Hermanson G. Bioconjugate Techniques, Second Edition
Подождите немного. Документ загружается.

Protocol
1. Prepare diazotized -aminobenzoyl biocytin by using the protocol outlined in Chapter 11,
Section 5, but instead of starting with 2 mg the biotinylation reagent dissolved in 40 l of
1 N HCl, use 9 mg in 180 l of 1 N HCl. Proportionally scale up the other reactant quanti-
ties used in the protocol. After the reaction is complete, immediately adjust the pH of the
fi nal solution to 9.
2. Add 1 g of single-stranded DNA to the above solution.
3. React for 30 minutes at room temperature.
4. Purify the biotinylated DNA probe by ethanol precipitation, gel fi ltration, n -butanol
extraction, or dialysis as discussed in previous sections.
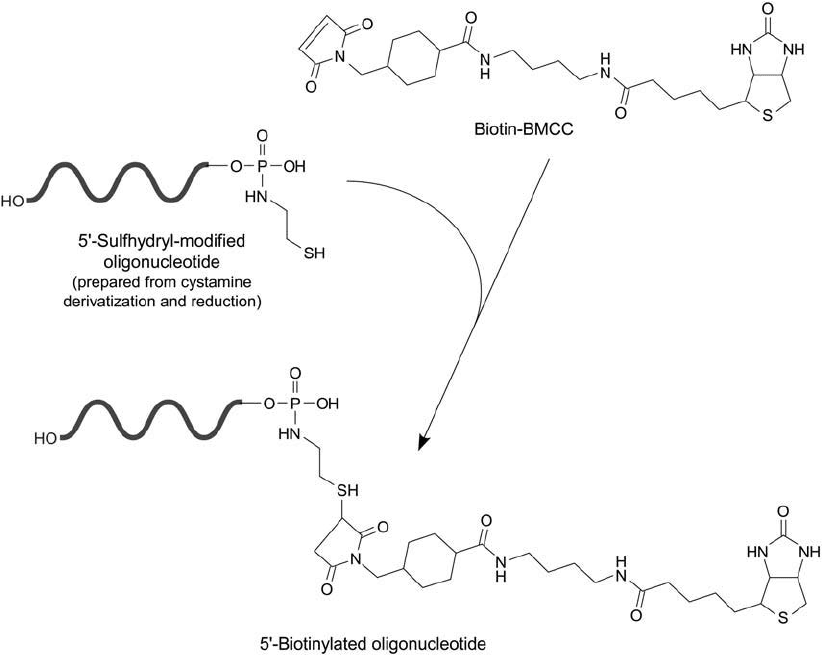
Reaction of Biotin–BMCC with Sulfhydryl-Modifi ed DNA
Biotin–BMCC is a sulfhydryl-reactive biotinylation reagent containing a maleimide group at
the end of an extended spacer arm. The long spacer (32.6 Å) provides enough distance between
modifi ed oligonucleotides and the bicyclic biotin end to allow effi cient binding of (strept)avidin
probes, even when hybridized to target sequences. The reagent may be used to add a biotin
label to DNA or RNA molecules after they have been modifi ed to contain thiol groups. For
instance, cystamine labeling at the 5 phosphate group of DNA via carbodiimide-mediated
phosphoramidate formation followed by disulfi de reduction (Section 2.2, this chapter) can cre-
ate the required sulfhydryl groups. Subsequent reaction with biotin–BMCC results in a deriva-
tive labeled only at an end of the DNA probe ( Figure 27.12 ), thus avoiding the potential for
hydrogen bonding interference in hybridization assays.
Since maleimide groups are highly specifi c for coupling to thiols in the pH range of 6.5 to
7.5, side reaction products can be avoided. The reaction is complete within two hours at room
temperature.
Protocol
1. Prepare 10–20 g of a sulfhydryl-containing oligonucleotide in 200 l of 50 mM sodium
phosphate, 10 mM EDTA, pH 7.2 (the methods outlined in Section 2.2 of this chapter
can be used to form the thiol group).
2. Dissolve biotin–BMCC in DMSO at a concentration of 5 mg/ml. Prepare fresh.
3. Add 50 l of the biotinylation solution to the oligo. Mix well.
4. React for 2 hours at room temperature.
5. Isolate the biotinylated probe by ethanol/salt precipitation as described in Section 1 for
nick translation (this chapter).
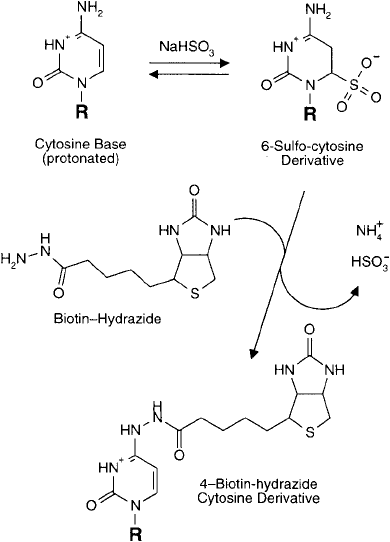
Biotin–Hydrazide Modifi cation of Bisulfi te-Activated Cytosine Groups
Biotin–hydrazide is the hydrazine derivative of D-biotin prepared using its valeric acid carboxy-
late (Chapter 11, Section 3). The hydrazide group typically is used to react with aldehyde and
ketone groups to give hydrazone linkages. However, the hydrazide compound also can undergo
transamination reactions with cytosine residues via catalysis with bisulfi te (Section 2.1, this chap-
ter) ( Figure 27.13 ). DNA or RNA probes containing cytosine groups may be modifi ed to contain
biotin labels using a simple, one-step procedure (Reisfeld et al., 1987). The detection limit of
DNA probes biotinylated using this technique can be less than 1 pg on blots, when analyzed
990 27. Nucleic Acid and Oligonucleotide Modifi cation and Conjugation
using a streptavidin–HRP conjugate with chemiluminescent detection. A longer chain analog of
biotin–hydrazide, biotin-LC-hydrazide, may be used to create an extended spacer between the
oligonucleotide and the bicyclic biotin group, thus increasing the binding effi ciency of avidin or
streptavidin conjugates. Alternatively, a hydrophilic biotin–PEG–hydrazide compound may be
used in this reaction to provide greater water solubility for the label (Chapter 18, Section 3).
Leary et al. (1983) reported that increasing the spacer arm length from 4 to 11 atoms when
biotinylating DNA probes can increase the detectability of the target DNA approximately 4-fold.
The following method is adapted from Reisfeld et al . (1987).
Protocol
1. Prepare 50 g of a single-stranded DNA probe in 300 l of 50 mM sodium acetate, pH 4.5.
2. Dissolve biotin–hydrazide in water at a concentration of 10 mg/ml.
3. Add 300 l of the biotin–hydrazide solution to the DNA solution.
4. Add sodium bisulfi te to obtain a fi nal concentration of 1 M in the reaction medium.
2. Chemical Modifi cation of Nucleic Acids and Oligonucleotides 991
Figure 27.12 Biotin–BMCC can be used to modify a reduced, cystamine derivative of DNA, forming a thioether
linkage.
5. React for 24 hours at 37 °C.
6. Remove excess reactants by dialysis against water at 4 °C.
2.4. Enzyme Conjugation to DNA
Enzymes useful for detection purposes in ELISA techniques (Chapter 26) also can be employed
in the creation of highly sensitive DNA probes for hybridization assays. The attached enzyme
molecule provides detectability for the oligonucleotide through turnover of substrates that can
produce chromogenic or fl uorescent products. Enzyme-based hybridization assays are perhaps
the most common method of nonradioactive detection used in nucleic acid chemistry today.
The sensitivity of enzyme-labeled probes can approach or equal that of radiolabeled nucleic
acids, thus eliminating the need for radioactivity in most assay systems.
The conjugation reactions involved in DNA–enzyme crosslinking are not unlike the methods
used to form antibody–enzyme conjugates (Chapter 20, Section 1). Bifunctional crosslinkers
can be used to couple a modifi ed oligonucleotide to an enzyme molecule using the same basic
principles effective in protein–protein conjugation. The only requirement is that the DNA mole-
cule be modifi ed to contain one or more suitable functional groups, such as nucleophiles like
amines or sulfhydryls. The modifi cation process used to create these functional groups can use
enzymatic (Section 1, this chapter) or chemical (Section 2, this chapter) means and it can result
992 27. Nucleic Acid and Oligonucleotide Modifi cation and Conjugation
Figure 27.13 Biotin–hydrazide may be incorporated into cytosine bases using a bisulfi te-catalyzed transamina-
tion reaction.
in random incorporation of modifi cation sites or be directed exclusively at one end of the DNA
molecule, such as in 5 phosphate coupling.
The following sections describe some of the more common procedures of preparing DNA–
enzyme conjugates.
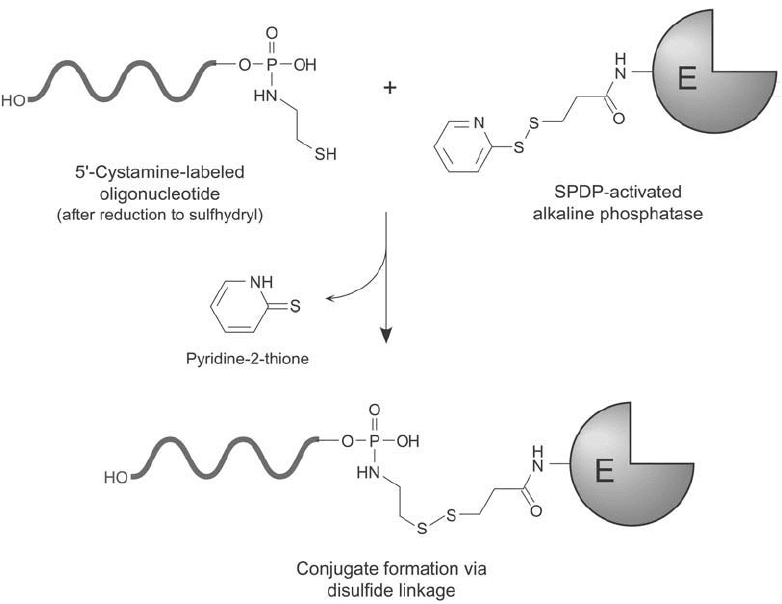
Alkaline Phosphatase Conjugation to Cystamine-Modifi ed DNA Using Amine- and
Sulfhydryl-Reactive Heterobifunctional Crosslinkers
A cystamine group added to the 5 phosphate of DNA molecules using a carbodiimide reaction
(Section 2.2, this chapter) can be used in a heterobifunctional crosslinking scheme to conju-
gate with alkaline phosphatase. Crosslinking agents containing an amine-reactive portion and
a sulfhydryl-reactive part work best in forming this type of conjugate. Perhaps one of the most
common heterobifunctional reagents used for DNA–enzyme formation is SPDP (Chapter 5,
Section 1.1). SPDP contains an NHS ester on one end able to create an amide bond linkage
with amino groups on protein molecules. After modifi cation of alkaline phosphatase with
this crosslinker, the enzyme is activated to contain pyridyl disulfi de groups for coupling to the
sulfhydryls on a cystamine-modifi ed DNA probe ( Figure 27.14 ). The reaction forms disulfi de
2. Chemical Modifi cation of Nucleic Acids and Oligonucleotides 993
Figure 27.14 An oligonucleotide modifi ed with cystamine and reduced to generate a free sulfhydryl may be
conjugated with an SPDP-modifi ed enzyme, forming a disulfi de linkage.
bonds between the oligonucleotide and the alkaline phosphatase enzyme. Since the crosslink
occurs only at the 5 end of the DNA strand, the presence of an enzyme molecule does not
adversely affect the ability of base pairing and hybridization to a target sequence.
The following protocol assumes that the labeling process used to create a sulfhydryl-modi-
fi ed DNA probe already has been done according to the method of Section 2.2 (this chap-
ter). The modifi cation procedure for activating alkaline phosphatase with SPDP may be done
according to the protocol described in Chapter 5, Section 1.1. To obtain effi cient labeling of
all the alkaline phosphatase added to the reaction medium, the modifi ed oligo is reacted in
a 10-fold molar excess. Reaction of the DNA probe in excess allows easy separation of not-
coupled oligo from conjugated probe, thus eliminating any potential interference in hybridi-
zation assays due to unlabeled oligonucleotide.
Protocol
1. Dissolve a 5 -sulfhydryl-modifi ed oligonucleotide in water or 10 mM EDTA at a con-
centration of 0.05–25 g/ l. Calculate the total nmoles of oligo present based upon its
molecular weight.
2. Prepare SPDP-activated alkaline phosphatase in 50 mM sodium phosphate, 0.15 M NaCl,
10 mM EDTA, pH 7.2. Add to the oligo solution, an amount of the activated enzyme
representing a 10-fold molar excess over the calculated amount of DNA present.
3. React at room temperature for 30 minutes with gentle mixing.
4. The alkaline phosphatase–DNA conjugate may be purifi ed away from excess oligo by
dialysis, gel fi ltration, or through the use of centrifugal concentrators. A simple way of
removing unreacted oligo is to use Centricon-30 concentrators (Amicon) which have a
molecular weight cutoff of 30,000. Since the enzyme molecular weight is approximately
140,000 and the conjugate is even higher, a relatively small DNA probe will pass through
the membranes of these units while the conjugate will not. To purify the conjugate using
Centricon-30 s, add 2 ml of the phosphate buffer from step 2 to one concentrator unit,
then add the reaction mixture to the buffer and mix. Centrifuge at 1,000 g for 15 minutes
or until the retentate volume is about 50 l. Add another 2 ml of buffer and centrifuge
again until the retentate is 50 l. Invert the Centricon-30 unit and centrifuge to collect
the retentate in the collection tube provided by the manufacturer.
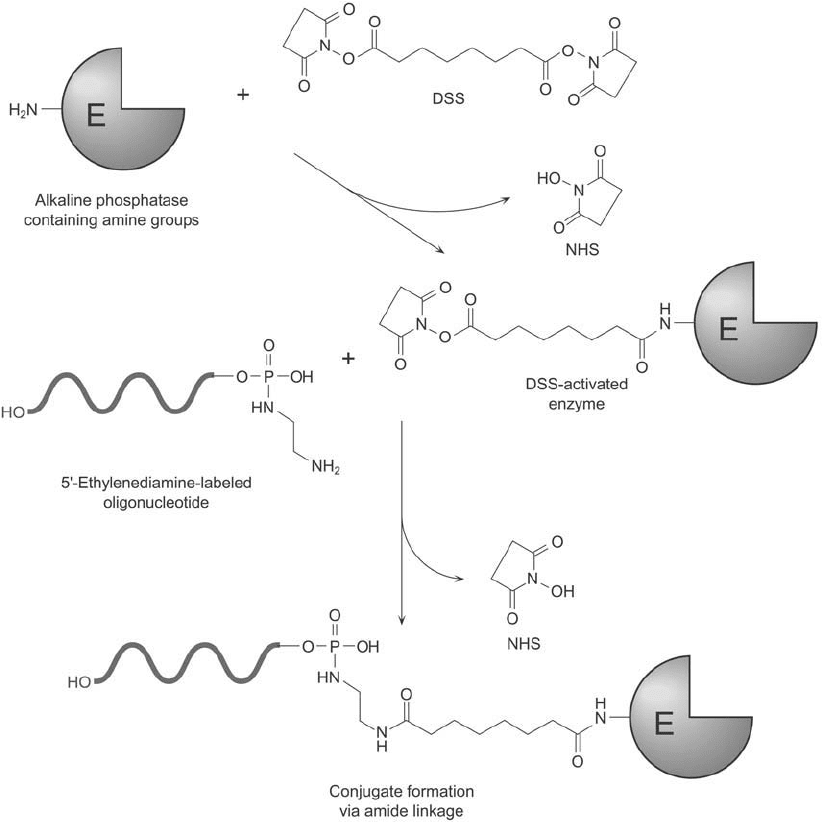
Alkaline Phosphatase Conjugation to Diamine-Modifi ed DNA Using DSS
Disuccinimidyl suberate (DSS) is a homobifunctional crosslinker containing an amine-reac-
tive NHS ester at both ends (Chapter 4, Section 1.2). Reaction of the reagent in excess with
diamine-modifi ed DNA probes creates an activated intermediate able to conjugate with enzyme
molecules through their available amine groups ( Figure 27.15 ) (Jablonski et al., 1986). The
coupling reaction produces stable amide linkages under mildly alkaline conditions. Although
the following protocol has been used to label oligonucleotides with success, it may be less effi -
cient than the previous protocol at forming the desired conjugate due to the homobifunctional
nature of the crosslinker. During the activation step, the modifi ed DNA must be purifi ed away
from excess DSS. Since this is done under aqueous conditions, hydrolysis of the free NHS ester
at the other end of the crosslinker takes place at the same time. Activity losses can be severe if
the separation step is not done rapidly. In fact, the original protocol called for several hours
994 27. Nucleic Acid and Oligonucleotide Modifi cation and Conjugation
of gel fi ltration chromatography and concentration before the conjugation reaction was done.
Farmer and Castaneda (1991) made a signifi cant improvement to this procedure by including
a faster separation step using alcohol extraction after activation of the oligo with DSS. The
purifi cation time decreased to minutes instead of hours. This does help to limit hydrolysis, but
cannot completely avoid it.
The following protocol is based on the methods of Farmer and Castaneda (1991), Kiyama
et al . (1992), and Ruth (1993).
2. Chemical Modifi cation of Nucleic Acids and Oligonucleotides 995
Figure 27.15 The homobifunctional crosslinker DSS may be used to conjugate an enzyme to a 5 -diamine-
modifi ed oligonucleotide. The NHS ester groups on DSS react with the amines to form amide bonds.
Protocol
1. Prepare an amine-modifi ed oligonucleotide according to any of the protocols discussed
in Section 2.1 (this chapter). Dissolve or buffer-exchange the oligo into 0.1 M sodium
borate, 2 mM EDTA, pH 8.25, at a concentration of 9 nmol (2.0 A
260 nm
units) in 15 l.
2. Dissolve DSS in dry DMSO at a concentration of 1 mg/100 l. Prepare fresh.
3. Add 30 l of the DSS solution to the oligo. Mix well.
4. React for 15 minutes at room temperature in the dark.
5. Immediately extract excess DSS and reaction by-products by the addition of 0.5 ml of
n-butanol. Mix vigorously by vortexing and centrifuge (1 minute, 15,000 rpm) to separate
the two phases. Carefully remove the upper layer and discard. Extract two more times
with n -butanol.
6. Chill the remaining sample on dry ice and lyophilize to remove the last traces of liquid.
The drying period will only take 15–30 minutes. The dried, DSS-activated DNA is stable
under anhydrous conditions.
7. Dissolve or dialyze alkaline phosphatase into 3 M NaCl, 30 mM triethanolamine, 1 mM
MgCl2, pH 7.6, at a concentration of 20 mg/ml.
8. Add 70 l of the alkaline phosphatase to the dried, DSS-activated DNA. Mix gently to
dissolve.
9. React overnight at 4 °C in the dark.
10. Remove unconjugated oligo by using a Centricon-30 centrifugal concentrator according
to step 4 of the protocol described in the previous section. Unconjugated enzyme may
be removed by ion-exchange chromatography using a MonoQ-10 (0.5 5 cm) FPLC col-
umn (GE Healthcare) or the equivalent. Binding buffer is 20 mM Tris, pH 8. Elute using
a linear gradient of 0–100 percent 20 mM Tris, 1 M NaCl, pH 8. Free enzyme will elute
before the more negatively charged oligo–enzyme conjugate.
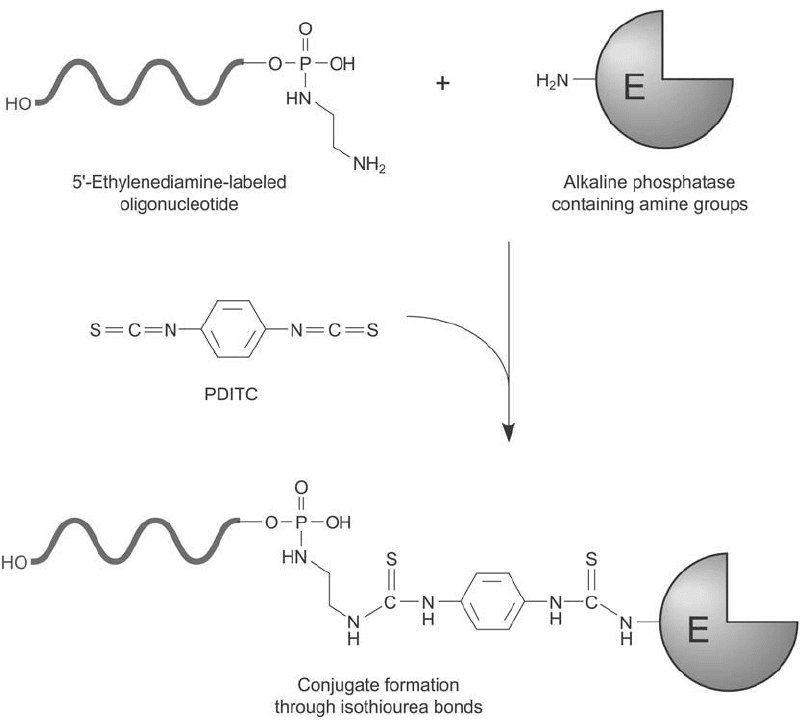
Enzyme Conjugation to Diamine-Modifi ed DNA Using PDITC
PDITC, 1,4-phenylene diisothiocyanate, is a homobifunctional crosslinker containing two
amine-reactive isothiocyanate groups on a phenyl ring. Reaction in excess with an amine-modi-
fi ed oligonucleotide results in the formation of a thiourea linkage, leaving the second isothio-
cyanate group free to couple with amine-containing enzymes or other molecules (Urdea et al .,
1988) ( Figure 27.16 ).
The following protocol is adapted from Keller and Manak (1989).
Protocol
1. Prepare an amine-modifi ed oligonucleotide using any of the methods of Sections 1 or 2.1
(this chapter). Dissolve or buffer exchange 70 g of the oligo into 25 l of 0.1 M sodium
borate, pH 9.3.
2. Dissolve 10 mg of PDITC (Aldrich) in 500 l of DMF. Add this solution to the oligo pre-
pared in step 1.
3. React for 2 hours at room temperature in the dark.
4. To extract excess reactant, add to the reaction medium, 3 ml of n-butanol and 3 ml of
water. Mix well. Centrifuge the mixture to separate the two phases. Discard the upper
996 27. Nucleic Acid and Oligonucleotide Modifi cation and Conjugation
yellow layer. Repeat the extraction process several times, and then dry the remain-
ing solution containing activated oligo using lyophilization or a rotary evaporator. The
PDITC-activated DNA is stable in a dried state.
5. Dissolve HRP (Chapter 26) in 200 l 0.1 M sodium borate, pH 9.3, at a concentration
of 10 mg/ml. If the HRP is supplied as an ammonium sulfate suspension, all ammonium
ions must be removed by extensive dialysis prior to the conjugation reaction. Add the
HRP solution to the activated oligo.
6. React overnight at room temperature in the dark.
7. Excess enzyme may be removed through isolation of the oligo–enzyme conjugate using
electrophoresis separation under nondenaturing conditions. The reaction solution is applied
to a 7 percent polyacrylamide gel using 90 mM Tris, 90 mM Boric acid, 2.7 mM EDTA, pH
8.3, as the running buffer. The conjugate appears as a brown band in the middle of the gel.
2. Chemical Modifi cation of Nucleic Acids and Oligonucleotides 997
Figure 27.16 The homobifunctional crosslinker PDITC may be used to conjugate an enzyme to a 5 -diamine-
modifi ed oligonucleotide, creating isothiourea linkages.
998 27. Nucleic Acid and Oligonucleotide Modifi cation and Conjugation
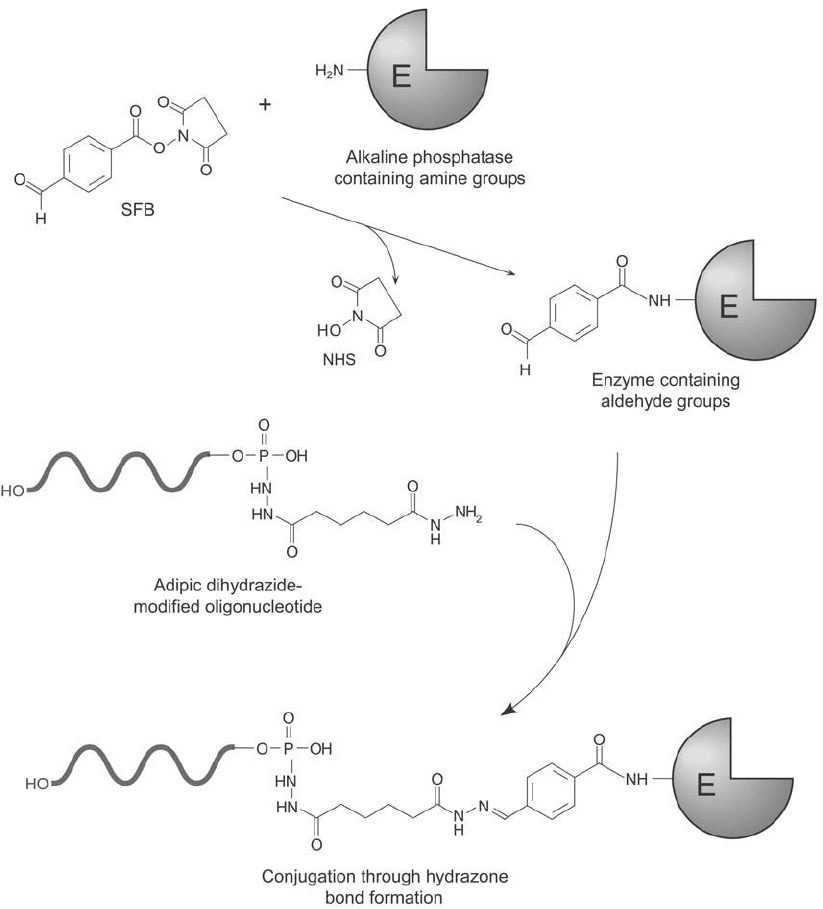
Conjugation of SFB-Modifi ed Alkaline Phosphatase to Bis-Hydrazide-Modifi ed Oligonucleotides
DNA probes modifi ed to contain a 5 -terminal hydrazide group (Section 2.1, this chapter)
can be conjugated to aldehyde-containing molecules, resulting in the formation of a hydra-
zone bond. The crosslinking agent succinimidyl p-formylbenzoate (SFB; Chapter 17, Section 2)
can be used to add aldehyde groups to proteins and other molecules that do not naturally
contain them (Chapter 1, Section 4.4). Reaction of SFB-modifi ed alkaline phosphatase with
a hydrazide–DNA derivative can produce a conjugate having excellent sensitivity for use as a
hybridization probe ( Figure 27.17 ). Other enzymes and detection molecules modifi ed to con-
tain aldehydes may be coupled to hydrazide–DNA probes using similar methods. Using an
SFB-modifi ed HRP (or periodate-oxidized HRP) that contains aldehyde groups to prepare the
DNA conjugate gives about a 40-fold less sensitivity in hybridization assays when compared to
an alkaline phosphatase conjugate in this procedure (Ghosh et al., 1989). However, this result
may change if chemiluminescent detection using an HRP conjugate is done, as this method can
result in femtogram detection limits.
The following protocol assumes the prior derivatization of an oligonucleotide at the 5 end
using a bis-hydrazide compound according to the protocol of Section 2.1 (this chapter) using a
carbodiimide-mediated reaction.
Protocol
1. Dissolve alkaline phosphatase at a concentration of 10 mg/ml in 0.1 M sodium bicarbon-
ate, 3 M NaCl, pH 8.5. Dialyze against this solution if the enzyme is already dissolved in
another buffer. This protocol requires at least 0.4 ml of the enzyme solution.
2. Dissolve SFB in acetonitrile at a concentration of 50 mM (12.35 mg/ml). Make at least 100 l.
3. Add 40 l of the SFB solution to the 0.4 ml alkaline phosphatase solution with mixing.
4. React for 30 minutes at room temperature.
5. Remove excess reactants by dialysis against 50 mM MOPS, 0.1 M NaCl, pH 7.5.
6. Add the aldehyde-derivatized alkaline phosphatase to 8 nmol of a 5 -hydrazide oligonu-
cleotide preparation made according to Section 2.1 (this chapter) using a carbodiimide
coupling protocol.
7. React overnight at room temperature.
8. Remove unconjugated oligonucleotide using gel fi ltration on a column of Bio-Rad P-100
(1.5 65 cm). Use 50 mM Tris, pH 8.5 as the chromatography buffer. Pool the enzyme
fractions and apply the sample to a 1 7 cm column of DEAE–cellulose equilibrated
with the same buffer. After washing the column with 0.1 M Tris, pH 8.5, a salt gradient
from 0 to 0.2 M NaCl in the same buffer is used to remove unconjugated enzyme. The
enzyme–DNA conjugate is then eluted with 0.1 M Tris, 0.5 M NaCl, pH 8.5.
2.5. Fluorescent Labeling of DNA
Oligonucleotide probes may be labeled with small fl uorescent molecules for detection of
hybridization by luminescence. Fluorescent probes are widely used in assay systems involving
bio-specifi c interactions (Chapter 9). Receptors for ligands may be localized in tissues or cells
by modifi cation of the ligand with the appropriate fl uorophore. Targeted molecules may be
quantifi ed through measurement or modulation of a fl uorescent signal upon binding of a tagged
ligand. The sensitivity of fl uorescent assays can approach that obtained using radiolabels.
Fluorescently labeled DNA probes can be used for detection, localization, or quantifi cation
of target DNA sequences. In situ hybridization mapping of genomic DNA sequences can be
Figure 27.17 SFB may be used to create aldehyde groups on enzyme molecules for subsequent conjugation to a
5 -bis-hydrazide-modifi ed oligonucleotide, forming hydrazone bonds.
2. Chemical Modifi cation of Nucleic Acids and Oligonucleotides 999
