Accardi L., Freudenberg W., Obya M. (Eds.) Quantum Bio-informatics IV: From Quantum Information to Bio-informatics
Подождите немного. Документ загружается.

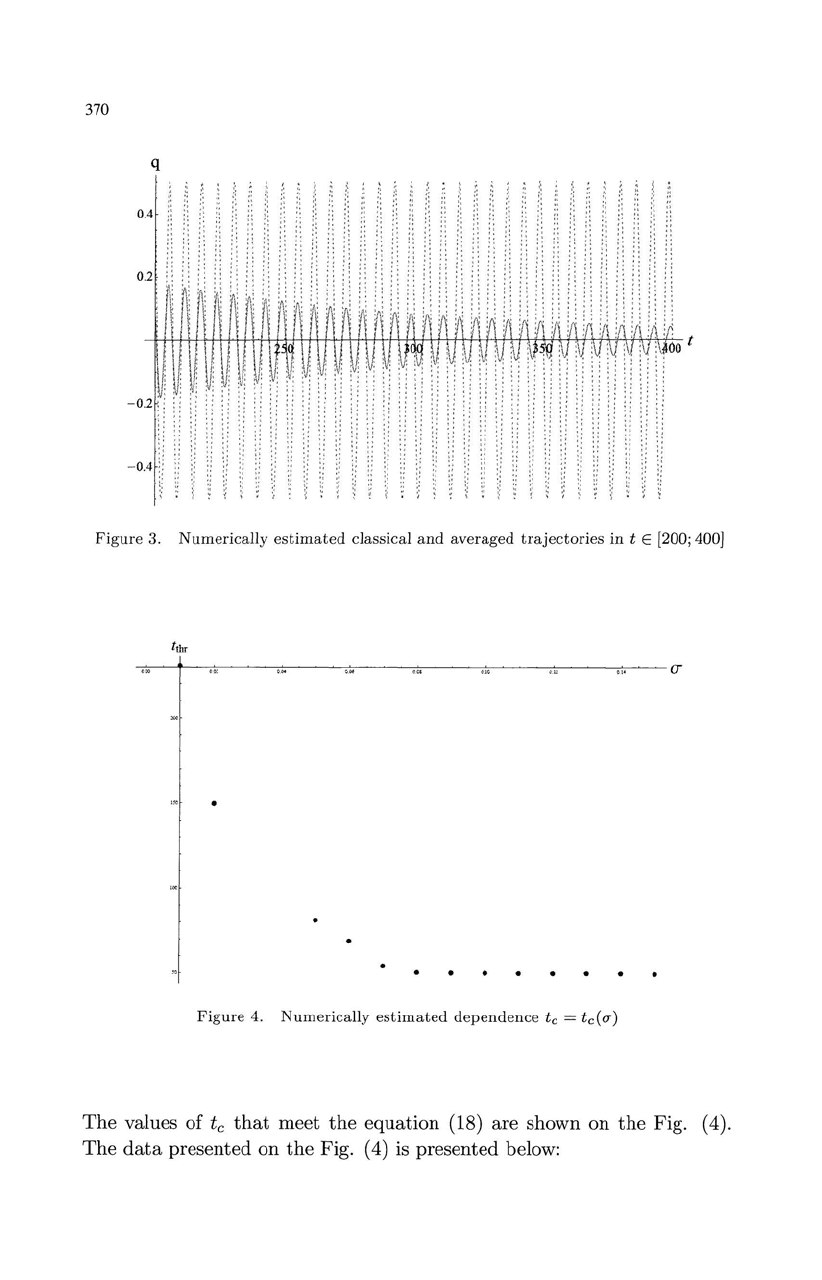
370
q
Figure
3.
Numerically
estimated
classical
and
averaged
trajectories
in t E [200; 400J
Figure
4.
Numerically
estimated
dependence
tc = tc
((T)
The
values of
tc
that
meet
the
equation
(18)
are
shown
on
the
Fig. (4).
The
data
presented
on
the
Fig. (4) is
presented
below:
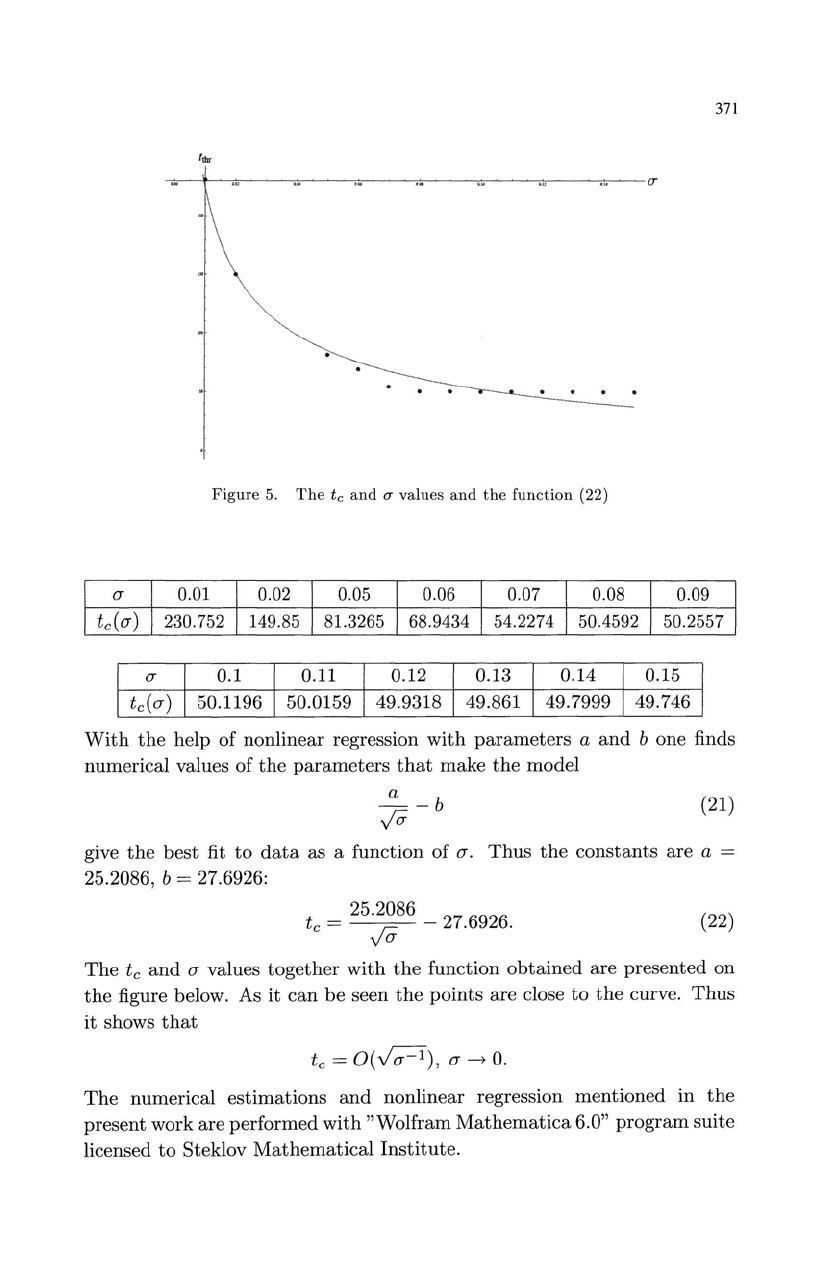
371
Figure
5.
The
tc
and
(]"
values
and
the
function
(22)
With
the
help
of
nonlinear regression
with
parameters
a
and
b one finds
numerical values
of
the
parameters
that
make
the
model
~-b
fo
(21)
give
the
best
fit
to
data
as a function of
(J".
Thus
the
constants
are
a =
25.2086, b = 27.6926:
25.2086
tc
=
fo
- 27.6926.
(22)
The
tc
and
(J" values
together
with
the
function
obtained
are
presented
on
the
figure below. As
it
can
be
seen
the
points
are
close
to
the
curve.
Thus
it
shows
that
The
numerical
estimations
and
nonlinear regression mentioned in
the
present work
are
performed
with
"Wolfram
Mathematica
6.0"
program
suite
licensed
to
Steklov
Mathematical
Institute.

372
Acknowledgements
This
work was
partially
supported
by
grants
RFBR-OS-OI-00727-a
and
RFBR-09-0l-12161-ofi-m,
and
also by
grants
NSh-3224.200S.1
and
by
Di-
vision of
Mathematics
of
RAS.
References
1.
LV. Volovich,
Randomness
in Classical Mechanics
and
Quantum
Mechan-
ics.
Foundations
of
Physics,
DOl
1O.1007/s10701-01O-9450-2;
Tim
e Irre-
versibility
Problem
and
Functional
Formulation
of
Classical Mechanics,
arXiv:0907.2445.
2.
V.V
. Kozlov,
Gibbs
ensembles
and
nonequilibrium
statistical
mechanics,
Moscow-Ijevsk (in
Russian),
2008.
3. LV. Volovich.
Functional
mechanics
and
time
irreversibility problem.
In
"Quantum
Bio-Informatics
III",
ed. L. Accardi,
W.
Freudenberg,
M. Ohya.
World Scientific, Singapore, 2010, pp. 393-404.
4. A.
S.
Trushechkin,
1.
V. Volovich,
Functional
Classical Mechanics
and
Ratio-
nal
Numbers,
P-Adic
Numbers,
Ultr
a
metric
Analysis
and
Applications, 1:4
(2009), 365-371; arXiv: 0910.1502.
5. A.
S.
Trushechkin,
Irreversibility
and
the
measurement
procedure
in
the
func-
tional
mechanics,
Theor.
Mathern.
Phys.
164:3 (2010), 435-440.
6.
LV. Volovich, Bogolyubov
equations
and
functional
mechanics,
Theor.
Mathern.
Phys.
164:3 (2010) 354-362.
7.
E.V
. Piskovskiy, A
study
of
some
model
systems
of
functional mechanics in
the
functional
mechanics framework,
Abst.,
The
Second
International
Con-
ference
on
Mathematical
Physics
and
Its
Applications,
Samara,
2010.
8. K. Inoue, M. Ohya, LV. Volovich, Semiclassical
properties
and
chaos degree
for
the
quantum
baker's
map,
Journal
of
Mathematical
Physics, 43, 734-755
(2002).
9. N.N. Bogolyubov, Y. A. Mitropolski,
Asymptotic
Methods
in
the
Theory
of
Nonlinear Oscillations.
Gordon
and
Breach, New York, 1961.
10. G. Bikhoff,
S.
Mac
Lane, A
Survey
of
Modern
Algebra,
4th
ed.,
Macmillan
Publishing
Co
Inc., New York, 1977.

Quantum
Bio-Informatics
IV
eds. L.
Accardi,
W.
Freudenberg
and
M.
Ohya
© 2011
World
Scientific
Publishing
Co.
(pp.
373-386)
QUANTILE-QUANTILE
PLOTS:
AN
APPROACH
FOR
THE
INTER-SPECIES
COMPARISON
OF
PROMOTER
ARCHITECTURE
IN
EUKARYOTES
KASPAR FELDMEIER, JOACHIM KILIAN, KLAUS HARTER,
DIERK WANKE* AND
KENNETH
W. BERENDZEN*
2MBP
Pfianzenphysiologie, Universitiit Tiibingen
Auf
der Morgenstelle 1, D-72076 Tiibingen,
Germany
* e-mail: dierk.wanke@zmbp.uni-tuebingen.de
* e-mail: kenneth.berendzen@zmbp.uni-tuebingen.de
Regulatory
non-coding
DNA
is
important
to
drive
gene
transcription
and
thereby
influence
mRNA
and
consequently
protein
abundance.
Therefore,
biologists
and
bioinformation
scientists
aim
to
extract
meaningful
information
from
these
se-
quence
regions,
in
particular
upstream
regulation
regions
called
promoters,
and
conclude
on
regulatory
sequence
function.
While
some
approaches
have
been
suc-
cessful for
single
genes
or
a single
genome,
it
is
an
open
question
whether
infor-
mation
on
promoter
function
can
readily
be
transferred
between
different species.
Thus,
it
is
useful
for
biologists
to
know
more
about
the
general
structure
and
composition
of
promoters
including
the
occurrence
of
cis
regulatory
DNA-elements
(CREs)
to
be
able
to
compare
promoter
architecture
between
organisms.
To
ap-
proach
this
task,
we
utilized
the
fully
sequenced
genomes
of
the
plant
model
or-
ganisms:
mouse-ear
cress
(Arabidopsis
thaliana),
western
balsam
poplar
(Populus
trichocarpa),
Sorghum
bicolor
and
rice
(Oryza
sativa).
For
the
interspecies
com-
parison
we
made
use
of
quantile-quantile
(QQ)-plots
of
the
variances
of
hexanu-
cleotides
or
known
functional
CREs
of
core-promoter
regions. Here, we
suggest
that
the
differences
in
promoter
architecture
correlate
with
the
sizes of
the
inter-
genic
space,
i.e. regions,
in
which
the
promoters
are
located.
In
contrast,
analysis
of
CREs
is
hampered
by
the
general
lack
of
well
characterized
transcription
factor-
CRE-relationships.
Keywords:
Promoter
code;
cis-regulatory
elements
CRE);
cross-species
pro-
moter
analysis;
promoter
evolution;
QQ-plot
of
variances.
1.
Introduction
Gene
expression
in
eukaryotes
is
controlled
by
several
intermingling
levels
of
regulation
that
involves
both
pre-
and
post-transcriptional
processes.
One
level
comprises
the
intermingling
of
proteins,
termed
transcription
factors
373

374
(TFs),
which
make
direct
physical
contact
with
specific
short
stretches
of
DNA
called
cis-regulatory
DNA-motif
elements
(CREs)
and
subsequently
act
on
the
transcription
machinery.
These
CREs
are
typically
positioned
upstream
of
the
DNA
sequence
encoding
the
RNA
transcript
of a gene
and
follow a
cryptic
regulatory
code
1.
The
interface
between
the
transcription
factors
and
their
regulatory
DNA
sequence
companions
harbor
essential
information
to
control
specific gene expression
and
integrate
information
derived from
upstream
signaling cascades
2.
From
an
evolutionary
point
of
view,
it
has
been
noted
that
the
same
or
highly
similar
TF
-CRE-
relations
exist
in
closely
related
species
and
control
the
expression
of
the
same
or
highly similar genes. However,
it
still re-
mains
to
be
elucidated,
how
TF-CRE-relations
change
or
are
retained
over
more
distant
evolutionary
time.
Previous
studies
have shown
that
several
DNA-motifs
are
conserved in
the
regulatory
promoter
sequences
between
many
eukaryote
genomes, e.g. Arabidopsis, Saccharomyces, Drosophila
and
Caenorhabditis,
and
this
is especially
true
for
the
core-promoter
region prox-
imal
to
the
genes
1.
To
draw
conclusions
on
promoter-
and
CRE-evolution
from
comparative
cross-species approaches,
it
is
of
importance
to
integrate
the
genomic sequences of closely
and
more
distantly
related
species in
the
same
analysis.
Plant
genomes
are
especially
suited
for
studies
on
promoter
sequences, as
most
CREs
are
located
upstream
of
the
transcription
start
site
(TSS) close
to
the
gene sequences
1,3.
Fortunately,
genomic sequences from several
plant
species
are
publicly
available which
one
can
use for interspecies comparisons.
One
of
the
best
studied
eukaryote
model
organisms
to
date
is
the
flowering
plant
Arabidop-
sis thaliana.
Its
physiological responses
and
its
lifecycle have
been
studied
for
more
than
100 years.
With
only
five chromosomes,
the
overall complex-
ity
of
its
genome is low
and
the
intergenic regions
that
harbor
the
promoters
are
small.
Thus
far,
Arabidopsis
has
risen
to
one
of
the
best
understood
eukaryote
model
organisms
with
the
best
annotated
genome sequence
of
all
eukaryotes
available
4.
While
Arabidopsis thaliana is a
modern
dicot
plant
of
little
agricultural
importance,
the
evolutionary
more
distantly
related
grasses
or
trees
are
of
a high economical value world wide
5.
A
major
drawback
for genetic ap-
proaches in grasses lies in
the
complex genomes
of
cereal
crops
that
have
multimerized
and
duplicated
during
the
breeding
and
inbreeding
processes
of
domestication.
Rice
has
a
small
size
and
relatively simple genomic or-
ganization
and
is favored in
laboratory
research as
it
is
the
second fully
sequenced
eukaryote
plant
species
6.
The
mono
cot
Sorghum bicolor, whose

375
genome sequence will be
about
to
be finished in
the
near future,
is
closer
related
to
rice
than
it is
to
the
dicot Arabidopsis
7.
Despite
their
econom-
ical importance, trees have a
great
disadvantage for genetic studies due
to
their
naturally
a long life cycle compared
to
cryptogams, making
laboratory
experiments tedious or
not
feasible
8.
Nevertheless,
the
genomic sequence
of
the
western balsam
poplar
(Populus trichocarpa) has been
submitted
to
public
databases
8
and
can
readily
been
analyzed by bioinformatics rou-
tines.
Here, we conduct
an
interspecies
promoter
motif
analysis
to
assess
whether
the
architecture of
the
core-promoters
and
CREs
distribution
therein
is evolutionary conserved. Therefore, we
extracted
upstream
se-
quences from Arabidopsis, rice, poplar
and
Sorghum
annotated
genes, which
contain
the
proximal promoters
and
most essential CREs.
The
variances
of all hexanucleotide motifs
and
known functional
CREs
were
computed
for these four species. For
the
pair-wise visualization of
the
interspecies
differences in
the
promoters, we employed quantile-quantile (QQ)-plots of
these variances.
This
approach disclosed
that
a higher information density
is
contained in
the
promoters of those species
with
more compact genomes.
2.
Material
and
Methods
2.1.
Plant
genome
information
The
genome sequence of
the
chromosome pseudomolecules for
the
plant
model organisms Arabidopsis thaliana, western
balsam
poplar
(Popu-
lus trichocarpa), Sorghum bicolor
and
rice (Oryza sativa) were retrieved
from GenBanks
Plant
Genomes
Central
(http://www
.
ncbi
. nlm.
nih.
gOY
/
genomes/PLANTS/PlantList
.html):
Arabidopsis thaliana
GenBank
ac-
cessions:
NC_003070.9, NC_003071.7, NC_003074.8, NC_003075.7
and
NC_003076.8; Populus trichocarpa
GenBank
accessions: NC_008467.1,
NC_008468.1, NC_008469.1, NC_008470.1, NC_008471.1, NC_008472.1,
NC_008473.1, NC_008474.1, NC_008475.1, NC_008476.1, NC_008477.1,
NC_008478.1, NC_008479.1, NC_008480.1, NC_008481.1, NC_008482.1,
NC_008483.1, NC_008484.1
and
NC_008485.1; Oryza sativa
GenBank
accessions:
NC_008394.1, NC_008395.1, NC_008396.1, NC_008397.1, NC_008398.1,
NC_008399.1, NC_008400.1, NC_008401.1, NC_008402.1, NC_008403.1,
NC_008404.1
and
NC_008405.1.
The
genomic sequences of Sorghum
bi-
color 1 v4 were retrieved from
the
Sorghum bicolor Genome
at
Plant
genome
data
base,
PlantGDB
(http://www
.
plantgdb
.
org/
/SbGDB/).
Promot-

376
ers were
extracted
as
1000
bp
5
of
the
annotated
start
for each genes
(without
redundancy)
using
Motif
Mapper
.NET
(5.1.1.39)
and
python
scripts (1.2)
(http://www.zmbp.uni-tuebingen.de/PlantPhysiology
I
ResearehGroups/harter/berendzen/programs.html).
2.2.
Hexanucleotide
Motifs
and
cisRegulatory
Elements
Known
CREs
were
retrieved
from
the
PLACE
databases
(http://www
.
dna.
afire.
go.
jp/PLACE/)
as
has
been
published
previously in
1.
Hexam-
ers were
generated
with
Motif
Mapper
All Oligos function.
2.3.
Variance
based
Promoter
Motif
Analysis
To
compute
the
variances for all DNA-motifs, frequency
distribution
curves
were compiled
with
the
Motif
Mapper
python
scripts. All frequency dis-
tribution
curves were
rooted
to
the
annotated
start
of
the
transcriptional
unit
of
the
genes
pointing
to
the
right. Subsequently,
the
variance for each
motif
was
computed
from
its
distribution
curve.
2.4.
Phylogenetic
Analysis
Comparison
of
information
from
the
Arabidopsis, Populus, Sorghum
and
rice (Oryza) genomes as well
as
correlation
data
has
been
compared
by
us-
ing
ClustalW
(http://www.ebi.ae
.
ukl
elustalw/).
Default
settings
were
used
to
calculate
the
dendrogram
files. Tree
graphs
were visualized using
Tree View software version 1.6.6
(http://taxonomy
.
zoology.
gla.
ae.
ukl
rod/treeview
.
html).
The
evolutionary
distance
was
computed
on
RbcL
and
Cytc
sequences.
Distance
trees
for
the
variances
of
DNA-motifs
in
the
promoter
were
computed
from
the
Euclidean
distance
of
the
variance vec-
tors
by
using
statistiXL
1.8 for MS Excel.
The
variances for each
of
the
motifs
and
all four organisms have
been
used
as
attribute
data
to
compute
a derived
distance
matrix
according
to
the
statistiXL
descriptions.
Next,
a correlation coefficient was calculated,
to
indicate
how similar
the
final
hierarchical
pattern
and
initial
distance
matrix
were. A
dendrogram
file
was provided
to
graphically
summarize
the
similarity
patterns.
2.5.
Interspecies
Quantile-Quantile
Plots
The
a-quantiles
of
the
datasets
were
computed
using
Math
Works
MATLAB
routines
9.
For each
of
the
DNA-motifs in each of
the
organisms
promoter

377
datasets
the
a-quantiles
have been assessed on
the
basis of
the
variances of
motif
occurrence. To conclude on
the
conservation of motifs in
the
distantly
related
plant
species,
the
quantiles of
the
datasets
were
plotted
against
each other.
In
a QQ-plot two variance
datasets
were compared by
their
quantiles.
The
same quantiles are calculated for
both
datasets
and
used
as x-
/y-coordinates
for
the
QQ-plot.
The
a-quantiles
used for QQ-plot
comparisons were as follows:
0.05, 0.1, 0.15, 0.2, 0.25, 0.3, 0.35, 0.4, 0.45,
0.5, 0.55, 0.6, 0.65, 0.7, 0.75, 0.8, 0.85, 0.9, 0.91, 0.92, 0.93, 0.94, 0.95, 0.96,
0.97, 0.98
and
0.99.
3.
Results
and
Discussion
3.1.
DNA-motif
variance
as
a
measure
of
information
content
Biologists
and
bioinformation scientists are working
handin-hand
to
gain
better
insight into how regulatory non-coding
DNA
the
promoter
sequences
is capable of mediating changes in gene expression.
The
approaches
taken
so far
to
compare gene regulation on a genomic scale are
hampered
by
the
varying occurrence of these small DNA-motifs between species
due
to
dif-
ferent genome sizes or DNA-base composition
(GC/AT
-content)
1.
On
the
other
hand,
the
positional frequency of a motif in promoters of one species
can
be graphed as frequency
distribution
curves for each motif, whereby
each
motif
has its own
trajectory
(Fig. 1A).
Strong
deviations from
the
mean
background frequency are characteristic for disequilibria, i.e. regions
within a
promoter
that
do
not
follow
the
average distribution,
and
presump-
tively are from information
states of
DNA-protein evolution.
It
has been
demonstrated
that
higher
order
information is indeed responsible for these
disequilibria since
many
of
these motifs are often DNA binding sites for pro-
teins
1.
Based
on
these observations, one
can
conclude
that
a positional bias
of a
motif
in promoters from a single genome reflects functional information.
A comparative approach of DNA-motif
distribution
curves
is
complicated,
as necessary normalization procedures will have a transforming effect
on
the
curves (Fig. 1B)
and
distort
the
biological
data.
Thus,
we
used
the
variance
of a motifs frequency-distribution within
the
promoter
as a measure of
its
information content. DNA-motifs
with
high variance will have distribution-
curve signatures
that
are distinct from
the
background
mean
frequency and,
hence,
harbor
more information
than
others.
By
using
the
variance of a
motif
as a measure for information content, no additional normalization
or correction
is
needed,
thus
preserving as much biological information as

378
possible.
This
approach provides us
with
several observations:
First,
two
different frequency-distribution curves for two different
DNA
motifs
with
different means
can
still have
the
same
variance (Fig.
lA);
the
variance is
invariant
und
er
translation
and
will
not
be altered, when
the
plot is shifted
along
the
y-axis. Second, normalized distribution curves display different
variances
with
different means (Fig. 1B). As a consequence,
we
can
also
eva
luat
e
rare
motives
and
omit compensating for DNA-base composition
between
the
different genomes of
the
organisms.
We
extracted
the
-1000
bp
upstream
of
the
annotated
start
sites of genes.
This
resulted in in 33238
promoter
sequences for
the
Arabidopsis genome,
10483 for Populus,
36250 for Sorghum
and
18328 for Oryza. Next,
the
frequency
distribution
curves for all 4096 hexanucleotides
and
426 known
CREs
were compiled
and
the
variances were calculated.
3.2.
Quantile-quantile
(QQ)-plots
for
interspecies
promoter
comparison
To compare
the
variances of
the
motifs of different species,
we
made use of
QQ-plots
9.
In
a QQ-plot, two
datasets
are
compared by
their
quantiles.
The
same quantiles were calculated for each
dataset
and
used as coordinates
for
the
QQ-plot. If, for example, two normal distributions
with
different
variances are
plotted
,
the
resulting QQ-plot will exhibit a linear function
with
a steepness
dependent
on
the
variances; if
they
have different means
the
resulting QQ-plot will be affine.
If
the
variances of
the
hexanucleotide
or
CRE
maps
show a similar distribution,
the
QQ-plots could have
an
affine
shape.
The
steepness would correlate
with
the
variance of
the
variances.
A higher consensus (positional bias) in one species could
hint
to
higher
average information
content
in
the
underlying
promoter
sequences.
First
we
examine
the
QQ-plots for hexanucleotides for all species. Ara-
bidopsis
and
Populus are
both
dicot species
and
are considered
to
be more
related
to
each
other
than
to
the
two mono cots Oryza
and
Sorghum.
When
the
quantiles of
the
variances of Arabidopsis
and
Populus were
QQ-plotted
against each
other
(Fig. 2A), a
graph
with
only a small steepness was
the
result.
This
can
be
explained by higher information content in
the
pro-
moters of Arabidopsis compared
to
Populus. However,
there
was a
strong
increase in
the
last quantile
[0.99]
in Populus, which
can
be
accounted
to
41
hexanucleotides
with
the
highest variance in
both
organisms. Among
these motifs, we identified several motifs of
putative
functionality, e.g.
the
TATA-box-like motifs
1.
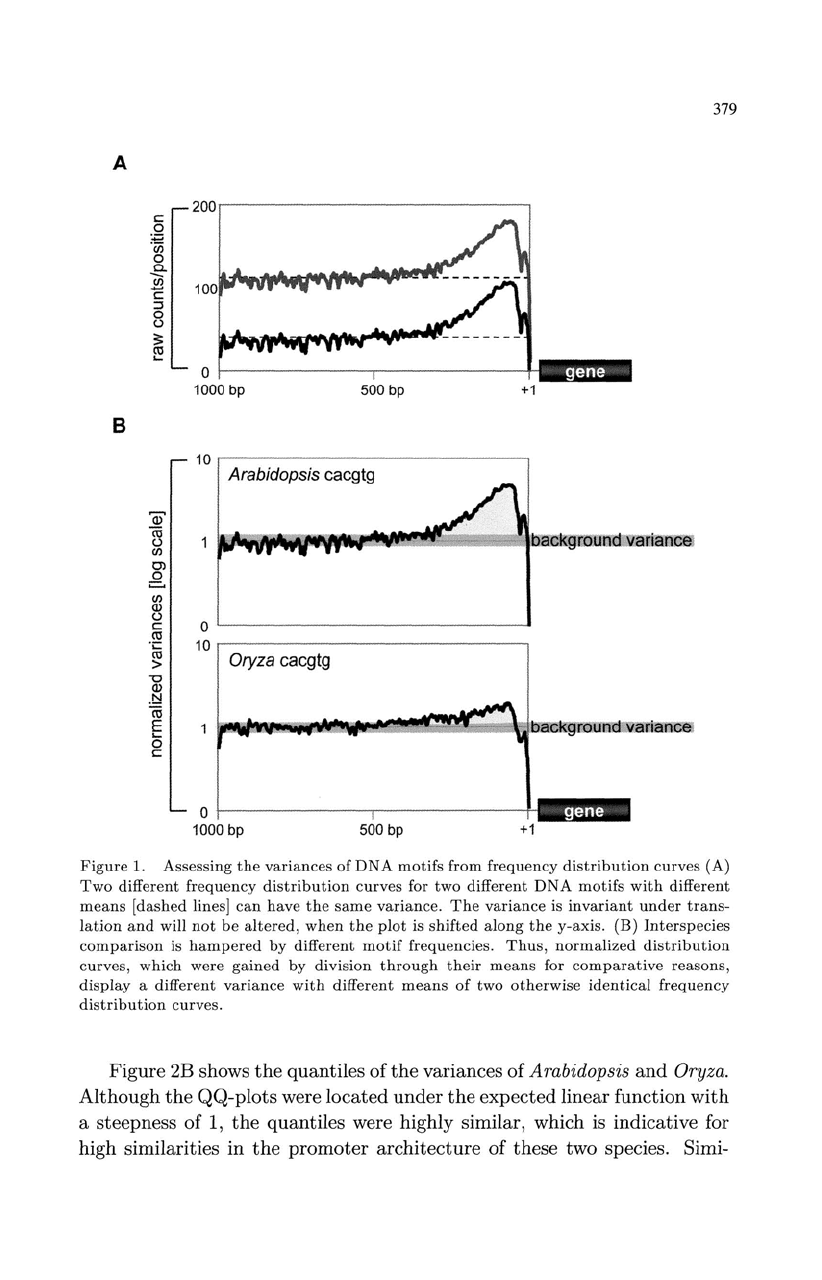
379
+1
+1
Figure
1. Assessing
the
variances
of
DNA
motifs
from
frequency
distribution
curves
(A)
Two
different
frequency
distribution
curves
for
two
different
DNA
motifs
with
different
means
[
dashed
lines]
can
have
the
same
variance.
The
variance
is
invariant
under
trans-
lation
and
will
not
be
altered,
when
the
plot
is
shifted
along
the
y-axis.
(B)
Interspecies
comparison
is
hampered
by
different
motif
frequencies.
Thus
,
normalized
distribution
curves,
which
were
gained
by
division
through
their
means
for
comparative
reasons,
display
a
different
variance
with
different
means
of
two
otherwise
identical
frequency
distribution
curves.
Figure
2B shows
the
quantiles
of
the
variances
of
Arabidopsis
and
Oryza.
Although
the
QQ-plots
were
located
under
the
expected
linear function
with
a steepness
of
1,
the
quantiles were highly similar, which is indicative for
high similarities in
the
promoter
architecture
of
these
two species. Simi-
