methionine and
35
S-cysteine (ICN
Biochemicals), 1 µL RNAsin
®
ribonuclease inhibitor, 1 µL plasmid, containing cDNA
(1 µg/µL CsCl-purified, or equivalent quality), and 15 µL nuclease-free water
2. Incubate mixture at 30°C for 90 min.
3. Analyze reaction products by (a) direct application to SDS-PAGE; (b) immunoprecipita-
tion followed by SDS-PAGE; or (c) purification on small metal chelating column fol-
lowed by SDS-PAGE (reaction mix must be exchanged into appropriate binding buffer,
e.g., PD-10 column from Amersham-Pharmacia Biotech).
4. Bands are visualized by standard autoradiography/fluorography. The predominant signal
should be the target protein of interest migrating at the predicted size for the unsubstituted
core protein.
5. If DNA sequencing does not show any errors, and cell-free translation generates a protein
product of the predicted size, then one can confidently move to the next step of generating
a recombinant virus from the plasmid.
3.2. Generating a Recombinant Virus
In this section, several protocols are outlined. The homologous recombination
allows transfer of the cDNA under control of the T7 promoter to be inserted into the
thymidine kinase (TK) locus of wild-type vaccinia virus. Disruption of the TK gene
allows for selection of positive recombinants by culture in the presence of bromo-
deoxyuridine, a lethal analog of deoxyuridine that is incorporated into replicating DNA
by an active TK gene. Plaques are selected and undergo three rounds of purification
to ensure clonal selection and remove contaminanting dormant wild-type virus. These
procedures require standard cell culture ware and tissue culture facilities. As indicated
above, these protocols assume a basic knowledge of virus manipulation. Extensive
protocols relating to vaccinia virus are available elsewhere (13). Cells required
are shown in Table 1, indicating the ATTC catalog number and the recommended
culture medium. Specialized equipment includes a sonicator (either probe or cup, e.g.,
Fisher Sonic Dismembranator 550), phase-contrast light microscope, and biohazard
containment.
Recombinant Proteoglycan Expression 209
3.2.1. Homologous Recombination
1. Day 1: Initiate culture of CV-1 cells. Seed a T-25 flask (one flask per construct) with
CV-1 cells at 1 × 10
6
cells/flask, in a total volume of 10 mL of DMEM + 10% FCS. Use
the next day when ~80% confluent (should be ~3 × 10
6
cells/flask).
2. Day 2: Infection and transfection. Thaw WR virus stock (titer of stock should be
~5 × 10
9
pfu/mL). Sonicate for 30 s, chill on ice for 2 min, and vortex vigorously for
30 s. Repeat once.
3. Trypsinize virus by mixing 20 µL of virus with an equal volume of trypsin (cell culture
grade). Incubate at 37°C for 30 min, vortexing briefly every 5 min. This is tube A, a
1/1 dilution (~2.5 × 10
9
pfu/mL).
4. Dilute A 1/100 in 2.5% DMEM (10 µL A + 990 µL 2.5% DMEM). This is tube B, a
1/200 dilution (~2.5 × 10
7
pfu/mL).
5. Aliquot a volume of B containing 1.5 × 10
5
pfu (~6 µL) and add to 1 mL of 2.5% DMEM.
This is tube C, final concentration of 1.5 × 10
5
pfu/mL.
6. Aspirate medium from the T-25 flask of CV-1 cells, and replace with 1 mL of tube C
(represents ~0.05 pfu per cell).
7. Incubate for 2 h at 37°C in 5% CO
2
, with gentle rocking every 15 min.
8. Set up transfection reagents 30 min before end of step 7. Note that all plasmids should be
at 1 µg/µL, and lipofectin is inhibited by serum proteins and penicillin.
9. Set up two sterile polystyrene tubes:
a. Tube A: 10 µL DNA + 200 µL DMEM SF
b. Tube B: 30 µLlipofectin (or lipofectamine) + 200 µL DMEM SF
10. Gently mix A and B. Incubate for 30 min at room temperature. Add 1.6 mL of DMEM SF
to the AB mixture.
11. Aspirate the virus inoculum from the T-25 flask. Add the DNA/lipofectin transfection
mixture (volume 2 mL) to the cells, ensuring good coverage of the cell layer.
12. Incubate for 5–6 h in a cell culture incubator.
13. Overlay transfection with 3 mL of 10% DMEM (do not aspirate transfection cocktail).
Continue culture for 2 d.
14. Day 4: Harvest homologous recombination. Cytopathic effect (CPE) should be clearly
visible by light microscopy. Scrape cell layer with a rubber policeman into a 15 mL coni-
cal tube. Pellet cells by centrifugation at 300g (3000 rpm), 10 min in a bench-top centri-
fuge. Aspirate and discard supernatant. Resuspend pellet in 0.5 mL of 2.5% DMEM.
15. Freeze and thaw lysate three times in a Dry Ice/ethanol bath.
16. Store at –80°C, or proceed with plaque assay (see Subheading 3.2.2.).
3.2.2. Plaque Assay (First Round)
1. Day 1: Initiate culture of 143B cells. Seed two 6-well plates (35-mm-diameter wells) per
recombination with 143B cells at a density of 5 × 10
5
cells/well in 10% MEM. Use the
next day when greater than 90% confluent (~1 × 10
6
cells per well). Note that a well-
defined monolayer is critical to the recognition of clearly defined plaques.
2. Day 2: Infection of 143B cells. Thaw the homologous recombination lysate (Subheading
5.1.2.) on ice. Sonicate for 30 s, chill on ice for 2 min, and vortex vigorously for 30 s.
Repeat once.
3. Incubate 100 µL of lysate with an equal volume of trypsin and incubate at 37°C for
30 min, vortex briefly every 5 min.
4. The trypsinized lysate is diluted into 2.5% MEM, and tested at a range of dilutions from
1/1000 through 1/10 000. An example of a serial dilution protocol is as follows:

210 McQuillan et al.
Virus solution Diluent Stock Dilution
200 µL trypsinized lysate + 1.8 mL 2.5% MEM = A (1/10)
200 µLA+1.8 mL 2.5% MEM = B (1/100)
400 µLB+3.6 mL 2.5% MEM = C (1/1000)
500 µLC+1.0 mL 2.5% MEM = D (1/2000)
400 µLC+1.6 mL 2.5% MEM = E (1/5000)
200 µLC+1.8 mL 2.5% MEM = F (1/10 000)
5. Aspirate media from 6-well plates, and infect with 0.5 mL of diluted lysate per well.
Infect with stock dilutions C, D, E, and F in triplicate.
6. Incubate for 2 h in the culture incubator, with gentle rocking every 15 min to ensure the
monolayer remains wet.
7. Prepare enough LMP agarose overlay to allow 2.5 mL per well. For example, for four
plates (i.e., one construct = four dilutions in triplicate = four 6-well plates), 32.0 mL 2%
LMP agarose (~45°C), 32.0 mL 5% MEM, and 0.36 mL BrdU (5 mg/mL).
8. Add 2.5 mL of agarose overlay (~45°C) solution per well. Do not aspirate virus inocu-
lum. Place plates at 4°C for 15 min to allow agarose to solidify, and then incubate plates
in culture incubator for 2 d.
9. Day 4: To visualize plaques easily, the 143B monolayer is stained overnight with neutral
red (a relatively nontoxic reagent). Prepare enough neutral red/agarose overlay to allow
2.0 mL per well. For example, for four plates, 32.0 mL 2% LMP agarose (~45°C), 32.0 mL
5% MEM, and 2.0 mL neutral red.
10. Add 2.0 mL of neutral red/agarose overlay (~45°C) solution per well. Place plates at 4°C
for 15 min to allow agarose to solidify, and then incubate plates in the culture incubator
overnight.
11. The first-round plaque assay will be complete on day 5; therefore, to reduce “down-time”,
143B cell cultures should be set up at this time for the second round (see Subheading 3.2.3.).
12. Day 5: Inspect plates by holding up to a light, and circle clearly defined, well-separated
plaques (these appear as a weakly stained “hole” in a red background). Wells from dilu-
tion C should contain an excessive number of plaques, and wells from dilution F are
likely to contain very few to no plaques. If this is not the case, then the homologous
recombination step has not worked and should be repeated.
13. Pick plaques using sterile glass Pasteur pipets to pull the agarose plug (containing virus)
with slight suction. The plug is then expelled into a sterile Eppendorf tube containing
500 µL of pre-warmed 2.5% MEM. Rinse the pasteur with the medium to recover all the
agarose. It is recommended that 24 plaques be picked.
14. Rapidly freeze and thaw the plaques three times in a Dry Ice/ethanol bath.
15. Store at –80°C, or proceed directly with the second-round plaque assay.
3.2.3. Second- and Third-Round Plaque Assays (
see
Note 2)
1. Day 1: Initiate culture of 143B cells. Seed one 6-well plate (35-mm-diameter wells) per
plaque, with 143B cells at a density of 5 × 10
5
cells/well in MEM 10%. Use the next day
when greater than 90% confluent (~1 × 10
6
cells per well).
2. Day 2: Infection of 143B cells. Thaw lysate from first-round plaque assay. Sonicate for
30 s, chill on ice for 2 min, and vortex vigorously for 30 s. Repeat once.
3. Incubate 250 µL of lysate with an equal volume of trypsin and incubate at 37°C for
30 min, vortex briefly every 5 min.

Recombinant Proteoglycan Expression 211
4. The trypsinized lysate is diluted into 2.5% MEM, and tested at a range of dilutions of 1/5,
1/50, and 1/500. An example of a serial dilution protocol is as follows :
Virus solution Diluent Stock Dilution
500 µL lysate-trypsin + 2.0 mL 2.5% MEM = A (1/5)
200 µLA+1.8 mL 2.5% MEM = B (1/50)
200 µLB+1.8 mL 2.5% MEM = C (1/500)
5. Aspirate media from 6-well plates, and infect with 0.5 mL of diluted lysate per well.
Infect with stock dilutions A, B, and C in duplicate.
6. Incubate for 2 h in the culture incubator, with gentle rocking every 15 min to ensure that
the monolayer remains wet.
7. Prepare enough LMP agarose overlay to allow 2.5 mL per well. For example, for eight
plates (i.e., one plate per first round plaque), 64.0 mL 2% LMP agarose (~45°C), 64.0 mL
5% MEM, and 0.72 mLBrdU (5 mg/mL),
8. Add 2.5 mL agarose overlay (≤ 45°C) solution per well. Place plates at 4°C for 15 min to
allow agarose to solidify, and then incubate plates in culture incubator for 2 d.
9. Day 4: The 143B monolayer is stained overnight with neutral red. Prepare enough neutral
red/agarose overlay to allow 2.0 mL per well. For example, for eight plates, 64.0 mL 2%
LMP agarose (~45°C), 64.0 mL 5% MEM, and 4.0 mL neutral Red.
10. Add 2.0 mL neutral red/agarose overlay (~45°C) solution per well. Place plates at 4°C for
15 min to allow agarose to solidify, and then incubate plates in the culture incubator
overnight.
11. The second-round plaque assay will be complete on d 5, therefore, to reduce “downtime”,
143B cell cultures should be set up at this time for the third round.
12. Day 5: Inspect plates by holding up to a light, and circle clearly defined, well-separated
plaques. Plaques are likely to be present in all wells if the first-round plaque is a genuine
TK- recombinant.
13. Pick at least 4 plaques per plate, and place into 500 µL of 2.5% MEM, as for the first
round.
14. Rapidly freeze and thaw the plaques three times in a Dry Ice/ethanol bath.
15. Store at –80°C, or proceed directly with the third-round plaque assay.
16. The third round is identical to the second round. Although you have picked four plaques
at the second round, take one plaque from each plate to the third round. After the third
round you should have eight recombinants that have been derived from eight distinct
plaques in the first-round plaque assay.
3.3. A Convenient Nomenclature: Keeping Track of All Those Plaques
There are a large number of samples that must be archived in the event that a puta-
tive positive drops out during selection, or a catastrophe (such as bacterial contamina-
tion) wipes out a third-round positive. Storage of original and subsequent rounds of
selection will allow one to rescreen at any stage. We have found the following num-
bering system to be useful. By example, selection for recombinant virus expressing
the proteoglycan “pg”:
1. Recombinantion step: pg recombination, 03-04-2000 (i.e., date of recombination).
2. Select first-round plaques: pg 1, pg 2, pg 3, …, pg 24.
212 McQuillan et al.
3. Select second-round plaques derived from pg 1, pg 2, pg 3, pg 4: pg 1.1, pg 1.2, pg 1.3, pg
1.4, pg 2.1, pg 2.2, …, pg 4.1, pg 4.2, pg 4.3, pg 4.4.
4. Select third-round plaques derived from pg 1.1, pg 2.1, pg 3.1, pg 4.1: pg 1.1.1, pg 1.1.2,
pg 1.1.3, pg 1.1.4, …, pg 4.1.1, pg 4.1.2, pg 4.1.3, pg 4.1.4 (see Note 3).
3.4. Screening Recombinants by PCR (
see
Note 4)
1. Day 1: Seed 6-well plates with 143B cells at a density of 5 × 10
5
cells/well in 10%MEM.
You will need one well per plaque. Use the next day when greater than 90% confluent
(~1 × 10
6
cells per well).
2. Day 2: Thaw third round plaque lysate on ice. Sonicate for 30 s, chill on ice for 2 min, and
vortex vigorously for 30 s. Repeat once.
3. Add 200 µL of lysate is added to 300 µL of 2.5% MEM to make the virus inoculum. Note
that there is no requirement for trypsinization at this step.
4. Add virus inoculum directly to the cell layer, followed by incubation for 2 h with gentle
rocking every 15 min.
5. Overlay cultures with 2 mL of 2.5% MEM and incubate a further 2 d.
6. Day 4: Harvest cells and media by scraping with a rubber policeman and transfer to a
centrifuge tube.
7. Pellet cells by centrifugation at 300g (3000 rpm) for 10 min. Resuspend pellet in 200 µL
of 1 M Tris-HCl, pH 9.0.
8. Set up proteinase K digestion for a final volume of 333.6 µL: 200 µL cell pellet, 16.7 µL
1 M Tris. HCl pH 7.8, 16.7 µL 10% SDS, 33.4 µL 60% sucrose, and 66.8 µL 10 mg/mL
proteinase K. Digest ~5 h at 37°C.
9. Extract two times with TE-saturated phenol.
10. Extract one time with phenol/chloroform
11. Ethanol-precipitate DNA overnight at –20°C
12. Resuspend pellet in 30 µL of TE buffer
13. Perform PCR reaction, including a “no DNA” and “plasmid DNA” controls. A typi-
cal reaction mix with a final volume of 100 µL is: 1 µL of extract, or 100 ng of
control DNA, DNA, dNTP’s (10 mM) 8 µL, forward primer (0.1 µg/µL) 1 µL, reverse
primer (0.1 µg/µL) 1 µL, 10 X Pfu buffer 10 µL, sterile water 78 µL, and Pfu poly-
merase 1 µL. Mix, spin briefly, and set in PCR machine. Do PCR as required for
specific primers.
3.5. Amplification of Recombinant Virus (
see
Note 5)
3.5.1. Plaque Amplification into 6-Well Plates
1. Day 1: Seed 6-well plates with 143B cells at a density of 5 × 10
5
cells/well in 10%
MEM. You will need one well per plaque. Use the next day, when greater than 90%
confluent.
2. Day 2: Thaw lysate from third-round plaque assay on ice. Sonicate for 30 s, chill on ice
for 2 min, and vortex vigorously for 30 s. Repeat once.
3. Add 250 µL of lysate to 250 µL 2.5% MEM to make virus inoculum. Make sure BrdU is
present at a final concentration of 25 µg/mL.
4. Add 0.5 mL virus inoculum directly to cell layer, followed by incubation for 2 h with
gentle rocking every 15 min.
5. Overlay cultures with 1.5 mL of 2.5% MEM and 25 µg/mL BrdU.
6. Day 4: Cytopathic effect (CPE) should be clearly visible when held under the light. Har-
vest cells and media by scraping into a 15-mL conical tube. Pellet cells by centrifugation

Recombinant Proteoglycan Expression 213
at 300g (3000 rpm) for 10 min. Aspirate and discard the supernatant. Resuspend cell
pellet in 500 µL 2.5% MEM.
7. Rapidly freeze and thaw plaques three times in a Dry Ice/ethanol bath. Store at –80°C.
3.5.2. Plaque Amplification into T-25 Flasks
1. Day 1: Seed T-25 flasks with 143B cells at 1 × 10
6
cells per flask. Use the next day, when
confluent. A single flask is needed for each individual clone.
2. Day 2: Thaw lysate from 6-well plate amplification (see Subheading 5.5.1.). Sonicate for
30 s, chill on ice for 2 min, and vortex vigorously for 30 s. Repeat once.
3. Add 300 µL of lysate to 700 µL of 2.5% MEM. Make sure BrdU is present at a final
concentration of 25 µg/mL.
4. Aspirate media from cells and infect with 1.0 mL of diluted lysate per flask.
5. Incubate for 2 h at 37°C with gentle rocking every 15 min.
6. Overlay each flask with 3.0 mL of 2.5% MEM and 25 µg/mL BrdU.
7. Day 4: CPE is clearly visible. Scrape cell layer into 15-mL conical tube. Pellet cells at
300g (3000 rpm) for 10 min. Aspirate supernatant and resuspend pellet in 500 µl of 2.5%
MEM.
8. Rapidly freeze and thaw plaques three times in a Dry Ice/ethanol bath. Store at –80°C if
required.
3.5.3. Plaque Amplification into T-175 Flasks
1. Day 1: Seed T-175 flasks with HeLa cells. Note that selection agent (BrdU) is no longer
required. A single flask is needed for each individual putative recombinant.
2. Day 2: Thaw lysate from T-25 amplification (see Subheading 5.5.2.). Sonicate for 30 s,
chill on ice for 2 min, and vortex vigorously for 30 s. Repeat once.
3. Add 250 µL of lysate to 1.75 mL of 2.5% DMEM.
4. Aspirate media from cells and infect with 2.0 mL of diluted lysate per flask.
5. Incubate for 2 h with gentle rocking every 15 min.
6. Overlay each flask with 25 mL of 2.5% DMEM.
7. Day 4: Cytopathic effects should be clearly visible. Scrape cell layer and media into a
50-mL conical tube. Pellet cells by centrifugation at 300g (3000 rpm) for 10 min. Aspi-
rate supernatant and resuspend pellet in 1.5 mL of 2.5% DMEM.
8. Rapidly freeze and thaw plaques three times in a Dry Ice/ethanol bath. Store at –80°C if
required.
3.6. Titration of Virus Stock (
see
Note 6)
1. Day 1: Seed 6-well plates with either UMR-106 or CV1 cells at 1×10
6
cells/well. (One
plate per recombinant virus). Use the next day, when confluent.
2. Day 2: Thaw virus stock (from Subheading 5.5.3.). Sonicate for 30 s, chill on ice for 2
min, and vortex vigorously for 30 s. Repeat once.
3. Incubate 10 µL of lysate with an equal volume of trypsin and incubate at 37°C for 30 min;
vortex briefly every 5 min.
4. The trypsinized lysate is diluted into 2.5% MEM and tested at a range of dilutions as
follows:
Virus solution Diluent Stock Dilution
20 µL trypsinized lysate + 1.8 mL 2.5% MEM = A (10
–3
)
200 µLA+1.8 mL 2.5% MEM = B (10
–4
)
