Hermanson G. Bioconjugate Techniques, Second Edition
Подождите немного. Документ загружается.

1020 28. Bioconjugation in the Study of Protein Interactions
5. Add the modifi ed bait protein to a sample containing potentially interacting prey proteins
and incubate for 1 hour protected from light. The sample may be cells (for cell–surface
interaction studies), cell lysate, or various extracts from cells, tissues, or biological fl uids.
6. Expose the sample to UV light to crosslink and capture interacting proteins via the pho-
toreactive group.
7. Recover interaction complexes by affi nity chromatography. This is typically done using
immobilized antibodies to the bait protein or using fusion tag binders, if the bait pro-
tein contains a fusion partner (e.g., immobilized glutathione for binding GST labeled bait
proteins).
8. Analyze the purifi ed complexes by SDS-PAGE, Western blotting, and mass spec.
3. Trifunctional Label Transfer Reagents
The earliest examples of label transfer reagents were cleavable heterobifunctional compounds
that incorporated a phenyl azide group, which also had a phenolic modifi cation on the ring.
The phenolic hydroxyl activates the ring for substitution reactions to occur ortho or para to its
position. These compounds thus can be radioiodinated using typical oxidation reagents such as
chloramine-T or Iodobeads. Iodination of the crosslinker with
125
I prior to its use results in a
radioactive label transfer reagent that can tag an unknown interacting protein with a radiola-
bel after cleavage of the crosslinker ’s spacer arm.
Subsequent designs of label transfer reagents used non-radioactive labels to avoid the safety
and regulatory issues posed by
125
I. Fluorescent constituents designed into cleavable photoreac-
tive crosslinkers make possible transfer of a fl uorescent label to an unknown interacting pro-
tein. An example of this type of reagent that incorporates a coumarin group is SAED, which
has been derivatized with an azido group on the aromatic ring. The reagent is non-fl uorescent
prior to exposure to UV light, but upon photoactivation and coupling to interacting proteins,
it becomes highly fl uorescent. The reagent also has a disulfi de bond that can be reduced, result-
ing in cleavage of the crosslinked proteins and transfer of the label to the unknown interacting
species. In this case, the fl uorescently labeled interacting proteins can be followed in cells to
determine the site of interactions or the fate of the proteins after interacting.
The most advanced type of bioconjugation reagent that is designed to study interacting pro-
teins is a trifunctional label transfer reagent. These compounds are a special category of trifunc-
tional crosslinkers (see Chapter 6), which possess two reactive groups and a third arm containing
a label. Typically, one of the reactive groups is thermoreactive and can be used to label a bait pro-
tein, while the second reactive group is usually photoreactive for selective activation and coupling
upon UV irradiation. The third arm of the reagent can be designed to have a fl uorescent group
for detection or terminate in a biotin group for both detection and purifi cation ( Figure 28.10 ).
An important feature of trifunctional label transfer reagents is the presence of a cleavable
cross-bridge on the arm containing the thermoreactive group. After a bait protein has been
conjugated with an interacting prey protein using this type of reagent, the complex then can be
cleaved, which releases the interacting proteins and transfers the label over to the prey protein.
The label can be used to detect prey proteins on a Western blot or to purify them from the sam-
ple solution. Unlike the heterobifunctional compounds discussed previously for studying pro-
tein interactions, label transfer reagents allow specifi c detection and purifi cation of unknown
interacting prey proteins for subsequent analysis.
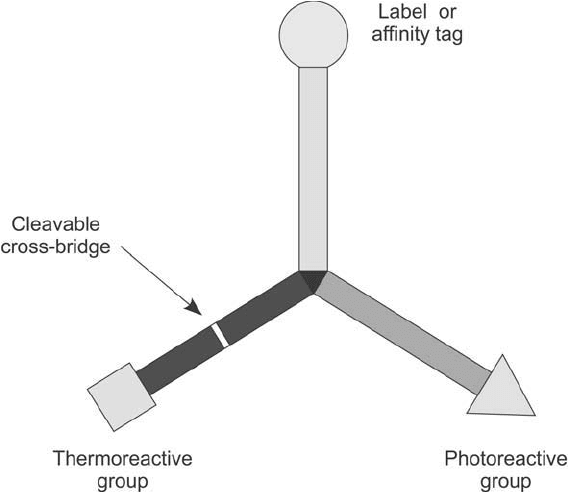
Figure 28.10 Trifunctional label transfer agents contain two arms with terminal reactive groups and a third
arm with a label or affi nity tag, such as a biotin group. One of the reactive groups typically is thermoreactive to
couple with bait proteins, while the second reactive group usually is photoreactive. The thermoreactive arm has
a cleavable cross-bridge to facilitate release of the captured protein and transfer of the label of affi nity tag to it.
The following sections describe three types of label transfer reagents, which are all built
with a biotin handle.
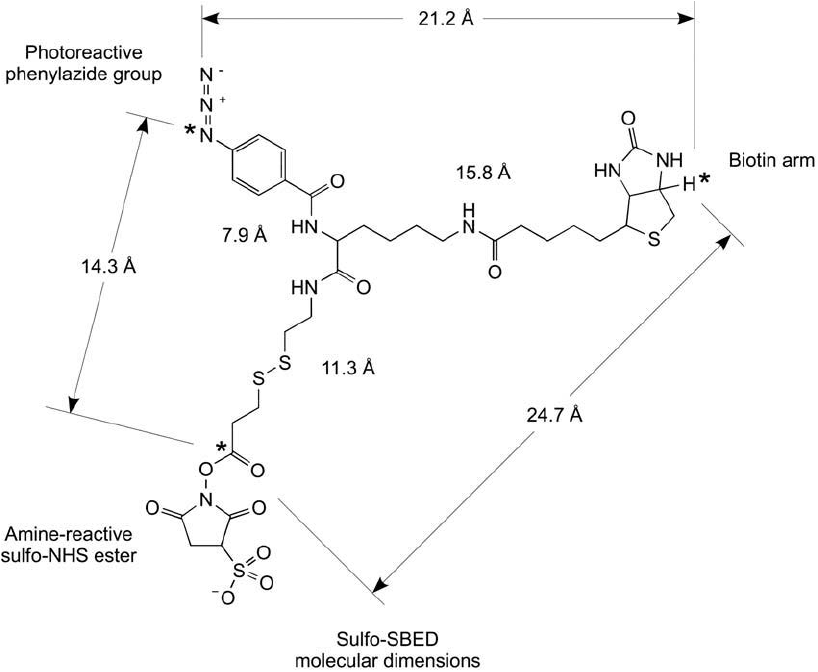
3.1. Sulfo-SBED
Sulfo-SBED is sulfosuccinimidyl-2-[6-(biotinamido)-2-( p-azidobenzamido) hexanoamido] ethyl-
1,3-dithiopropionate), a trifunctional reagent containing an amine reactive sulfo-NHS ester
on one arm, a photoreactive phenyl azide group on the second arm, and a biotin handle on
its third arm ( Figure 28.11 ; see Chapter 6, Section 2 for a general description and use). This
compound has found great utility in capturing protein interactions and subsequently isolating
them for analysis. A purifi ed bait protein initially is labeled with Sulfo-SBED by its sulfo-NHS
ester to form amide linkages through reaction with lysine or N-terminal amines. The modifi ed
bait then is incubated with a sample and biomolecules are allowed to interact with it. Finally,
the sample is exposed to UV light and the photoreactive group is able to couple with nearby
amine groups on any interacting proteins. The resultant complex can be purifi ed, identifi ed, or
detected through specifi c binding to the biotin groups ( Figures 28.12 and 28.13 )
Sulfo-SBED is soluble in organic solvents, such as DMSO (125 mM), DMF (170 mM), and
methanol (12 mM), or to a lesser degree in pure water ( 5 mM). The solubility of Sulfo-SBED in
3. Trifunctional Label Transfer Reagents 1021
1022 28. Bioconjugation in the Study of Protein Interactions
buffered aqueous solutions may vary from about 0.1 mM to 3 mM (e.g., 1 mM in 0.1 M PBS).
However, to dissolve Sulfo-SBED at higher concentrations in a reaction buffer, fi rst dissolve
it in a water-miscible organic solvent such as DMSO or DMF and transfer an aliquot to the
aqueous buffer solution. Use no more than about 1–10 percent of solvent in the fi nal buffered
reaction mixture to prevent buffer precipitation and minimize the denaturation of proteins.
Bait proteins modifi ed with Sulfo-SBED may precipitate if the level of modifi cation is too high,
primarily due to the hydrophobic nature of the crosslinker and the biotin handle. To prevent
precipitation or at least minimize it, adjust the molar excess of Sulfo-SBED over the bait pro-
tein to a level where the protein remains in solution. Some precipitation may be removed by
centrifugation or fi ltration prior to use.
A derivative of Sulfo-SBED containing a thiol-reactive pyridyl disulfi de group on its thermo-
reactive arm has been reported for modifi cation of bait proteins containing a cysteine residue.
Figure 28.11 Sulfo-SBED is a label transfer agent that contains a water-soluble sulfo-NHS ester to label bait
proteins and a phenyl azide group for photoreactive capture of a prey protein. The biotin label can be used for
detection or isolation of protein–protein conjugates using (strept)avidin reagents. The stars indicate the atoms
that were used to measure the indicated molecular dimensions.
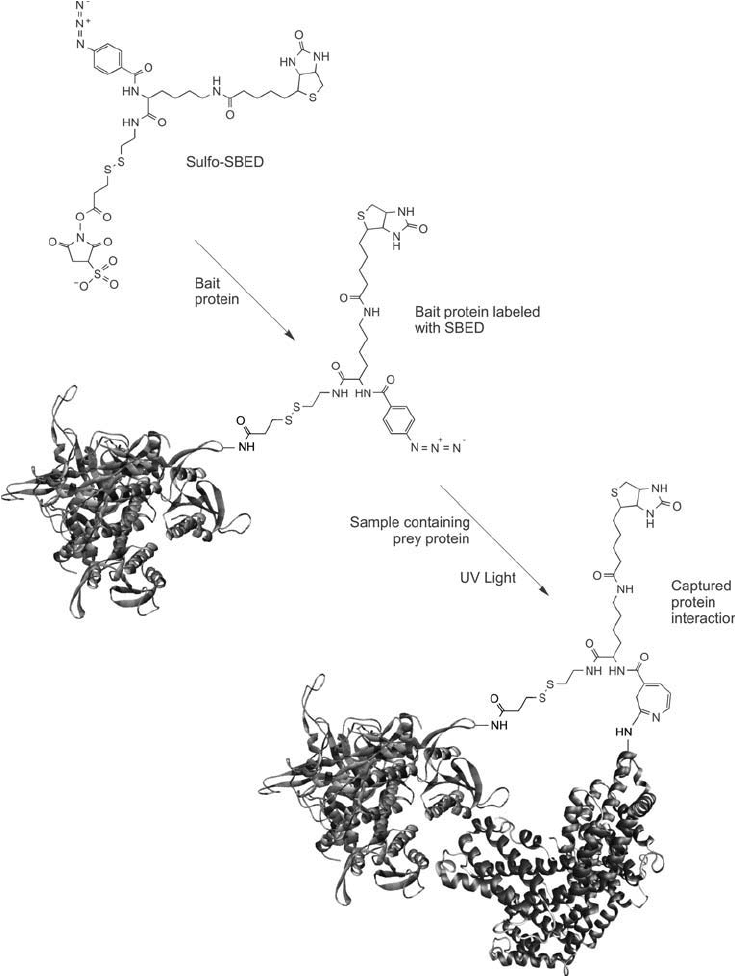
Figure 28.12 Sulfo-SBED fi rst is used to label a bait protein through reaction of the sulfo-NHS ester with avail-
able amine groups on the protein, yielding an amide bond linkage. This labeled bait protein then is added to a
sample containing proteins that potentially could interact with the bait. After an incubation period, the sample
is exposed to UV light to photoactivate the phenyl azide group. This reaction causes any interacting prey pro-
teins to be crosslinked with the bait protein, forming a complex containing a biotin affi nity tag.
3. Trifunctional Label Transfer Reagents 1023
1024 28. Bioconjugation in the Study of Protein Interactions
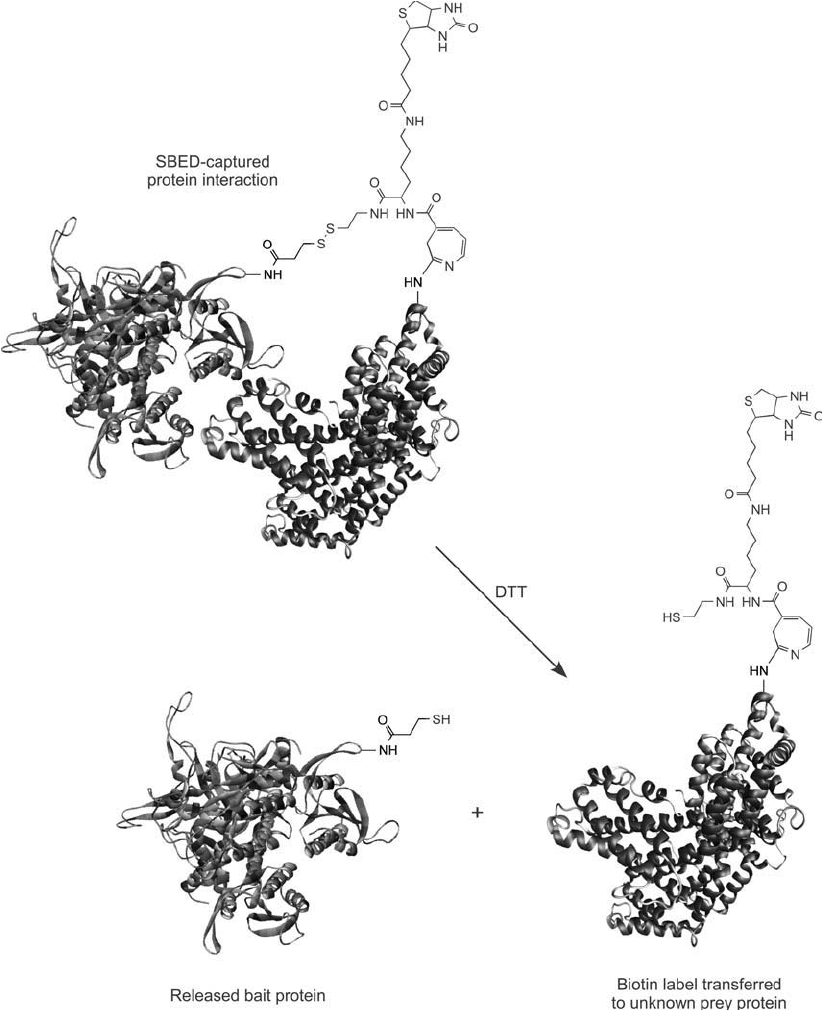
Figure 28.13 A sulfo-SBED-captured protein interaction can be released using DTT to cleave the disulfi de within
the cross-bridge leading to the bait protein. The result transfers the biotin label to the unknown interacting pro-
tein. The biotin tag thus allows the interacting protein to be detected or isolated using (strept)avidin reagents.
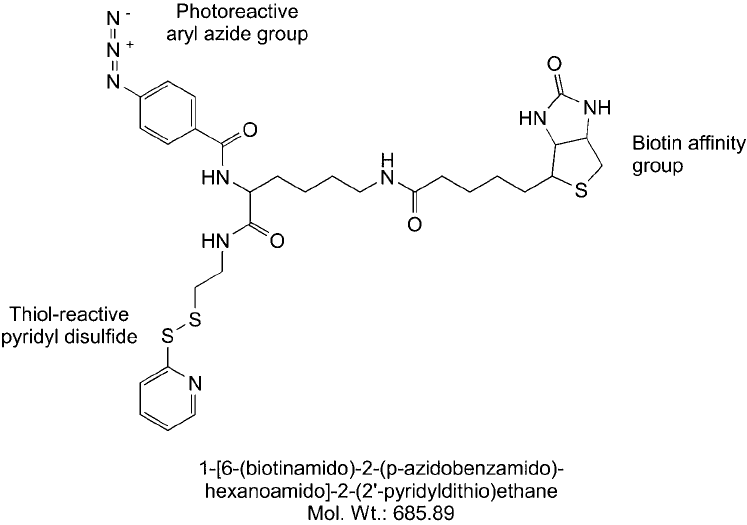
Figure 28.14 A trifunctional label transfer reagent containing a thiol-reactive pyridyl disulfi de group, a photo-
reactive phenyl azide, and a biotin affi nity tag. This compound can be used to label bait proteins through avail-
able thiol groups and capture interacting prey proteins by photoreactive conjugation.
Reaction of this compound with a thiol results in the formation of a reversible disulfi de linkage.
After capturing interacting proteins with this reagent, reduction of the disulfi de transfers the
biotin label to the unknown prey protein(s). This compound was prepared from Sulfo-SBED
by reduction of the disulfi de in its sulfo-NHS ester arm followed by reaction with 2,2 -
dithiodipyridine to give the thiol-reactive compound, 1-[6-(biotinamido)-2-( p-azidobenzamido)-
hexanoamido]-2-(2-pyridyldithio)ethane ( Figure 28.14 ) (Alley et al., 2000; Ishmael et al.,
2001, 2002; Trakselis, 2005). This derivative has most of the features of Sulfo-SBED in terms
of being a trifunctional label transfer agent containing biotin, but extends its application to
sulfhydryl-containing bait proteins.
Sulfo-SBED has been used to investigate protein–protein interactions in the following appli-
cations: studying the bacteriophage T4 replisome (Ishmael et al., 2003), cell–surface antigens
of mycoplasma species bovine group 7 in binding and activation of plasminogen (Bower et al .,
2003), bacterium–host protein–carbohydrate interactions (Ilver et al., 2003), transcription acti-
vator interactions with multiple SWI/SNF subunits (Neely et al., 2002), binding of protein D/E
to the surface of rat epidermal sperm (Tubbs et al., 2002), for the study of the gp41–gp59 com-
plex in bacteriophage T4 helicase (Ishmael et al., 2002), identifi cation of a region in alcohol
dehydrogenase that binds to -crystallin during chaperone action (Santhoshkumar and Sharma,
2002), regions of the mouse CD14 molecule required for toll-like receptor 2- and 4-mediated
3. Trifunctional Label Transfer Reagents 1025
1026 28. Bioconjugation in the Study of Protein Interactions
activation of NF-kB (Muroi et al., 2002), active site residues of cyclophilin A that are important
for signaling via CD147 (Yurchenko et al., 2002), mapping protein–protein interactions in
the bacteriophage T4 DNA polymerase holoenzyme (Alley et al., 2000), import of adenovirus
DNA involving the nuclear pole complex receptor CAN/Nup214 and histone H1 interactions
(Trotman et al., 2001), insulin-like growth factor (IGF)-1 interaction with IgG-binding pro-
teins (Horney et al., 2001), 3F3/2 anti-phospho-epitope antibody binding to the mitotically
phosphorylated anaphase-promoting complex/cyclosome (Daum et al., 2000), SH3 binding
sites of ZG29p in its interaction with amylase (Kleene et al., 2000), studying the proteasome
activator PA28 in Hsp90-dependent protein refolding (Minami et al., 2000), investigating
functional elements in -crystallin (Sharma et al., 2000), Helicobacter pylori adhesin bind-
ing to fucosylated histo-blood group antigens (Ilver et al., 1998), identifi cation of low abun-
dance proteins by electrophoresis and MALDI-TOF MS (Bergstrom et al., 1998), molecular
probes for muscarinic receptors (Jacobson et al., 1995), in the quantitation of triple-helix for-
mation (Geselowitz and Neumann, 1995), studying prion protein interactions with its recep-
tor (Santuccione et al., 2005), the activation of Hsp70 chaperones (Steel et al., 2004), effect
of oxidized B3-crystallin peptide on lens L-crystallin and its interaction with B 2-crystallin
(Udupa and Sharma, 2005), the effect of oxidized B3-crystallin peptide on the thermal aggre-
gation of bovine lens gamma-crystallins (Udupa and Sharma, 2005), for the mass spectromet-
ric detection of affi nity purifi ed crosslinked peptides (Hurst et al., 2004), in mapping protein
interfaces combined with MALDI-TOF and ESI-FTICR mass spectrometry (Sinz et al., 2005),
studying the activation of the antioxidant enzyme 1-CYS peroxiredoxin and its requirement
for glutathionylation mediated by binding with GST (Manevich et al., 2004), investigating the
recruitment of HAT complexes by direct activator interactions with the ATM-related Tra1 sub-
unit (Brown et al., 2001), the identifi cation of annexin A2 heterotetramer as a receptor for the
plasmin-induced signaling in human peripheral monocytes (Laumonnier et al., 2005), and in
the role of the proteasome activator PA28 in the Hsp90-dependent protein refolding (Minami
et al ., 2000).
In addition to the use of Sulfo-SBED to capture unknown prey proteins interacting with
labeled bait proteins, the reagent also may be used to study the interaction interfaces between
two known proteins that specifi cally interact. Sinz et al. (2005) used this approach to identify
the interaction surfaces between calmodulin and M13, which is a short peptide from skeletal
muscle light chain kinase. In this application, calmodulin was labeled with Sulfo-SBED and
allowed to interact with purifi ed M13. After an incubation period, the solution was exposed
to UV light and the two proteins were crosslinked together. Next, the conjugated proteins were
proteolytically digested with trypsin and the resultant biotinylated peptides were enriched on
a column of immobilized monomeric avidin (Thermo Fisher). The crosslinked peptides come
from one peptide segment of calmodulin and one peptide segment of M13. Finally, these bioti-
nylated and crosslinked peptides were identifi ed using nano-HPLC separation into a nano-
ESI-FTICRMS with software-facilitated deconvolution of the peptide identities. Such analysis
provides insight into the interaction surfaces involved with the binding event between the two
proteins.
The following protocol represents a suggested method that will work well for many pro-
teins. It is a blend of protocols used in the literature and recommended by Thermo Fisher in
the Sulfo-SBED instruction manual. Modifi cations to reaction conditions may be necessary in
certain cases to maintain protein stability or solubility, depending on the properties of the par-
ticular bait protein being used.
Protocol
1. Dissolve a purifi ed bait protein in 0.1 M sodium phosphate, 0.15 M NaCl, pH 7.2, or a
similar buffer at neutral pH, which doesn ’t contain any competing amines (i.e., avoid Tris
or Imidazole). The bait protein may be at a concentration of anywhere from 0.1 mg/ml
to 10 mg/ml. Prepare the solution in a dark tinted vial or wrap the vessel in foil to prevent
light exposure when the crosslinker is added.
2. Prepare a solution of Sulfo-SBED in dry DMF or DMSO at a concentration of 40 g/ml.
Protect from light.
3. Add a quantity of the Sulfo-SBED solution to the bait protein solution so that a 1- to
5-fold molar excess of crosslinker over the bait protein results in the reaction mixture. Mix
well. Using greater quantities of Sulfo-SBED to the bait protein may result in precipitation
due to the hydrophobic nature of crosslinker. In addition, over modifi cation of the bait
protein with the crosslinker may block sites of protein interaction, thus preventing com-
plex formation. As a practical example, Horney et al. (2001), used a 1:1 molar ratio of
Sulfo-SBED to the bait protein IGF-1 with success.
4. React for 30 minutes at room temperature or 2 hours at 4°C. Continue to protect the
solution from light.
5. Remove excess crosslinker and reaction by-products by dialysis or size exclusion chroma-
tography. For small quantities of bait proteins, dialysis may be the better choice, because
gel fi ltration columns often bind nonspecifi cally enough protein to make recoveries unac-
ceptably low.
6. Add the labeled bait protein to a sample containing the putative interacting prey pro-
teins. The quantity of bait protein to be added to a given sample should be within the
same concentration level as the amount of prey proteins present. The optimal level of
addition may have to be determined by varying the amount of bait protein concentra-
tions in a number of sample solutions to decide which concentration results in the best
interaction complexes being formed.
7. Incubate the sample mixture for at least 1 hour at room temperature or under conditions
optimal for the interaction to be studied.
8. Expose the sample to UV light to photoactivate the aryl azide and cause the conjuga-
tion reaction to occur. For best results, use a UV lamp that irradiates in the range of
300–370 nm. Irradiate for 2–15 minutes, using a briefer exposure when using UV lamps
of greater wattage.
9. Interaction complexes that were captured by the crosslinking reaction can be recovered or
analyzed using the biotin groups. An immobilized streptavidin support, for instance, can
be used to purify the conjugates away from other sample proteins. Alternatively, a strepta-
vidin–HRP conjugate can be used to probe a Western blot after electrophoresis separa-
tion of the sample. Reduction of the conjugate may be done by cleavage of the disulfi de
bond with DTT. The typical addition of DTT to the electrophoresis sample buffer will
facilitate disruption of the complexes and transfer of the biotin label to the interacting
prey proteins. This reduction step also can be done using 50 mM DTT or 50 mM TCEP
in aqueous solution to permit recovery of the biotinylated prey proteins on immobilized
streptavidin. Alternatively, the use of an affi nity support having milder elution character-
istics for biotinylated proteins, such as immobilized monomeric avidin, would facilitate
isolation of the biotinylated complexes or prey proteins under non-denaturing conditions.
3. Trifunctional Label Transfer Reagents 1027
1028 28. Bioconjugation in the Study of Protein Interactions
3.2. MTS-ATF-Biotin and MTS-ATF-LC-Biotin
MTS-ATF-Biotin is 2-[N2-(4-Azido-2,3,5,6-tetrafl uorobenzoyl)-N6-(6-biotinamidocaproyl)- L -
lysinyl]ethyl methanethiosulfonate and MTS-ATF-LC-Biotin is 2-{N2-[N6-(4-Azido-2,3,5,6-
tetrafl uorobenzoyl)-6-aminocaproyl]-N6-(6-biotinamidocaproyl)- L-lysinylamido}]ethyl meth-
anethiosulfonate. Both reagents are trifunctional crosslinkers similar in design to Sulfo-SBED
discussed previously, but in addition to the biotin handle, they contain a thiol-reactive group
and an enhanced photoreactive, perfl uorinated phenyl azide group. The two reagents differ
in the length of the cross-bridge in the photoreactive arm, with the LC version containing an
extended aminocaproyl spacer. Relative to the spacing possible between the reactive groups on
these compounds, the LC version therefore provides nearly twice the maximal molecular dis-
tance over its shorter analog (21.8 Å versus 11.1 Å) ( Figure 28.15 ). Thus, interacting proteins
may be captured either through use of a long or short crosslink, depending on the optimal dis-
tances between the proteins–or at least to the nearest thiol on the bait protein.
Both MTS-ATF-Biotin and MTS-ATF-LC-Biotin contain a methanethiolsulfonate group (MTS)
on their thermoreactive arm, which is able to couple with the sulfhydryl on a cysteine residue.
This reaction proceeds with loss of the methyl sulfonate leaving group (sulfi nic acid) to pro-
duce a disulfi de linkage ( Figure 28.16 ). Unlike a pyridyl disulfi de group, however, which also
reacts with thiols to form disulfi de linkages, the MTS group is unstable to hydrolysis in aqueous
solution, especially if other strong nucleophiles are present. Therefore, most MTS compounds
dissolved or brought into PBS buffer at physiological pH will hydrolyze with a half-life on the
order of 10–15 minutes. However, they also have very rapid reactivities with thiols (Stauffer
and Karlin, 1994; Holmgren et al., 1996; Liu et al., 1996). If the parent MTS reagent is fully
water-soluble, the reaction with a thiol on a bait protein can take place with high yields in just a
matter of minutes. However, both of these trifunctional label transfer compounds are hydropho-
bic, so their MTS reactivity in the aqueous phase may be somewhat slower than corresponding
hydrophilic MTS reagents.
For use in studying protein interactions, these compounds fi rst are reacted with a thiol on a
purifi ed bait protein to form a disulfi de bond. Since the reagents are water-insoluble, they must
be dissolved in an organic solvent such as DMF or DMSO and then an aliquot added to the bait
protein in an aqueous buffer to initiate the reaction. Once modifi ed, the biotinylated bait protein
then is incubated with a sample containing potential interactive prey proteins. After an incuba-
tion period, initiating the photoreaction by exposure to UV irradiation captures the interacting
proteins. Any interaction complexes thus formed can be isolated or detected using the biotin
handle. In addition, the disulfi de bond formed with the bait protein during the crosslinking
reaction can be reduced to cleave the conjugates and transfer the biotin label to the unknown
interacting prey proteins. This is a powerful way of labeling unknown interacting proteins for
subsequent analysis.
Although MTS-ATF-Biotin and MTS-ATF-LC-Biotin are available commercially (Thermo
Fisher and Toronto Research), they are relatively new and don ’t have the publications or appli-
cations backing up their use as has Sulfo-SBED. Therefore, the following protocol should be
used as a suggested starting point to develop optimized methods for studying specifi c protein
interactions. The bait protein to be modifi ed with MTS-ATF-Biotin or MTS-ATF-LC-Biotin
should be purifi ed and contain at least one accessible thiol for reaction with the MTS group.
Ellman’s reagent (Chapter 1, Section 4.1) may be used to determine if a sulfhydryl is present
and able to react with the crosslinkers. A free thiol also may be created by reduction of internal
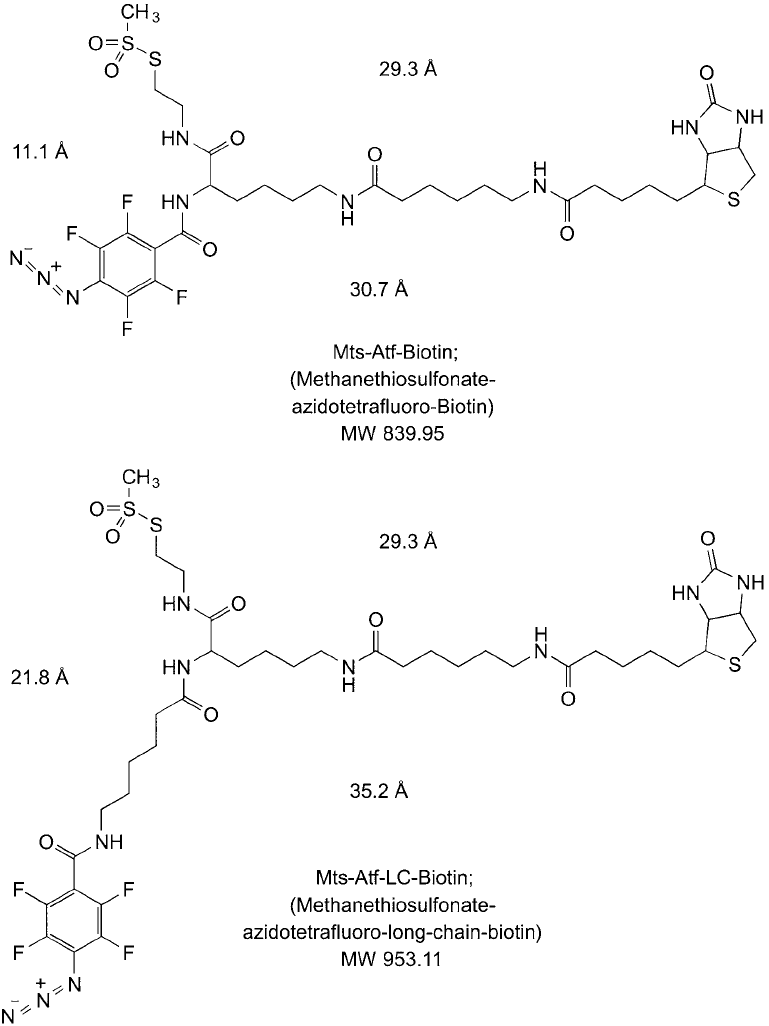
Figure 28.15 Two similar label transfer reagents containing a thiol-reactive methanethiolsulfonate group to
label bait protein through available sulfhydryls, a tetrafl uorophenyl azide group for high-effi ciency photoreac-
tive conjugation with interacting prey proteins, and a long biotin affi nity tag.
3. Trifunctional Label Transfer Reagents 1029
