Castilho Leda R., Moraes Angela Maria (Ed.) Animal Cell Technology: From Biopharmaceuticals to Gene Therapy
Подождите немного. Документ загружается.


ent cell lines and experimental conditions (operation mode, media formu-
lation, initial conditions, etc.) may influence the fitting of parameters and,
consequently, different parameter values are expected for the same phe-
nomena. Table 8.2 presents parameter values fitted for models of Table 8.1
from specific experimental conditions. These are indicative of the range of
values they can assume for animal cell systems. However, some of these
values may be considered inappropriate. This is the case of the value fitted
for
X,max
in Equation 40 – 0.125 h
–1
– surely a value not representative
for an animal cell system.
Kinetic formulations for animal cell death
Necrosis and apoptosis, the two mechanisms of cell death, are discussed
extensively in Chapter 7, and are considered in more detail in this section.
Cell death is particularly important for animal cell systems, considering
their intrinsic fragility in the imposed culture conditions, mainly as a result
of hydrodynamics shear stress and media composition. This scenario is
very different from that of microbial systems, for which it is rare to find
formulas representing death processes. When establishing culture condi-
tions for animal cells, there is always much concern in minimizing injury
to cells, to insure high viabilities for long periods. Nevertheless, modifica-
tions in the environment, mainly in media composition, occur during the
culture that normally lead to loss of viability. That explains why many
Table 8.2 Fitted parameters for models in Table 8.1
Eq.
X,max
(h
–1
)
k
GLC
(mM)
k
GLN
(mM)
k
i,LAC
(mM)
k
i,NH3
(mM)
Other parameters
(37) 0.036 – 0.06 – – –
(38) 0.045 1 0.30 – – –
(39) 0.033 0.278 – 13.9 – –
(40) 0.125 – 0.8 8.0 1.05 –
(41) 0.063 0.15 0.5 140 20 –
(42) – – 0.0309 – –
X,min
¼ 0.013 h
–1
;
GLC
Limiar
¼ 0.0303 mM
(43) 0.056 0.06 ––––
(44) 0.055 – 0.15 – –
(k
Serum
)
0
¼26.5;
¼0.21;
k9
i,NH
3
¼ 26 mM
2
(45) – – – – –
a
1
¼ 0.043 h
–1
;
a
2
¼ 0.07 g/L
(46) – – – – –
d
0
¼ 0.0029 h
–1
;
d
1
¼ 0.0195 h
–1
;
(47) 0.045 to 0.072 0 to 0.0094 0 to 0.040 – – 0.0043< Æ
1
< 0.0092
(L/10
9
cel.h)
202 Animal Cell Technology
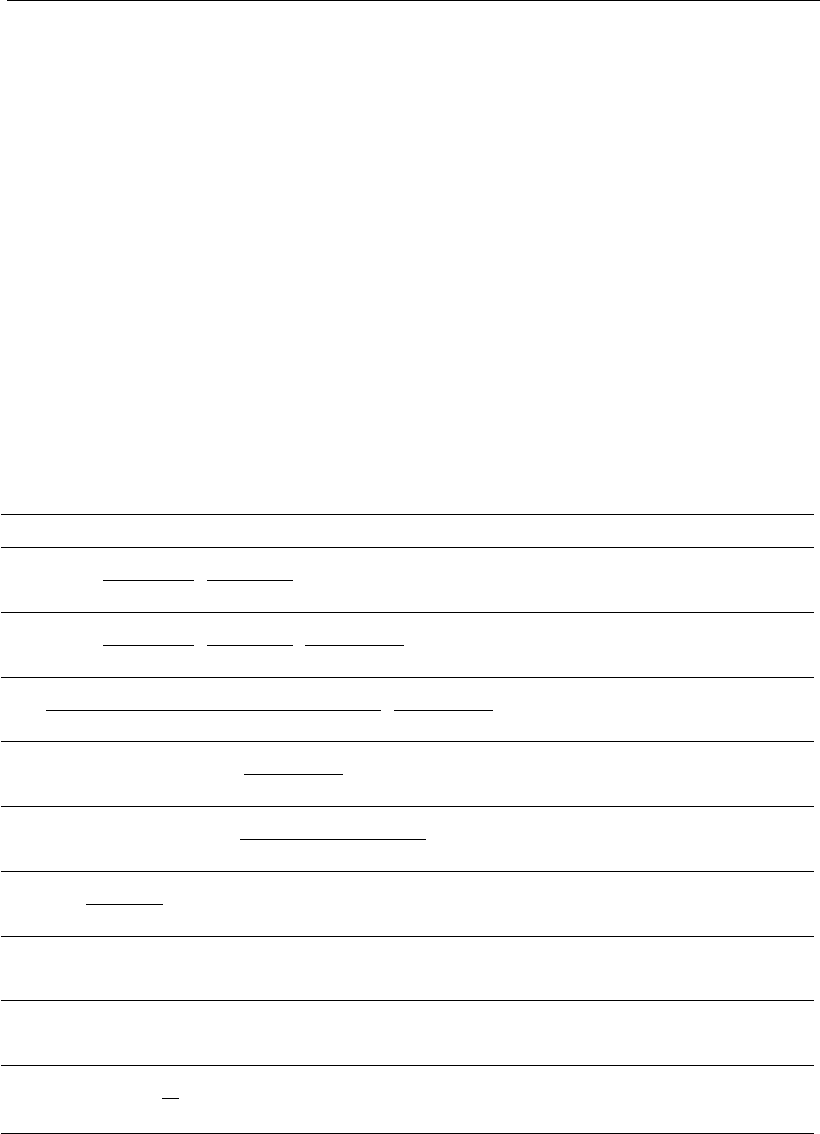
models incorporate the influence of substrates and products in their equa-
tions to simulate death kinetics (Table 8.3).
Similar to what has been shown in Table 8.1 for specific growth rate,
many mathematical expressions listed in Table 8.3 employ Monod-type
structures for limiting phenomena, and Aiba and Shoda-type structures
for inhibitory behavior. Limiting components for cell death are lactate and
ammonia, that is, the presence of these byproducts increases the specific
cell death rate. On the other hand, substrates, such as glucose and
glutamine, inhibit cell death (Equations 48 to 52).
Similar to what was shown for growth, some models establish a linear
relationship between the specific death rate (k
d
) and an autoinhibitory
product synthesis (Lee et al., 1995). This autoinhibitory product is
represented by the expression X
v
=D, where X
v
is a viable cells concentra-
tion, measured in terms of cell number per volume, and D is the specific
feed rate that plays the part of substrate supply to the culture. By setting
k
d
=
X
as a function of X
t
=D (where X
t
is a total cell concentration) it is
possible to build up a more robust model that can fit a larger amount of
experimental data (Equation 56) (Zeng et al., 1998).
Table 8.3 Kinetic expressions for specific cell death rate in animal cell systems
Formulations References Eq.
k
d
¼ k
d,max
NH
3
k
d
NH
3
þ NH
3
LAC
k
d
LAC
þ LAC
Batt and Kompala, 1989 (48)
k
d
¼ k
d,max
NH
3
k
d
NH
3
þ NH
3
LAC
k
d
LAC
þ LAC
k
d
i,GLN
k
d
i,GLN
þ GLN
Bree et al., 1988 (49)
k
d
¼
k
d,max
(
X,max
k
d
LAC
LAC) (
X,max
k
d
NH
3
NH
3
)
k
d
i,GLN
k
d
i,GLN
þ GLN
de Tremblay et al., 1992 (50)
k
d
¼ k
d,min
þ (k
d,max
k
d,min
)
k
d
i,GLN
k
d
i,GLN
þ GLN
Dalili et al., 1990 (51)
k
d
¼ (
X,min
D
min
) k
d,max
(GLC GLC
thres
)
k
d
GLC
þ (GLC GLC
thres
)
Frame and Hu, 1991a. (52)
k
d
¼ b
1
þ
b
2
GLN þ b
3
Po¨rtner et al., 1996 (53)
k
d
¼ c
1
e
c
2
X
Glacken et al., 1989 (54)
k
d
¼ d
0
e
(d
1
=
X
)
Linardos et al., 1991 (55)
k
d
¼ (
0
þ
1
X
)
X
t
D
Zeng et al., 1998 (56)
Adapted from Po¨rtner and Scha¨fer, 1996). X t, total cell concentration measured as total cell number per volume
(10
6
cell/ml).
Mathematical models for growth and product synthesis in animal cell culture 203

Table 8.4 lists parameter values fitted to models presented in Table 8.3.
Formulations for substrate uptake
Generally, the expressions proposed for the kinetics of glucose or gluta-
mine uptake do not differ significantly from each other. Table 8.5 (for
formulations) and Table 8.6 (parameters) sum up the kinetic models for
the description of substrate uptake rates.
Pirt’s formulations, or variations of them (see Equation 26), were
employed in many models shown in Table 8.5. In one of them (Equation
59) the term e
k
d
is added to better fit data obtained at low specific
growth rate (Linardos et al., 1991). Other propositions introduced a
minimum specific growth rate (
X,min
) to fit the non-linearity observed at
the same low specific growth rate (Frame and Hu, 1991a, 1991b).
For Equations 62 and 63, the maintenance coefficient appears limited by
substrate through a Monod-type formulation.
Finally, in Equation 64, Zeng (1996a) modified his own original model
(presented in Equations 28 to 30), by eliminating parameter S* that
assumes low values for hybridomas culture. The influence of nutrients is
dependent, not on nutrient concentration, but on available mass of
nutrient per cell unit S=X
v
.
Formulations for product synthesis
Table 8.7 (mathematical expressions) and Table 8.8 (parameters) review
the main models found in the literature for the representation of product
Table 8.4 Fitted parameters for models of Table 8.3
Eq. k
d,max
(h
–
1)
k
d
NH
3
(mM)
k
d
LAC
(mM)
k
d
i,GLN
(mM)
Other parameters
(48) 0.08 1.44 311 –
(49) 0.0833 1.44 15 5.10
–4
–
(50) 0.0288 – – 0.02
k
d
NH
3
¼ 0.0025 (1/mM.h);
k
d
LAC
0.00042 (1/mM.h)
(51) 0.08 – – 5.10
–6
k
d,min
¼ 0.05 h
–1
(52) 0.00628 – – –
D
min
¼ 0.00636 h
–1
;
min
¼ 0.013 h
–1
;
k
d
GLC
¼ 0.240 mM
(53) ––––
b
1
¼ 0.002 h
–1
;b
2
¼ 6.10
5
g/L.h;
b
3
¼ 0.0025 g/L
(54) 0.051 – – – c
1
¼ 0.051 h
–1
;c
2
¼ –101.2 h
(55) – – – d
0
¼ 0.0029 h
–1
;d
1
¼ 0.0195 h
–1
(56) ––––
0.0<
0
< 4.6.10
–5
h
–1
0.0<
1
< 0.0038 L/10
9
.cel.h
204 Animal Cell Technology
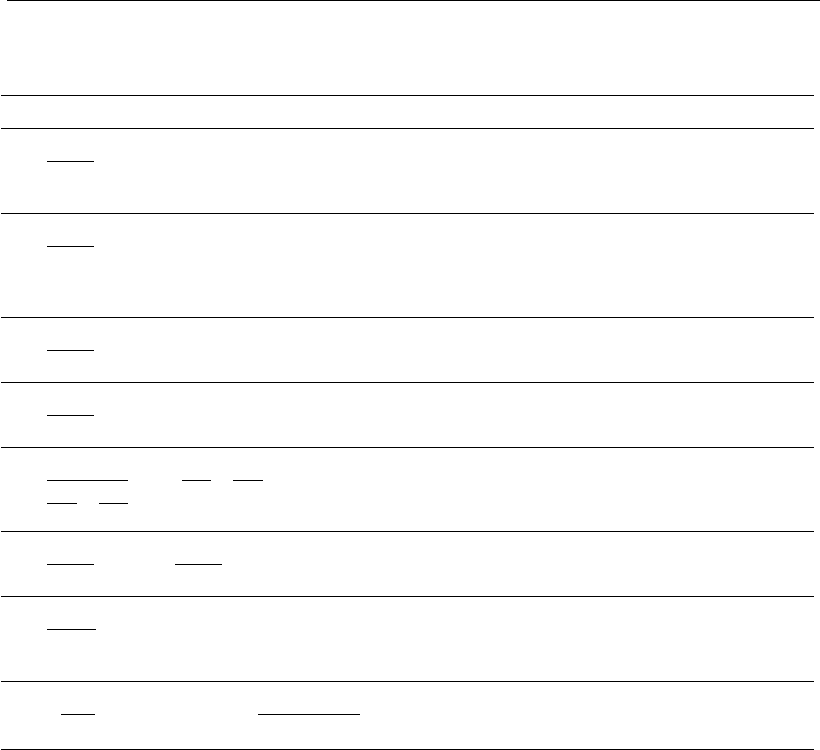
(proteins) synthesis. Once more, the diversity of mathematical structures
reflects the variability of response in such systems, as well as the lack of
knowledge of intracellular mechanisms.
Some models utilize the concept of Luedeking and Piret (1959) for
growth associated and/or non-growth associated production (Equations
65 to 67). However, in some circumstances, the best fit occurs when the
specific death rate k
d
is considered, instead of the specific growth rate
X
(Equations 68, 70 and 71) (Linardos et al., 1991; Zeng, 1996a, 1996b). This
statement follows the observation that antibody production increases
when cells are under conditions of stress.
Considering the influence of media components on specific production
rate, only one expression makes serum relevant (Equation 69), while
glutamine appears as a limiting substrate in three formulas (Equations 69
to 71), always with a Monod-type structure. Glucose shows either an
inhibitory effect over production (Equations 71 and 72), or a limiting and
inhibitory pattern simultaneously (Equations 70). Instead of the classical
Table 8.5 Kinetic parameters for specific uptake rates in hybridoma systems: equations are
valid for both substrates – GLC and GLN
Formulations References Eq.
S
¼
1
(Y
max
X=S
)
X
de Tremblay et al., 1992;
Hiller et al., 1991
(57)
S
¼
1
(Y
max
X=S
)
X
þ m
S
Harigae et al., 1994; Hiller et al.,
1991; Miller et al., 1988;
Kurokawa et al., 1994
(58)*
S
¼
1
(Y
max
X=S
)
X
þ m
S
e
k
d
Linardos et al., 1991 (59)
S
¼
1
(Y
max
X=S
)
(
X
min
) Frame and Hu, 1991a (60)
S
¼
1
1
Y
X=S
º
Y
P=S
X
þ
Æ
2
Y
P=S
1
Y
X=S
X,min
Frame and Hu, 1991b (61)
S
¼
1
(Y
max
X=S
)
X
þ m
S
S
k
S
S
þ S
de Tremblay et al., 1992 (62)
S
¼
h
1
S
h
2
þ S
Po¨rtner et al., 1996;
Gaertner and Dhurjati, 1993
(63)
S
¼
1
Y
max
S
X
þ m
S
!
þ ˜
m
S
S
S þ (X
v
k
S#
S
)
Zeng, 1996a (64)
*Same formulation proposed in Equation 26.
Mathematical models for growth and product synthesis in animal cell culture 205

Table 8.6 Fitted parameters for models of Ta ble 8.5
Eq. References S Y
max
X=S
(10
8
cel/mmol)
m
S
(10
–10
mmol/cel.h)
Other parameters
(57) de Tremblay et al., 1992 GLN 3.80 – –
(58) Hiller et al., 1991 GLC 1.02 –
Miller et al., 1988 GLC 2.8 0.50 –
Harigae et al., 1994 GLC 5.88 0.51 –
GLN 3.44 0.08 –
Hiller et al., 1991 GLN 16.9 1.56
(59) Linardos et al., 1991 GLC 5.93 1.96 –
GLN 6.30 0.29 –
(60) Frame and Hu, 1991a GLC 0.355 mg cel/mg GLC
(p/,0.0508 h
–1
):
0.101 mg cel/mg GLC
(p/.0.0508 h
–1
)
p/,0.0508 h
–1
:
min
¼ 0.00630 h
–1
or
p/.0.0508 h
–1
:
min
¼ 0.391 h
–1
(61) Frame and Hu, 1991b GLC
1.82
(p/,0.00429 h
–1
)
0.52
(p/.0.00429 h
–1
)
–
Æ
2
¼ 0.00672 mg/cel.h;
º ¼ 9.31.10
–9
mg/cel;
X,min
¼ 0.013 h
–1
;
Y
P=S
¼ 73.4 mg/mM
(62) de Tremblay et al., 1992 GLC 1.09 0.00708 k
S
S
¼ 1.0 mM
(63) Po¨rtner et al., 1996 GLN – –
h
1
¼ 0.68 10
–10
mmol/cel.h
h
2
¼ 0.2 mmol
Gaertner and Dhurjati, 1993 GLC – –
h
1
¼ 2.70 10
–10
mmol/cel.h
h
2
¼ 0.34 mmol
(64) Zeng, 1996a GLC 1.72 0.086
˜
m
GLC
¼ 0.094 L/10
9
cel.h
k
s9
GLC
¼ 0.29 g/cel
GLN 9.52 0.0093
˜
m
GLN
¼ 0.033 L/10
9
cel.h
k
S#
GLN
¼ 0.089 g/cel
206 Animal Cell Technology

formulation for the limitation or inhibition of a substrate (Equations 21
and 22), a hyperbolic structure is employed to represent inhibition, and
the parameter GLC
is introduced to represent the minimum glucose
concentration that guarantees product synthesis.
In Equation 70, Zeng (1996b) considers also the loss of production
capacity within the cell population – in the term (F
1
þ e
F
2
˜t
). This is a
characteristic common to many hybridomas that are expressed in long-
term culture. For Zeng (1996b), any cell population has two categories of
individuals, one characterized with high protein productivity, and another
with low protein productivity. The loss of producing capacity is attributed
to mutation or loss of genetic materials, and is irreversible. The parameter
‘‘F
2
’’ in Equation 70 represents the transformation rate from a producing
to a non-producing population. The parameter ‘‘F
1
’’ states a relationship
between the specific production rates of these two populations, each one
of them homogeneous in its productivity. Finally, an exponential function
–e
F
2
˜t
– simulates the loss of producing capacity within a cell popu-
lation.
If a process is operated in perfusion with high cell densities, Equation 70
might need an adjustment, as indicated in Equations 71 and 72. This can
occur by taking the relation (S=X
V
) for the main state variable, which
Table 8.7 Kinetic expressions for specific monoclonal synthesis rate
Formulations References Eq.
MAb
¼ Po¨rtner et al., 1996;
Hiller et al., 1991
(65)
MAb
¼ Æ
x
þ Frame and Hu, 1991b (66)
MAb
¼
Æ
0
k
þ
X
X
þ de Tremblay et al., 1992 (67)
MAb
¼ Æ
1
k
d
þ
1
Linardos et al., 1991 (68)
MAb
¼
2
(Serum)
GLN
k
MAb
GLN
þ GLN
Dalili et al., 1990 (69)
MAb
¼ (Æ
1
k
d
þ
1
)
GLN
k
MAb
GLN
þ GLN
k
Mab
i,GLC
GLC GLC
þ k
MAb
i,GLC
(F
1
þ e
F
2
˜t
) Zeng, 1996b (70)
MAb
¼ (Æ
1
k
d
þ
1
)
GLN
k
MAb#
GLN
X
v
þ GLN
k
MAb#
i,GLC
N
v
GLC þ k
MAb#
i,GLC
X
v
Zeng, 1996a (71)
MAb
¼
D
Per
D
Per
þ k
MAb
D
Per
k
MAb
i,GLC
GLC þ k
MAb
i,GLC
Zeng, 1996a (72)
D
Per
¼ specific perfusion rate (h
1
).
Mathematical models for growth and product synthesis in animal cell culture 207

includes the influence of substrate and high cell density (Equation 71); or
by considering the stimulation of productivity caused by the specific
perfusion rate (D
Per
).
Tables 8.9 and 8.10 show examples of kinetic expressions for byproduct
synthesis and the parameter values fitted to them.
Frequently, the specific byproduct formation rate is presented as a
function of specific substrate consumption rate and substrate-to-product
yield (see Equation 12), but other structures can be assumed. The specific
production rate can be limited by a precursor substrate and modeled by a
Monod-type expression, as in Equation 73, or it may be inhibited by a
substrate that is not, in principle, linked to its production, as in Equation
Table 8.8 Fitted parameters for models of Table 8.7
Eq. References Æ
(mg/mg
cel
)
(mg/10
9
cel.h)
Other parameters
(65) Po¨rtner et al., 1996 – 0.0167 –
Hiller et al., 1991 – 0.708 –
(66) Frame and Hu, 1991b –0.0266 0.00672
(mg/mg
cel
.h)
–
(67) de Tremblay et al., 1992 0.146
Æ
0
¼ 0.115 mg/10
8
cel
k
¼ 8.33.10
–4
h
–1
(68) Linardos et al., 1991 – –
Æ
1
¼ 273 g/10
6
cel
1
¼ 0.0375 g/10
6
cel.h
(69) Dalili et al., 1990 – –
2
¼ 0.4 mg/10
8
cel.h
k
MAb
GLN
¼ 0.1 mM
(70) Zeng, 1996b – –
Æ
1
¼ 0 – 51.6 g/10
9
cel
1
¼ 0.5 – 1.21 mg/10
9
cel.h
k
MAb
GLN
¼ 4.42 – 9.01 mM
k
MAb
i,GLC
¼ 4.42 – 9.01 mM
GLC* ¼ 0.1 mM
F
1
¼ 0.37 – 1.18
F
2
¼ 0 – 0.018 h
–1
(71) Zeng, 1996a
Æ
1
¼ 0.092 mg/10
9
cel
1
¼ 1.33 mg/10
9
cel.h
k
MAb#
GLN
¼ 0.0079 mmol/10
9
cel
k
MAb#
i,GLC
¼ 12.65 mmol/10
9
cel
(72) Zeng, 1996a
¼ 4.16 mg/10
9
cel.h
k
MAb
D
Per
¼ 0.115 h
–1
k
MAb
i,GLC
¼ 13.97 mM
208 Animal Cell Technology
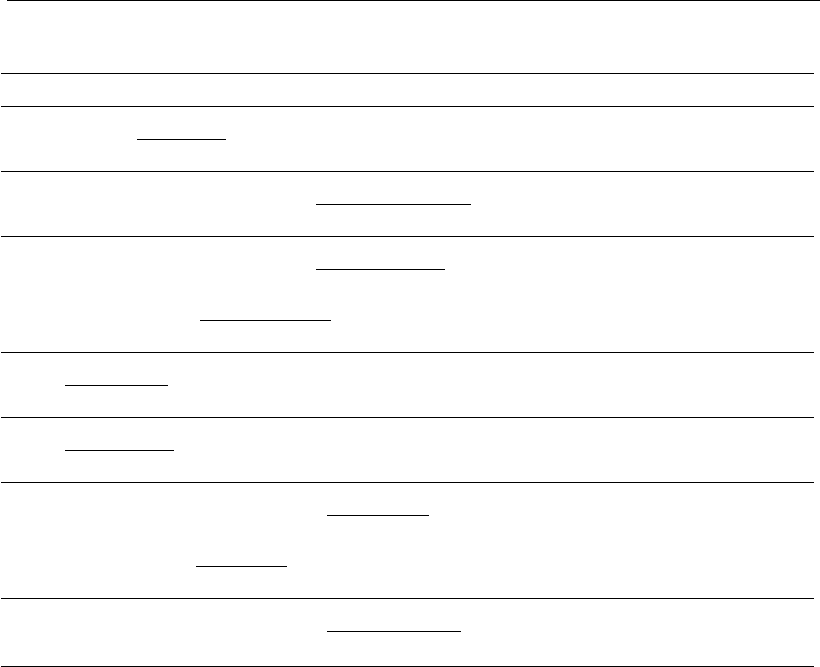
76 and 77. For example, glucose inhibits ammonia synthesis, an assump-
tion that takes account of the complex regulation of dual-substrate
metabolism (GLC and GLN) in animal cell systems (see Chapter 4).
In Equations 74 and 78 data fit to a model that considers the concept of
excess substrate in culture, as set previously in Equation 36 (Zeng, 1995).
However, this formulation needs modification in order to include high cell
density conditions. In Equations 75 and 79, as seen before, the mass of
nutrient available per cell (S=X
V
) is the state variable to take this into
account.
8.3.3 Parameter fitting in models
Unstructured models, as detailed in Sections 8.3.1 and 8.3.2, are formu-
lated by a series of kinetic and differential non-linear equations that
represent the dynamics of all the state variables during the process. Thus,
to simulate a model that consists of parameters and state variables, it is
necessary to attribute values to the parameters.
The initial parameters values can be estimated in different ways:
(i) from the stoichiometric relationship, utilizing experimental or theor-
etical data;
Table 8.9 Kinetic formulations for synthesis of byproducts (ammonia and lactate)
Formulations References Eq.
LAC
¼
LAC,max
GLC
k
LAC
GLC
þ GLC
Gaertner and Dhurjati, 1993 (73)
LAC
¼ Y
LAC=X
X
þ m
LAC
þ ˜
max
LAC, GLC
GLC GLC
GLC GLC
þ k
LAC
GLC
Zeng, 1995 (74)
LAC
¼ Y
LAC=X
X
þ m
LAC
þ ˜
max
LAC, GLC
GLC
GLC þ k
LAC#
GLC
X
v
þ ... Zeng, 1996a (75)
...þ ˜
max
LAC, GLN
GLN
GLN þ k
LAC#
GLN
X
v
NH
3
¼
E
1
þ E
2
GLC
E
3
þ GLC
Gaertner and Dhurjati, 1993 (76)
NH
3
¼
G
1
þ G
2
LAC
G
3
þ LAC
Gaertner and Dhurjati, 1993 (77)
NH
3
¼ Y
NH
3
=X
X
þ m
NH
3
þ ˜
max
NH
3
, GLN
GLN3
GLN3 þ k
NH
3
GLN
þ ... Zeng, 1995 (78)
... þ˜
max
NH
3
, GLC
GLC
GLC þ k
NH
3
GLC
NH
3
¼ Y
NH
3
=X
X
þ m
NH
3
þ ˜
max
NH
3
, GLN
GLN
GLN þ k
NH
3
#
GLN
X
v
Zeng, 1996a (79)
Mathematical models for growth and product synthesis in animal cell culture 209

(ii) by linearization and, sometimes, simplification of kinetic equations
and by fitting the experimental data by linear regression;
(iii) by utilizing parameter values available in the literature.
Item (i) above is largely employed to obtain substrate-to-cell and
substrate-to-product yield factors. When the substrate consumption is
diverted to several different products, an estimate of the substrate-to-
product yield factors can be based on the stoichiometric relationships
obtained from the biochemical reactions.
Table 8.10 Fitted parameters for models of Table 8.9
Eq. Byproducts
(bP)
Y
bP=X
mmol
sP
10
9
cel
m
bP
mmol
sP
10
9
cel h
˜
max
bP,S
mmol
sP
10
9
cel h:
k
bP
S
(mM)
Other parameters
(73) LAC 0.34
LAC,max
¼ 2.7
(mmol/10
10
cel.h)
(74) LAC 10,42 0.050 0.355 0.43
(75) LAC 2553 888 1322 (S ¼ GLN)
3663 (S ¼ GLC)
k
LAC#
GLN
0.013
(mmol/10
9
cel)
k
LAC#
GLC
¼ 17.54
(mmol/10
9
cel)
(76) NH
3
E
1
¼ 2.0
(mmol
2
/10
10
cel.L.h)
E
2
¼ 0.35
(mmol/10
10
cel.h)
E
3
¼ 3.3 mM
(77) NH
3
G
1
¼ 6.2
(mmol
2
/10
10
cel.L.h)
G
2
¼ 0.22
(mmol/10
10
cel.h)
G
3
¼ 14 mM
(78) NH
3
0.21 0.00068 0.032 (p/S ¼ GLN)
0.0041 (p/S ¼ GLC)
0.56
(p/GLN)
0.96
(p/GLC)
.
(79) NH
3
0.73 – 0.025 (S ¼ GLN)
k
NH
3
#
GLN
0.071
(mmol/10
9
cel)
210 Animal Cell Technology
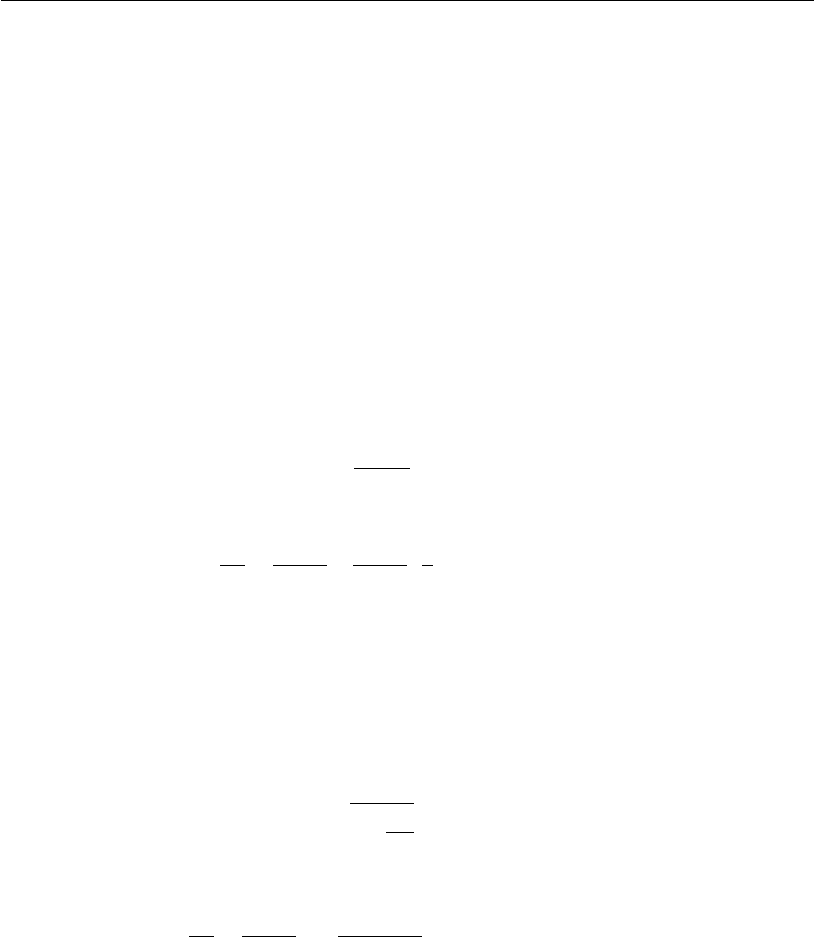
Item (ii) is largely employed in obtaining kinetic parameters such as the
substrate limitation and inhibition constants for cellular growth and death
(k
S
and k
i,S
, respectively) and substrate inhibition constants for production
(k
P
i,S
); maximum specific growth and death rates (
X,max
and k
d,max
), as well
as the global yield factors (Y
X=S
and Y
P=S
), that may be associated or not to
the cellular growth.
Simplification of the equations involves separating the parameters to be
estimated.
Besides the simplification, a subset of data must be selected, from the
available data set, that is more adequate for the determination and fitting
of the parameters.
Andrew’s kinetic model (Equation 21) (Andrews, 1968), for example,
relates the specific growth rate (
X
) with the substrate concentration S,
applying three parameters (
X,max
,k
S
and k
i,S
).
The initial parameter estimations of
X,max
and k
S
can be based on a
simplification of Equation 21, by disregarding the inhibition term for the
substrate. Thus, Equation 21 becomes Equation 80.
X
¼
X,max
S
k
S
þ S
(80)
Re-arranging this equation results in its linearization.
1
X
¼
1
X,max
þ
k
S
X,max
1
S
(81)
Using data extracted from the test conducted with non-inhibiting
substrate concentrations, the construction of the graph 1=
X
¼
a þ b (1=S) allows a fit of values for ‘‘a’’ and ‘‘b’’ and, from these, an
estimate of
X,max
and k
S
(Equation 81).
An initial estimate for parameter k
i,S
can be obtained by the simplifica-
tion of Equation 21, disregarding the limitation for the substrate term, S,
forming Equation 82:
X
¼
X,max
S
S þ
S
2
k
i,S
(82)
Re-arranging this equation results in its linearization:
1
X
¼
1
X,max
þ
1
X,max:
k
i,s
!
S (83)
Using data extracted from the test conducted with inhibiting substrate
concentrations, the construction of the graph 1=
X
¼ a þ b S allows a fit
of values for ‘‘a’’ and ‘‘b’’, and, from these, an estimate of
X,max
and k
i,S
(Equation 83). Others ways of estimating parameters can be found in
Bonomi and Schmidell (2001).
Once the initial values of the parameters are estimated, the model
becomes dependent only on the state variable. From now on, forecasts can
be made with the model and the results compared to the experimental data
for different culture conditions (model simulation). Thus, the quality of
Mathematical models for growth and product synthesis in animal cell culture 211
