Iozzo Renato V. Proteoglycan Protocols
Подождите немного. Документ загружается.

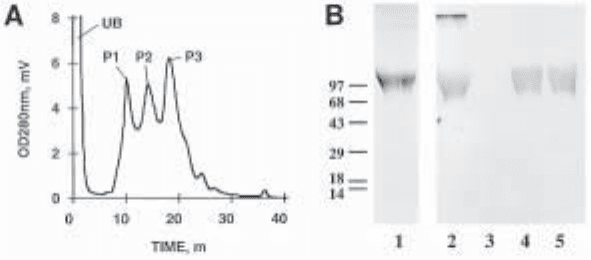
Purification of Recombinant Decorin 225
2. Apply this material to a C4 hydrophobic interaction column (previously equilibrated with
2.5 M ammonium sulfate in 50 mM phosphate buffer, pH 6.3) at room temperature with a
slow flow rate (e.g., 1.0 mL/min for a 50 by 4.6 mm column) and wash the column with
the equilibration buffer until a baseline signal is achieved (see Note 10).
3. Remove bound decorin with a reverse linear gradient of ammonium sulfate from 2.5 to 0 M
in 50 mM phosphate buffer, pH 6.3, while simultaneously applying an increasing linear
gradient of ethylene glycol from 0 to 50% in 50 mM phosphate buffer, pH 8.3. When
monitored by absorbance at 280 nm, we typically observe three major peaks eluting from
the column (Fig. 1A; see Note 11). Coomassie blue/toluidine blue-staining SDS-polyacryla-
mide gels and immunoblotting using a polyclonal antibody directed against the amino-ter-
minal end of human decorin (7) reveal that the second and third peaks in the chromatogram
contain highly purified decorin, with no other molecules detected (Fig. 1B).
4. Collect decorin containing peaks and remove ethylene glycol and ammonium sulfate by
dialysis using a 30-kDa MWCO dialysis membrane (Spectrapor). We typically dialyze
against phosphate-buffered saline before storing the final decorin product.
3.4. Prokaryotic Expression of Decorin Sequences
as Anthranilate Synthase Fusion Proteins
1. Cloning of human decorin subdomains into modified pATH3 vector: PCR amplify the
desired decorin core protein sequence(s) using synthetic oligonucleotide primers that are
designed to incorporate appropriate restriction sites for subcloning the product into
the modified pATH3 vector in the correct reading frame. Restrict PCR product with
appropriate restriction enzymes (e.g., BamHI and XbaI) and subclone into the modified
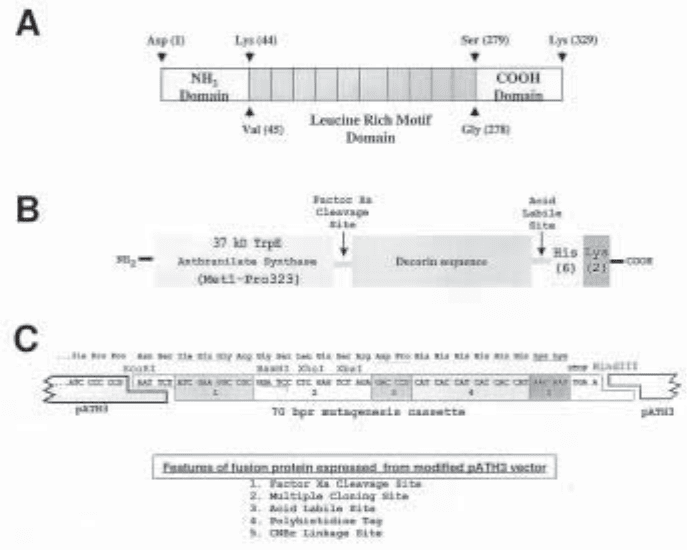
pATH3 vector. Figure 2 illustrates the boundaries of the decorin subdomains that we have
subcloned into our modified pATH3 vector and expressed as fusion proteins, as well as the
sequence of the salient features of this expression vector and resultant fusion polypeptides.
Fig. 1. Purification of human recombinant decorin by hydrophobic interaction chromatog-
raphy. (A) Representative chromatogram resulting from C4-hydrophobic interaction column
loaded with pooled DEAE-Sepharose purified decorin. Note the three predominant peaks
detected in the eluted material, P1-P3. (B) SDS-PAGE analysis of three peaks fractionated by
HIC: lane 1, Coomassie blue toluidine blue staining of the starting material loaded on the HIC
column; lanes 2–5, immunoblot analysis of the starting material (lane 2) and the eluted peaks,
P1-P3 (lanes 3–5, respectively), using a polyclonal antibody directed against the amimo
terminal end of human decorin (7).
226 McBain and Mann
2. Expression of fusion proteins in E. coli WM6: Use E. coli WM6 cells to express the re-
combinant fusion protein. Prepare a freshly seeded plate with WM6 transformants and
pick a single colony for overnight expansion in LB medium containing 100 µg/mL ampi-
cillin (37°C while shaking at 225 rpm). Dilute the overnight culture with 4 volumes of
M9 minimal medium containing casamino acids supplemented with 100 µg/mL ampicil-
lin (i.e., cell density diluted to 20% that of the overnight culture) (see Note 12). Incubate
the diluted culture for 3 h at 37°C with shaking and then induce fusion protein expression
by adding indoylacrylic acid (IAA) to a final concentration of 10 µg/mL. Incubate cul-
tures in IAA containing medium for 5 h at 37°C with shaking at 225 rpm to accumulate
high levels of expressed fusion protein (see Note 13). Add PMSF to the cultures to a final
concentration of 0.2 mM. At this stage the total bacterial culture can be analyzed for the
expression of the fusion protein. Remove 10 µL of culture, dilute with 2× SDS-PAGE
sample buffer (reducing), and analyze total protein content by SDS-PAGE (see Note 14).
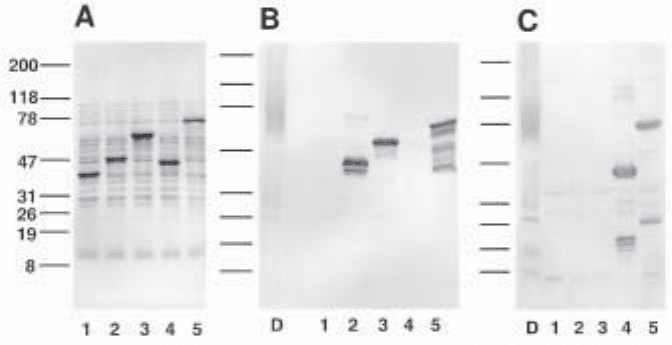
Figure 3 shows the profile of total proteins present in the WM6 cultures (cell + medium)
and their immunoreactivity with rabbit polyclonal antisera raised against either human
decorin (7) or a carboxy-terminal peptide of human decorin (8).
Fig. 2. Structure of decorin core protein and prokaryotic expression vector used to produce
anthranilate synthase-decorin hybrid proteins. (A) Delineation of human decorin subdomains
that have been expressed as hybrid proteins. (B) Model of hybrid fusion protein illustrating the
salient features introduced by expression from our modified pATH3 vector. (C) Sequence of
the fusion site in modified pATH3 vector illustrating position of specific engineered features.
Purification of Recombinant Decorin 227
3. Isolation and solubilization of fusion protein inclusion bodies: Pellet WM6 cells by cen-
trifugation at 5000g for 15 min at 4°C and determine the weight of the cell pellet. Resus-
pend the cell pellet in lysis buffer using 3 mL of lysis buffer per gram of wet cell pellet
weight. While shaking cell suspension at 25°C, add 4 mg of deoxycholic acid per gram of
original cell pellet weight. Incubate lysate at 37°C until it becomes viscous (10–30 min)
and then add 20 µL of DNase I solution per gram cell pellet weight. Incubate at 25°C until
no longer viscous (30–60 min). Centrifuge at 12,000g for 15 min at 4°C. Decant superna-
tant and resuspend pellet in 9 vol of lysis buffer containing 0.5% Triton X-100 and
10 mM EDTA, pH 8.0. Incubate for 5 min at 25°C before centrifuging for 15 min at
12,000g, 4°C. Decant supernatant and solubilize the residual pellet in lysis buffer con-
taining 8 M urea and 2 mM PMSF. Analyze all supernatants and urea insoluble pellet by
SDS-PAGE to determine the distribution of the fusion protein.
4. Notes
1. Serum-containing medium can be processed as per steps 5–7 under Subheading 3.1.,
but, the decorin produced from this material can contain low levels of bovine proteoglycans
derived from the serum. By collecting and processing conditioned medium made under
serum-free conditions, the levels of contaminating proteoglycans from the bovine serum
are undetectable.
Fig. 3. Expression of anthranilate synthase-decorin hybrid proteins in E. coli WM6. Frac-
tionation of 5–20% SDS-polyacrylamide gel of total proteins present in combined media and
cell pellet: (A) Ponceau S staining of a nitrocellulose membrane before immunodetection; (B,C)
immunoblot analysis using rabbit polyclonal antisera raised angainst either human decorin
proteoglycan or a carboxy-terminal peptide of the core protein of human decorin (8), respec-
tively. Total proteins derived from WM6 cultures transformed with (lane 1) modified pATH3
vector only (no decorin sequence inserted); (lane 2) modified pATH3 vector with decorin NH
2
domain insert (Asp1-Lys44); (lane 3) modified pATH3 vector with decorin LRM domain
insert (Val45-Gly278); (lane 4) modified pATH3 vector with decorin COOH domain insert
(Ser279-Lys329); (lane 5) modified pATH3 vector with whole decorin sequence insert
(Asp1-Lys329). As a positive control, lane D contains the proteoglycan from of human decorin.
228 McBain and Mann
2. We have found that 16 h is an optimal period to allow cultures to condition their serum-
free medium with decorin. Longer incubations tend to increase DNA contamination in
the decorin preparations, due to increased cell lysis, while shorter incubations yield lower
concentrations of decorin.
3. It is important to add protease inhibitors before the centrifugation of conditioned medium in
order to reduce degradation of decorin core protein caused by proteases released from any
lysed cells.
4. Increasing the ionic strength of the conditioned medium prior to performing ion-exchange
chromatography reduces binding of weak polyanionic molecules to the DEAE-Sepharose
matrix and thus increases the binding capacity of the column and the purity of the isolated
decorin.
5. For handling of large volumes of conditioned medium we typically use the 5-gal plastic
bottles from common water coolers (each will hold 18–19 L of conditioned medium).
6. Occasionally it may be necessary to siphon off the top 1–2 mm of the matrix from the guard
column, as this may become clogged with a floculant precipitate.
7. To reduce loss of decorin in the wash buffer of the DEAE-Sepharose column, the total wash
time should not exceed 4 h. The fast flow rates necessary can be achieved under a gravity-
driven flow by positioning the connected wash bottle approximately 7 ft (2 m) above the
column.
8. We typically analyze the purity of eukaryotic expressed decorin during the purification
scheme by electrophoresing samples under reducing conditions on 5–20% SDS-polyacry-
lamide gradient gels. These gels are subsequently stained with Coomassie blue to detect any
protein contaminants and then with 0.1% toluidine blue (w/v) in 0.1 M acetic acid, to
detect proteoglycans.
9. The DEAE column flow-through conditioned medium can be reapplied to the ion-exchange
columns for an approx 10% additional yield of decorin.
10. We use the Hydrocell C4-1000, 50 mm × 4.6 mm (BioChrom Labs) on a TOSOH HPLC
system (TSK 6011) equipped with a TSK 6041 UV detector and a GM8010 gradient
monitor; however, a Pharmacia FPLC system (or other equivalent) can be used.
11. Preliminary preparative runs using the larger, 150 mm x 21 mm, Hydrocell C4-1000 column
indicate that approx 600 mg of recombinant human decorin can be bound and recovered
from this column.
12. Although the pATH3 expression vector uses a fairly tight promoter, if low yields of the
fusion protein are observed expression can sometimes be improved by depleting tryptophan
levels in the cultures for 2–3 h before inducing expression with IAA. For example, the
overnight LB cultures are separated from the tryptophan-containing spent medium by cen-
trifugation and resuspended at 20%, their overnight density in tryptophan-free M9 medium
containing casamino acids.
13. The optimal expression period must be determined emperically for each particular fusion
molecule. For example, depending on the stability and accumulation of individual proteins,
the induction period can range from 2 to 12 h.
14. To determine whether the induction step has been successful, it is advised that a duplicate
culture not induced by IAA be prepared and analyzed by SDS-PAGE.
References
1. Moscatello, D. K., Santra, M., Mann, D. M., McQuillan, D. J., Wong, A. J., and Iozzo, R. V.
(1998) Decorin suppresses tumor cell growth by activating the EGF-receptor. J. Clin. Invest.
101, 406–412.
Purification of Recombinant Decorin 229
2. Yamaguchi, Y., Mann, D. M., and Ruoslahti, E. (1990) Transforming growth factor is
negatively regulated by a proteoglycan. Nature 346, 281–284.
3. Koerner, T. J., Hill, J. E., Myers, A. M., and Tzagoloff, A. (1991) High-expression vectors
with multiples cloning sites for construction of trpE fusion genes: pATH vectors. Meth.
Enzymol. 94, 477–490.
4. Mandecki, W., Powell, B. S., Mollison, K. W., Carter, G. W., and Fox, J. L. (1986) High-
level expression of a gene encoding the human complement factor C5a in Escherichia
coli. Gene 43, 131–138.
5. Yamaguchi, Y. and Ruoslahti, E. (1988) Expression of human proteoglycan in Chinese
hamster ovary cells inhibits cell proliferation. Nature 336, 244–246.
6. Choi, H. U., Johnson, T. L., Pal, S., Tang, L-H, Rosenberg, L., and Neame, P. J. (1989)
Characterization of the dermatan sulfate proteoglycans, DS-PGI and DS-PGII, from bovine
articular cartilage and skin isolated by octyl-sepharose chromatography. J. Biol. Chem.
264, 2876–2884.
7. Mann, D. M., Yamaguchi, Y., Bourdon, M. A., and Ruoslahti, E. (1990) Analysis of
glycosaminoglycan substitution in decorin by site-directed mutagenesis. J. Biol. Chem.
265, 5317–5323.
8. Roughly, P. J., White, R. J., Magny, M-C., Liu, J., Pearce, R. H., and Mort, J. S. (1993)
Non-proteoglycan forms of biglycan increase with age in human articular cartilage.
Biochem. J. 295, 421–426.

Prokaryotic Expression of Proteoglycans 231
231
From:
Methods in Molecular Biology, Vol. 171: Proteoglycan Protocols
Edited by: R. V. Iozzo © Humana Press Inc., Totowa, NJ
22
Prokaryotic Expression of Proteoglycans
Alan D. Murdoch and Renato V. Iozzo
1. Introduction
For molecules with such extensive posttranslational modifications, it may not be
immediately obvious why one would wish to express the protein cores of proteoglycans
in prokaryotic systems, where none of this modification can occur. However, there are
several cases where this is not a problem, and some where there is a positive advantage
to expressing a core protein with none of the normal eukaryotic modifications.
Bacterially expressed proteoglycan core proteins have been used successfully to
raise polyclonal antisera (1,2) and as both immunization and screening agents in mono-
clonal antibody production, notably in the production of domain specific monoclonals
(3,4). Bacterial fusion proteins have also been used to probe core protein domain–
carbohydrate interactions (5), as substrate for modification enzymes such as core pro-
tein–UDP-xylose xylosyltransferase (6,7), and to gain at least some structural and
functional information about important extracellular matrix molecules (8,9).
Bacterial fusion systems can be divided roughly into two main groups depending
on the size of the vector-encoded fusion partner. Those with small tags such as
polyhistidine (6 × His), which allow purification on immobilized metal ions (e.g.,
Qiagen, Clontech), FLAG (Sigma), or c-myc peptide tags (e.g., Invitrogen), which
allow detection and purification with specific antibodies, and the small (4-kDa)
calmodulin-binding peptide, which allows purification on calmodulin resin
(Stratagene), have the advantage that the tag can be left on the protein of interest with
minimal or no interference with the structure or function. Larger tags such as
glutathione S-transferase, protein A, maltose-binding protein, or cellulose-binding
domain (Amersham Pharmacia Biotech, Novagen, New England Biolabs) may help
with fusion proteins that have solubility problems, but the large fusion partner can
present its own problems. There are many methods available for enzymatic or autocata-
lytic cleavage of fusion partners, but in many cases, such as monoclonal antibody
production or binding studies, it is not necessary to remove the fusion partner, as
232 Murdoch and Iozzo
experimental design can control for the presence of the partner. The choice of fusion
partner may be largely empirical, since not all proteins will express in the first system
of choice, and several may need to be tested before satisfactory expression levels are
achieved.
The pMAL system from New England Biolabs fuses the protein of interest to mal-
tose-binding protein (MBP), the product of the Escherichia coli malE gene (10,11).
MBP is normally secreted into the bacterial periplasmic space; the pMAL-p2 vector
can be used to take advantage of this and produce secreted periplasmic fusion pro-
teins. The pMAL-c2 vector has the signal peptide sequence of the malE gene removed
and can be used for high-level cytoplasmic expression of fusion proteins. In both
cases, the fusions can be purified by virtue of the affinity of MBP for maltose using a
column of immobilized amylose (a maltose polymer). In this chapter we present details
of pilot-scale experiments for cytoplasmic and periplasmic expression of MBP fusion
proteins, with an example of cytoplasmic expression, and a protocol for affinity puri-
fication. Further details about the system can be found in the pMAL system handbook
(available on the NEB website at http://www.neb.com). In addition, since many fusion
proteins will form insoluble inclusion bodies in many of the common E.coli strains,
we include a convenient method for solubilization of protein from these inclusion
bodies in a form suitable for affinity purification.
2. Materials
1. pMAL-c2 and pMAL-p2 vectors, components of the pMAL system. General molecular
biology supplies: restriction endonucleases, T4 DNA ligase, etc., facilities for running
horizontal agarose gels, water baths, DNA sequencing facilities. PCR reagents, including
proofreading polymerases such as Pfu or Tli polymerases if necessary. SDS-PAGE gel
equipment, columns, pumps, and fraction collector for affinity chromatography.
2. Luria-Bertani (LB) growth medium: 10 g of tryptone, 5 g of yeast extract, 5 g of NaCl per
liter. Adjust pH to 7 and autoclave.
3. Glucose. 1 M glucose stock: Dissolve 18 g of glucose and make up to 100 mL. Filter
sterilize and add 10 mL to cooled sterile LB for rich growth medium (see Note 1).
4. Ampicillin 1000 × stock: Dissolve 0.5 g of ampicillin in 5 mL of H
2
O. Filter sterilize into
aliquots and store at –20°C.
5. X-gal: Dissolve at 40 mg/mL in dimethylformamide. IPTG 1 M stock: Dissolve 2.38 g of
IPTG in 10 mL of H
2
O, filter sterilize into aliquots. Store both X-gal and IPTG at –20°C.
6. Column buffer: 20 mM Tris-HCl, pH 7.4, 200 mM NaCl, 1 mM EDTA. Dissolve 2.42 g of
Tris base, and 11.7 g of NaCl per liter and add 2 mL of 0.5 M EDTA stock. Adjust pH to 7.4.
In addition, the column buffer used in the example shown in Fig. 1 also contained 1 mM
sodium azide (1 mL of 1 M stock per liter) and 10 mM β-mercaptoethanol (0.7 mL/L).
7. Cold osmotic shock wash buffer: 30 mM Tris-HCl, pH 8.0, 20% sucrose. Dissolve 3.63 g
of Tris base and 200 g of sucrose per liter and adjust pH to 8.0.
8. For the cold osmotic shock: 5 mM MgSO
4
. Dissolve 60 mg of anhydrous MgSO
4
in
100 mL of H
2
O.
9. Anti-MBP serum, component of the pMAL system.
10. Amylose resin, component of the pMAL system.
11. Inclusion body lysis buffer: 50 mM Tris-HCl, pH 8.0, 100 mM NaCl, 1 mM EDTA. Dis-
solve 3.03 g of Tris base, and 2.92 g of NaCl and add 1 mL of 0.5 M EDTA for 500
mL of buffer and adjust the pH to 8.
Prokaryotic Expression of Proteoglycans 233
12. Inclusion body wash buffer: as per lysis buffer above, but add 10 mL of 0.5 M EDTA and
2.5 mL of Triton X-100 per 500 mL.
13. Inclusion-body solubilization buffer: Lysis buffer containing 8 M urea and 0.1 mM PMSF.
Dissolve 48 g of urea by adding 50 mL of H
2
O and then making the solution up to 100
mL. Deionize by adding 1 g of mixed-bed resin TMD-8 (Sigma) and stir for 1 h. Filter the
beads out of the solution through a sintered glass filter. Add to the solution 0.61 g of Tris
base, 0.58 g of NaCl, 0.2 mL of 0.5 M EDTA and adjust pH to 8. Add PMSF just before
use to 0.1 mM (see Note 2).
14. Alkaline pH buffer: 50 mM KH
2
PO
4,
pH 10.7, 50 mM NaCl, 1 mM EDTA. Dissolve 3.4 g of
KH
2
PO
4
and 1.46 g of NaCl and add 1 mL of 0.5 M EDTA per 500 mL. Adjust the pH to 10.7.
3. Methods
3.1. Subcloning and Screening
1. Subclone the cDNA encoding the gene or open reading frame of interest into both the
pMAL-c2 and pMAL-p2 vectors for small-scale expression. The translational reading
frame of the insert must be the same as the malE gene from the vector. If no vector-
derived amino acids are wanted in a factor Xa-cleaved protein, then the open reading
frame should be cloned into the XmnI site in the vector. Bear in mind that the first encoded
amino acid in this case should not be proline or arginine, since this will inhibit factor Xa
cleavage. Otherwise, the remaining restriction sites can be used, preferably for direc-
tional cloning. If using the PCR to generate insert, proofreading polymerases will pro-
duce suitable blunt-ended fragments for XmnI cloning, and downstream primers should
include a translational stop codon. A number of cloning strategies are also presented in
the pMAL system manual.
2. Ligate 25–50 ng of suitably digested pMAL-c2 and -p2 with 1–4 times the molar amount
of insert in a standard ligation. For directionally cloned inserts, include a vector-only
control reaction.
3. Make competent TB1 or other E. coli strain (or purchase ready competent cells). We have
found the CaCl
2
method works well for TB1.
4. Mix the ligation reaction with 50 µL of competent cells, incubate on ice for 30 min, and
heat shock at 42°C for the length of time suitable for the strain used (2 min for TB1).
5. Recover the cells by adding 100–200 µL of LB and incubating at 37°C for 30 min. Spread
all of the transformation on 2 or 3 LB plates containing 100 µg/mL of ampicillin and
incubate overnight at 37°C.
6. If the insert was directionally cloned into the vector, and the vector control plate has few
or no colonies, pick colonies into 3 mL of LB with 100 mg/mL of ampicillin and shake
overnight at 37°C for plasmid minipreps (see Note 3).
7. If directional cloning was not possible and blue/white screening is necessary, either rep-
lica plate the colonies onto a fresh plate containing 100 µg/mL amp, 80 µg/mL of X-gal,
and 0.1 mM IPTG, or pick colonies with sterile toothpicks and stab or patch them onto an
LB amp plate and an LB amp/X-gal/IPTG plate and incubate at 37°C overnight. White
colonies on the X-gal plate show which colonies should be picked from the LB amp plate
for overnight liquid culture in 3 mL of LB with 100 µg/mL amp (see Note 3). Do not pick
colonies directly from plates containing IPTG.
8. Prepare miniprep DNA from the overnight cultures. Cut the DNA with suitable restric-
tion enzyme(s) and run on agarose gels to check for insert. Plasmids that appear to con-
tain insert should be sequenced to check for appropriate insert sequence and that the
reading frame across the MBP/fusion partner junction has been retained. Glycerol stocks
234 Murdoch and Iozzo
should be prepared for archival purposes and cultures restreaked on LB plates containing
100 µg/mL ampicillin for small-scale expression testing.
9. Grow a small (5-mL) culture from a single bacterial colony in LB + glucose + ampicillin,
shaking with good aeration at 37°C, to an A
600
of ~0.5. Take a 1-mL sample and pellet the
cells in a microfuge (top speed, 1 min). Aspirate the supernatant, resuspend the pellet in
50 µL of SDS-PAGE sample buffer and store at –20°C until needed (uninduced sample).
10. Add IPTG to the remaining 4 mL to 0.3 mM, and continue shaking at 37°C for 2 h.
Remove a 0.5-mL sample, microfuge as before and resuspend in 100-µL SDS-PAGE
sample buffer (induced sample).
11. Boil the uninduced and induced samples for 5 min and run 20-µL samples on an SDS-PAGE
gel of a suitable percentage for the expected fusion protein size and check for an induced band
of the correct size following staining with Coomassie blue (see Fig.1, lanes 2 and 3).
3.2. Pilot-Scale Expression—Cytoplasmic Expression
1. Grow an overnight culture of cells (5 mL) in LB + glucose + amp from a single colony of a
stock identified as expressing the fusion protein (above).
2. Add 0.8 mL of overnight culture to 80 mL (1/100) of fresh growth medium and grow as
before to an A
600
of ~0.5. Take an uninduced sample as before, add IPTG to a final con-
centration of 0.3 mM, and shake for a further 2 h (see Note 4). Take an induced sample as
before and centrifuge the remaining culture at 4000 g for 10 min. Resuspend the pellet in
10 mL of column buffer (see Note 5).
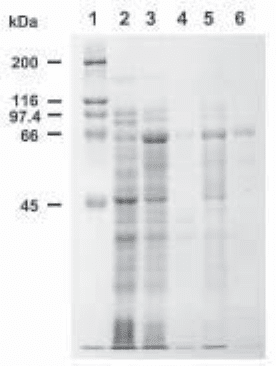
Fig. 1. Purification scheme for a maltose-binding protein/perlecan domain II fusion protein.
A cDNA encoding the entire low-density lipoprotein-like region (domain II) of the basement
membrane heparan sulfate proteoglycan perlecan was subcloned into the pMAL-c2 vector (3)
and a pilot-scale expression was performed as described under Subheading 3.1. Samples were
run on a 10% SDS-PAGE gel and stained with Coomassie brilliant blue. Lane 1, molecular
weight markers. Lane 2, uninduced crude total lysate. Lane 3, induced crude total lysate.
Lane 4, crude soluble extract. Lane 5, crude insoluble material. Lane 6, amylose resin sample:
material from the crude soluble extract that bound to the amylose resin in the small batch bind-
ing procedure detailed under Subheading 3.1.6.
Prokaryotic Expression of Proteoglycans 235
3. Freeze the suspension at –20°C (–70/80°C can also be used). Thaw in cold water.
4. Sonicate the cells to disrupt them and release protein. Prevent the suspension from overheat-
ing by using an ice-water bath and sonicating in short pulses. Protein release can be moni-
tored with the Bradford assay. Sonicate until no more protein is being released.
5. Centrifuge the sonicated suspension at 9000 g for 20 min. Remove the supernatant (crude
soluble extract) and store on ice. Resuspend the pellet in 10 mL of column buffer (crude
insoluble material).
6. Add approx 200 µL of settled amylose resin as supplied and microfuge briefly. Remove
supernatant and wash the resin twice by resuspension in 1.5 mL of column buffer and
microfuging. Resuspend the resin in 200 µL of column buffer and add 50 µL of resus-
pended slurry to 50 µL of the crude soluble extract. Incubate on ice for 15 min, microfuge
for 1 min, wash pellet with 1 mL of column buffer, microfuge again, and resuspend the
resin pellet in 50 µL of SDS-PAGE sample buffer.
7. Take 5 µL of the crude soluble extract and the crude insoluble material and add 5 µL 2×
SDS-PAGE sample buffer to each. Boil these and the uninduced, induced, and amylose
resin samples for 5 min. Microfuge the tubes to pellet the amylose resin and run 20 µL of the
supernatant along with 20 µL of the uninduced and induced total lysate samples and all of
the crude soluble and insoluble samples on an SDS-PAGE gel (see Fig.1).
3.3. Pilot-Scale Expression—Periplasmic Expression
1. Proceed as above to grow and induce a culture of cells. After centrifuging for 10 min at
4000 g (see Subheading 3.1., step 2) resuspend the pellet in 10 mL of 30 mM Tris-HCl,
pH 8.0; 20% sucrose.
2. Add EDTA to a final concentration of 1 mM (20 µL of 0.5 M EDTA to 10 mL) and
incubate at room temperature with shaking for 5–10 min.
3. Centrifuge at 8000 g at 4°C for 10 min. Decant all the supernatant and resuspend the pellet
in 10 mL of ice-cold 5 mM MgSO
4
. Shake for 10 min in an ice-water bath.
4. Centrifuge again at 8000 g for 10 min at 4°C. The supernatant is the cold osmotic shock
extract. Samples of this extract (10 µL plus 10 µL 2× SDS-PAGE sample buffer) can be
run on SDS-PAGE gels. Depending on the level of expression, immunoblotting with the
anti-MBP serum supplied with the kit may be necessary.
3.4. Affinity Chromatography
1. The maltose-binding protein will bind to the amylose resin in a variety of buffer systems at
around pH 7 and in varying ionic strengths. Intracellular proteins extracted in column buffer
are ready to apply to the resin, following dilution if necessary (see below). Proteins in
cold osmotic shock extracts should be adjusted to 20 mM Tris-HCl, pH 7.4 by the addi-
tion of 1 M Tris-HCl pH 7.4. Proteins from cytoplasmic extracts that contain intramo-
lecular disulfide bridges (such as the perlecan domain II fusion in Fig. 1) may need to be
purified over the column in a reducing environment (e.g., 10 mM β-mercaptoethanol) to
prevent loss from incorrect disulfide bonding and aggregation. A refolding protocol may
then be necessary for the downstream application (see Note 6).
2. Pour an amylose resin column suitable for the amount of protein to be purified. The pMAL
system handbook suggests a 2.5 × 10 cm column; we have found K9/15 and XK16/20
columns from Pharmacia and 1 × 10 cm econocolumns from Bio-Rad to be satisfactory.
Column size and bed volume should be adjusted to the expected yield given a binding
capacity of ~3 mg/mL packed resin. We have generally used 10-mL columns (~30 mg
binding capacity is more than enough for many purposes, and larger bed volumes only
need longer washing steps). Wash the column with 8 vol of column buffer.
