Accardi L., Freudenberg W., Obya M. (Eds.) Quantum Bio-informatics IV: From Quantum Information to Bio-informatics
Подождите немного. Документ загружается.

380
A
500 1000 1500 2000
Arabidopsis
B
500 1000 1500 2000
Arabidopsis
c
O
..
··~·
···
-
··
·
···
-·
·
o 500 1000 lS00 2000 2500
Arabidopsis
o
15001
'
ff)
.
::I
"3 I
§'1000t
CL
'
SOO~'"
o
o
E
F
500 1000 1500 2000
Oryza
SOD
1000
1S00
2000 2500
Oryza
2500r ... -
••
_-_
..
.
....
. .
2000'
E ,
.E
1500+
g'
i
en
1000·
500
..
j
o
..
"
...
.
......
.....
....
...
.
.......
,
...
. '
o 500 1000 lS00 2000 2500
Populus
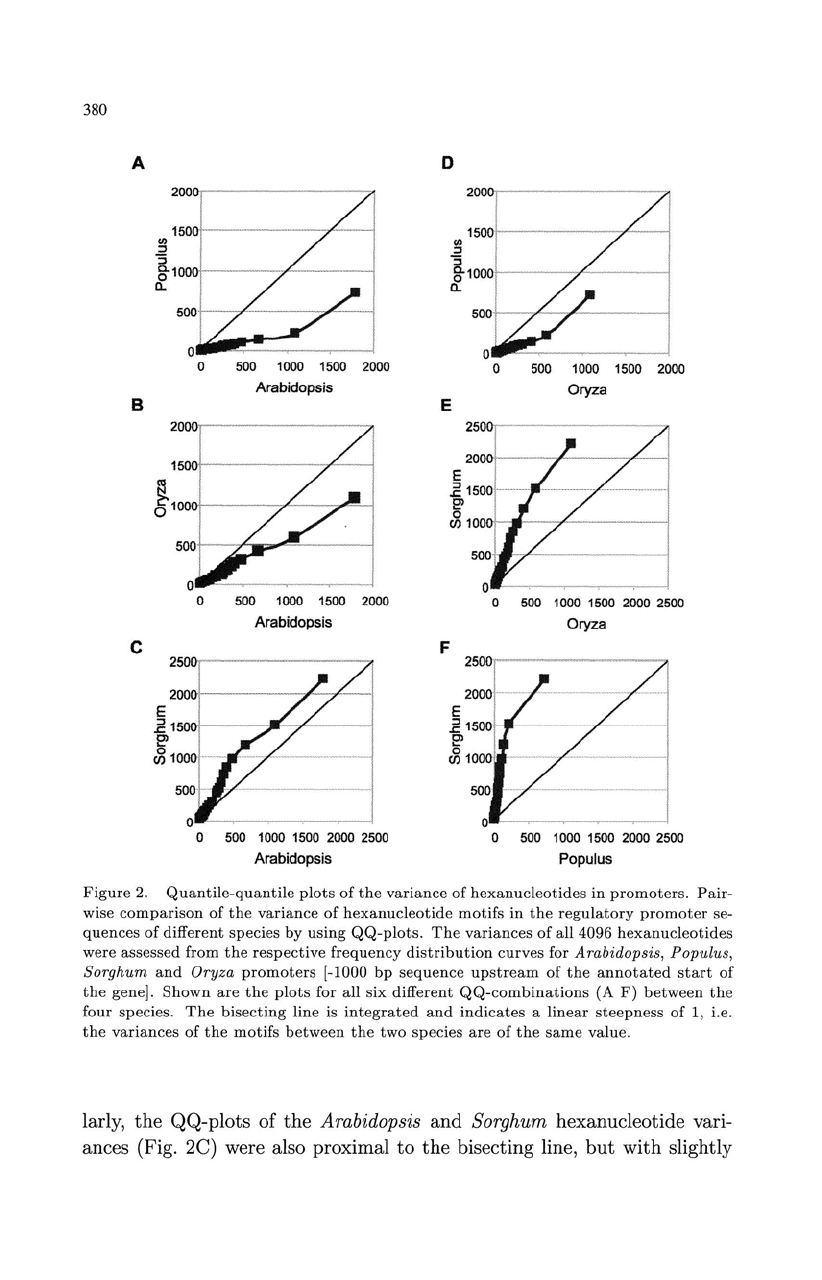
Figure
2.
Quantile·quantile
plots
of
the
variance
of
hexanucleotides
in
promoters.
Pair·
wise
comparison
of
the
variance
of
hexanucleotide
motifs
in
the
regulatory
promoter
se·
quences
of
different
species
by
using
QQ·plots.
The
variances
of
all 4096
hexanucleotides
were
assessed from
the
respective
frequency
distribution
curves
for Arabidopsis, Populus,
Sorghum
and
Oryza
promoters
[·1000
bp
sequence
upstream
of
the
annotated
start
of
the
gene].
Shown
are
the
plots
for all six different
QQ-combinations
(A
F)
between
the
four
species.
The
bisecting
line is
integrated
and
indicates
a
linear
steepness
of 1, i.e.
the
variances
of
the
motifs
between
the
two
species
are
of
the
same
value.
larly,
the
QQ-plots of
the
Arabidopsis
and
Sorghum hexanucleotide vari-
ances (Fig. 2C) were also proximal
to
the
bisecting line,
but
with
slightly

381
higher
variances
in
Sorghum. Here again,
the
majority
of
difference was
found in
the
quantiles between 0.90
and
0.96.
This
can
not
readily
be
ex-
plained
by
e.g. different sizes
of
the
promoter
dataset
-
both
species
had
a
similar
number
of
annotated
promoters.
However,
it
might
be
possible
that
the
Sorghum genome
data
or
the
dataset
contains
a
certain
degree
of
re-
dundancy
and,
thus,
there
were
more
highly conserved motifs
with
a
higher
variance found
that
lead
to
higher quantiles. Therefore,
further
research
on
the
nature
of
these
elements
and
on
the
Sorghum
dataset
needs
to
be
done
to
clarify
this
matter.
The
comparison
ofthe
hexanucleotides for Populus
and
Oryza (Fig. 2D)
displays overall similarities
and
trends
to
the
Populus
and
Arabidopsis com-
parison
(Fig. 2A). Hence,
the
Populus
promoter
dataset
contains
unique
features
that
account
for
these
differences.
This
observation
can
not
be
ex-
plained
by
differences in
the
sizes
of
the
dataset
as
the
number
of
promoters
contains in
the
Oryza
and
Populus
dataset
are
of a similar
magnitude,
while
Arabidopsis is larger. Nevertheless,
the
same
trends
were observed for all
comparisons
with
the
Populus
promoters.
Figure
2E
displays
the
compar-
ison
of
hexanucleotide variances
of
Sorghum
and
Oryza. Interestingly,
the
quantiles
of
Sorghum were
much
higher
than
those
of
Oryza. As
has
been
noted
previously,
an
explanation
would
be
that
the
underlying
promoter
re-
gions in
Sorghum have a higher
amount
of high-consensus hexanucleotides
-
probably
hinting
to
a higher
number
of
regulatory
active sequences
or
to
redundance
within
the
dataset.
In
Figure
2F, Sorghum is
compared
to
the
Populus
dataset.
While
Populus displayed
the
lowest values
in
its
quan-
tiles,
the
Sorghum
dataset
contains
the
highest values
in
its quantiles
of
our
organisms
and,
hence,
the
resulting
plot
has
a very high steepness
due
to
the
high
amount
of high-consensus hexanucleotides (those
with
specific
positional bias)
in
the
Sorghum
promoters.
While
we consider
the
variance in hexanucleotides a
measure
for over-
all
promoter
architecture
and
motif
composition, we
expected
that
the
variances
of
CREs
would provide us
with
information
on
conserved core-
promoters
and
retained
CRE
function
in
gene regulation.
Figure
3 gives
QQ-plots
of
the
CRE-motives
for
the
four
plant
species
under
investigation.
For
comparative
reasons, we also included
the
data
of
the
hexanucleotide
variances [in grey] from
the
previous
study
in
the
CRE-plots.
This
will pro-
vide us
with
the
information,
whether
the
variances
of
the
CREs
differ from
the
general
promoter
architecture.
Since
the
CRE-motives
harbor
known
regulatory
information, we could
expect
from
them
to
have high variances
in
the
promoter
regions
and,
thus,
harbor
a
higher
order
of
information.
382
A
B
c
o
2000 4000
Arabidopsis
6000
2000 4000 6000
ArabidopsiS
2000 4000 6000
Arabidopsis
o
o
2000 4000 6000
Oryza
E
o
2000 4000 6000
Oryza
F
o
:2000
4000 6000
POpulus
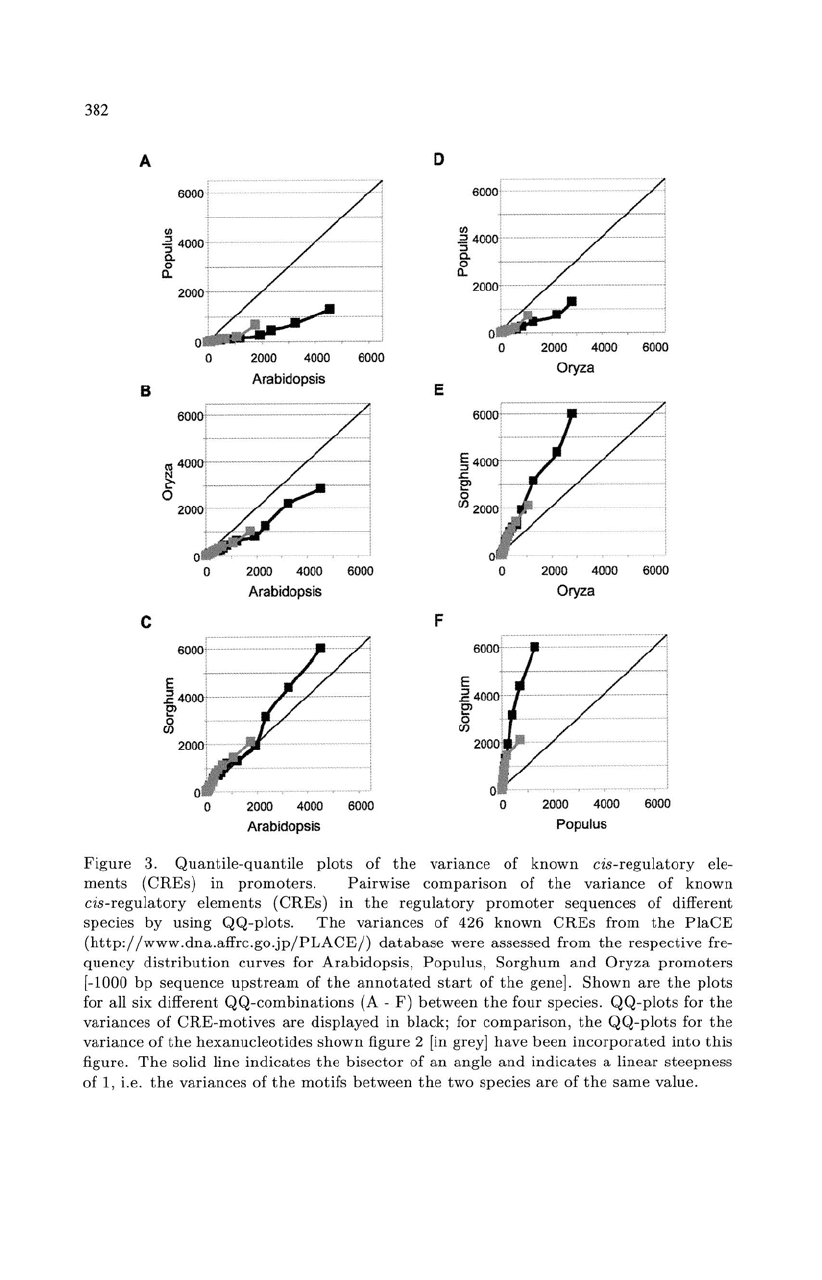
Figure
3.
Quantile-quantile
plots
of
the
variance
of
known
cis-regulatory
ele-
ments
(CREs)
in
promoters.
Pairwise
comparison
of
the
variance
of
known
cis-regulatory
elements
(CREs)
in
the
regulatory
promoter
sequences
of
different
species
by
using
QQ-plots.
The
variances
of
426
known
CREs
from
the
PlaCE
(http://www.dna.affrc.go.jp/PLACE/)
database
were
assessed
from
the
respective
fre-
quency
distribution
curves
for
Arabidopsis,
Populus,
Sorghum
and
Oryza
promoters
[-1000
bp
sequence
upstream
of
the
annotated
start
of
the
gene].
Shown
are
the
plots
for
all
six
different
QQ-combinations
(A
-
F)
between
the
four
species.
QQ-plots
for
the
variances
of
CRE-motives
are
displayed
in
black; for
comparison,
the
QQ-plots
for
the
variance
of
the
hexanucleotides
shown
figure 2 [in grey]
have
been
incorporated
into
this
figure.
The
solid
line
indicates
the
bisector
of
an
angle
and
indicates
a
linear
steepness
of
1, i.e.
the
variances
of
the
motifs
between
the
two
species
are
of
the
same
value.

383
In
Figure
3A
the
quantiles
of
Populus were
plotted
against
Ambidop-
sis.
Similar
to
the
hexanucleotide comparison,
the
quantiles
of
Populus
were
much
lower
than
those
of
A mbidopsis.
On
the
one
hand
this
find-
ing could
be
explained
by
the
on
average smaller sizes of
promoters
with
higher
information
content
in
Ambidopsis.
On
the
other
hand,
the
Pop-
ulus
dataset
is
of
a
poor
quality
due
to
mis-annotation
or
problems
to
curate
the
genome information.
Next,
the
quantiles
of
the
CRE
variances
of
Oryza were
compared
to
Ambidopsis (Fig. 3B).
It
was obvious
that
the
variances were
of
approximately
of
the
same
values for
the
promoters
of
both
species
and,
thus,
close
to
the
bisecting line. Here, also
the
steepness
of
the
QQ-plot
for
the
CRE
variances was similar
to
that
of
the
QQ-plot
of
the
hexanucleotides, which
points
towards
the
fact,
that
the
amount
of
relative
information
between
two species is similar for hexanucleotides
and
CRE-motifs.
Figure
3C
shows
the
QQ-plot
of the
CRE
variances
of
Sorghum
and
Ambidopsis.
The
quantiles
of
the
variances were
very
similar for
both
species, which is indicative
of
similar frequency
distribution
curves for
the
CREs
in
the
promoters
of
the
two species. Especially for
the
lower quantiles
the
graph
follows
the
dissecting line. Hence, one
can
conclude
that
the
motif
composition in
the
promoters
of
the
two species
and
their
architectures
are
highly similar in general. However,
this
similarity was highest for
the
known
functional
CREs.
Only
for
the
quantiles
0.97 - 0.99,
the
variances diverged.
The
simplest
explanation
for
this
observation
was
the
presence
of
longer
CRE-consensi
or
naturally
rare
motifs, which
occur
slightly
more
in one
genome
than
the
other
and,
thus,
have
exorbitantly
high
variance over a
very low
mean
frequency.
The
QQ-plots
of
the
CRE
variances
of
Populus
against
the
Oryza
(Fig. 3D)
or
Sorghum (Fig. 3F)
datasets
revealed a very low
information
content
for Populus. Again,
the
values
in
the
quantiles of Populus were
the
smallest in
the
whole
dataset
and,
therefore, Populus also displayed a
low
CRE
consensus.
The
same
trend
was seen for
the
Sorghum
promot-
ers.
In
Figure
3E,
the
quantiles
of
CRE
variance in Sorghum were
plotted
against
the
Oryza
dataset;
these
quantiles
displayed similarities for
both
species
and,
thus,
the
distribution
of
the
known
functional elements
in
the
promoters
likely follows a similar
trend
in frequency. Since
the
quantiles
of Sorghum have also
been
similar
to
that
of
Ambidopsis (Fig. 3C), we
could conclude
that
CRE
distribution
in
the
promoters
might
indeed
be
conserved between
the
three
species. Moreover,
these
similarities could
be
detected
with
QQ-plots
of
the
variance
as
a
measure
irrespective
of
the
384
A
B c
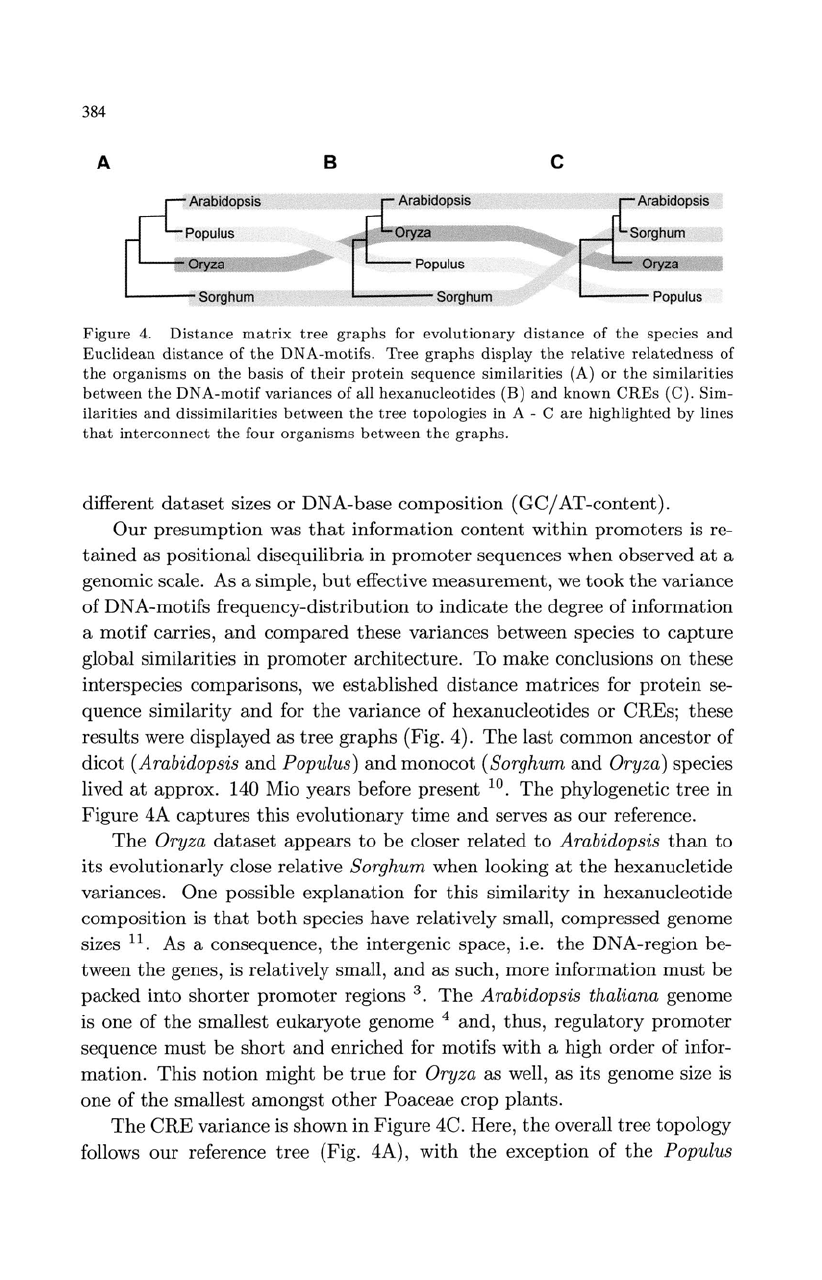
Figure
4.
Distance
matrix
tree
graphs
for
evolutionary
distance
of
the
species
and
Euclidean
distance
of
the
DNA-motifs.
Tree
graphs
display
the
relative
relatedness
of
the
organisms
on
the
basis
of
their
protein
sequence
similarities
(A)
or
the
similarities
between
the
DNA-motif
variances
of
all
hexanucleotides
(B)
and
known
CREs
(C).
Sim-
ilarities
and
dissimilarities
between
the
tree
topologies
in
A - C
are
highlighted
by
lines
that
interconnect
the
four
organisms
between
the
graphs.
different
dataset
sizes
or
DNA-base composition
(GC/AT-content).
Our
presumption
was
that
information
content
within
promoters
is re-
tained
as positional disequilibria
in
promoter
sequences
when
observed
at
a
genomic scale. As a simple,
but
effective measurement, we
took
the
variance
of
DNA-motifs frequency-distribution
to
indicate
the
degree
of
information
a
motif
carries,
and
compared
these
variances between species
to
capture
global similarities in
promoter
architecture. To make conclusions
on
these
interspecies comparisons, we established distance matrices for
protein
se-
quence similarity
and
for
the
variance
of
hexanucleotides
or
CREs;
these
results were displayed as
tree
graphs
(Fig. 4).
The
last
common ancestor
of
dicot (Arabidopsis
and
Populus)
and
monocot (Sorghum
and
Oryza) species
lived
at
approx. 140 Mio years before present
10.
The
phylogenetic
tree
in
Figure
4A
captures
this
evolutionary
time
and
serves as
our
reference.
The
Oryza
dataset
appears
to
be
closer
related
to
Arabidopsis
than
to
its
evolutionarly close relative Sorghum when looking
at
the
hexanucletide
variances.
One possible
explanation
for
this
similarity in hexanucleotide
composition is
that
both
species have relatively small, compressed genome
sizes
11.
As a consequence,
the
intergenic space, i.e.
the
DNA-region be-
tween
the
genes, is relatively small,
and
as such, more information
must
be
packed
into
shorter
promoter
regions
3.
The
Arabidopsis thaliana genome
is one
of
the
smallest eukaryote genome 4
and,
thus,
regulatory
promoter
sequence
must
be
short
and
enriched for motifs
with
a
high
order
of
infor-
mation.
This
notion
might
be
true
for Oryza as well, as
its
genome size is
one
of
the
smallest
amongst
other
Poaceae crop plants.
The
CRE
variance is shown
in
Figure
4C. Here,
the
overall
tree
topology
follows
our
reference
tree
(Fig. 4A),
with
the
exception
of
the
Populus

385
dataset,
which could likely
be
considered as
an
outlier.
The
reasons for
the
aberrant
Populus set
might
be
manifold,
but
most
likely
originate
from a
low
quality
in
the
annotation
of
the
gene
starts.
4.
Conclusions
We have shown
that
promoter
architecture
is
strongly
dependent
on
the
sizes of intergenic regions.
The
observation
that
the
CRE
variances for
Arabidopsis, Oryza
and
Sorghum were highly similar
supports
the
idea
that
the
changes
in
protein
sequence
or
in cis-regulatory
DNA
over
time
might
be
proportional.
Thus,
known functional elements display
the
same
evo-
lutionary
concepts
as
are
considered for genome
or
protein
evolution.
To
further
support
and
refine
our
findings, well
annotated
genome
information
of
more
species is needed
and
has
to
be
integrated
in
our
analysis.
The
sequence
information
of species
with
different genome sizes
and
larger evo-
lutionary
distances
would
be
of
special
interest.
Moreover,
the
analysis
of
the
CRE
variances is
hampered
by
too
little
information
on
transcription
factor
(TF)-CRE
relationships. Hence,
our
approach
is well
worth
to
be
repeated
with
updated
information
on
both,
TF-CREs
and
a larger
number
of
promoter
datasets.
Acknowledgements
We
thank
Jochen
Supper
for his
constant
input
and
critical discussions.
This
work was
supported
by
the
DFG
(HA2146/11-1).
References
1.
K.
W.
Berendzen, K.
Stueber,
K.
Harter
and
D. Wanke,
Bmc
Bioinformatics
7(NOV
30 2006).
2.
J.
L.
Riechmann,
J.
Heard,
G.
Martin,
L.
Reuber,
C.
Jiang,
J.
Keddie,
L.
Adam,
O.
Pineda,
O.
J.
Ratcliffe, R. R.
Samaha,
R.
Creelman,
M. Pilgrim,
P.
Broun,
J.
Z.
Zhang, D.
Ghandehari,
B. K.
Sherman
and
G. Yu, Science
290,
2105(Dec 2000).
3. D.
Walther,
R.
Brunnemann
and
J.
Selbig,
PLoS
Genet
3,
p.
ell(Feb
2007).
4.
Arabidopsis
Genome
Initiative,
Nature
408,
796(Dec 2000).
5. Y.
Yamazaki
and
P. Jaiswal, Plant Cell
Physiol46,
63(Jan
2005).
6.
International
Rice
Genome
SequencingProject,
Nature
436,
793(Aug 2005).
7.
X.
Wang,
G.
Haberer
and
K.
F.
X. Mayer,
BMC
Genomics
10,
p. 284 (2009).
8. G.
A.
Tuskan,
S.
Difazio,
S.
Jansson,
J.
Bohlmann,
I.
Grigoriev, U. Hell-
sten,
N.
Putnam,
S.
Ralph,
S.
Rombauts,
A. Salamov,
J.
Schein,
L.
Sterck,
A.
Aerts,
R. R.
Bhalerao,
R. P.
Bhalerao,
D. Blaudez,
W.
Boerjan,
A.
Brun,

386
A.
Brunner,
V. Busov, M.
Campbell,
J.
Carlson, M.
Chalot,
J.
Chap-
man,
G.-L.
Chen,
D.
Cooper,
P. M.
Coutinho,
J.
Couturier,
S.
Covert,
Q.
Cronk,
R.
Cunningham,
J.
Davis,
S.
Degroeve, A.
Dejardin,
C.
Depam-
philis,
J.
Detter,
B. Dirks,
I.
Dubchak,
S.
Duplessis,
J.
Ehlting,
B. Ellis,
K. Gendler, D.
Goodstein,
M. Gribskov,
J.
Grimwood, A. Groover, L.
Gunter,
B.
Hamberger,
B. Heinze, Y.
Helariutta,
B.
Henrissat,
D. Holligan,
R.
Holt,
W.
Huang,
N. Islam-Faridi,
S.
Jones, M.
Jones-Rhoades,
R.
Jorgensen,
C. Joshi,
J.
Kangasjarvi,
J.
Karlsson, C. Kelleher,
R.
Kirkpatrick,
M.
Kirst,
A. Kohler, U. Kalluri,
F.
Larimer,
J.
Leebens-Mack, J.-C. Leple, P. Locas-
cio, Y. Lou,
S.
Lucas,
F.
Martin,
B.
Montanini
, C. Napoli, D.
R.
Nelson,
C. Nelson, K. Nieminen, O. Nilsson, V.
Pereda,
G.
Peter,
R.
Philippe,
G.
Pi-
late,
A. Poliakov,
J.
Razumovskaya, P.
Richardson,
C. Rinaldi, K.
Ritland,
P. Rouze, D. Ryaboy,
J.
Schmutz,
J.
Schrader,
B. Segerman, H. Shin, A. Sid-
diqui,
F.
Sterky, A. Terry,
C.-J.
Tsai,
E.
Uberbacher,
P.
Unneberg,
J.
Vahala,
K.
Wall,
S.
Wessler, G. Yang,
T.
Yin, C. Douglas, M.
Marra,
G.
Sandberg,
Y. Van de
Peer
and
D.
Rokhsar,
Science
313,
1596(Sep 2006).
9.
Y.-Y. Ho, L. Cope, M.
Dettling
and
G.
Parmigiani,
Methods Mol
Biol408,
171 (2007).
10. S.-M. Chaw, C.-C.
Chang,
H.-L.
Chen
and
W.-H. Li, J Mol
Evol58,
424(Apr
2004).
11. M.
Spannagl,
O.
Noubibou,
D. Haase, L. Yang, H.
Gundlach,
T.
Hindemitt,
K. Klee, G.
Haberer,
H. Schoof
and
K.
F.
X. Mayer, Nucleic Acids Res
35,
DS34(Jan
2007).

Quantum
Bio-Informatics
IV
eds. L.
Accardi,
W .
Freudenberg
and
M.
Ohya
© 2011
World
Scientific
Publishing
Co. (pp.
387-4
01)
ENTROPY
TYPE
COMPLEXITIES
IN
QUANTUM
DYN
AMICAL
PROCESSES
NOBORU WATANABE
Department
of
Information Sciences,
Tokyo University
of
Science,
Noda
City, Chiba 278-8510, Japan
E-mail: watanabe@is.noda.tus.ac.jp
In
a
study
of
several
systems,
we
are
int
eres
ted
to
examin
e (1)
the
dynamics
of
state
change
and
(2)
th
e
comp
lexity
of
states
.
Ohya
introdu
ced
in
1991 (ref.[21])
a
general
idea
so-called a
Information
Dynamics
(ID),
which
const
itute
s a
theory
under
a
frame
of
the
ID
by
sy
nthesizin
g
the
formaliti
es
of
the
investigati
ons
of
(1)
and
(2).
There
are
two
kind
of
complexities
in ID.
One
is a
complexity
of
state
describing
system
itself
and
anoth
er is a
transmitted
complexity
betw
een
two
systems.
Entropies
of
classical
and
quantum
systems
are
the
example
of
these
complexiti
es.
In
order
to
treat
a flow
of
dynamics
proce
ss,
dynamical
entropies
were
introduced
in
not
only
classical
but
also
quantum
systems
.
The
main
purpose
of
this
paper
is
to
compare
with
these
mean
entropies
and
the
complexiti
es
in
rD,
we calc
ulate
these
mean
entropies
for
some
simple
models
to
discuss
the
complexity
of
information
transmission
for
OOK
and
PSK
modulations.
1.
Introduction
In
several complicated systems,
it
is
important
to
study
(1)
the
dynamics
of
state
change
and
(2)
the
complexity of
states
of
systems.
Information
Dynamics (ID)
introduced
by
Ohya
is a general
idea
constructing
a
theory
by synthesizing
the
research schemes
of
(1)
and
(2)
under
a frame
of
the
ID.
In
ID,
there
are
two
type
of complexities,
that
is, (a) a complexity
of
state
describing
system
itself
and
(b) a
transmitted
complexity between
two systems. Entropies
of
classical
and
quantum
information
theory
are
the
example
of
the
complexities
of
(a)
and
(b).
In
quantum
information theory,
Ohya
introduced
a
compound
state
and
defined
Ohya
mutual
entropy
18
based
on
quantum
relative
entropy
of
Umegaki 31
in
1983, he
extended
it
19
to
general
quantum
systems
by using
the
relative
entropy
of
Araki
5
and
Uhlmann
32.
Based
on
the
quantum
mutual
entropy,
he
quantum
capacity
is discussed in
24,25,28.
One
can
discuss
the
coding theorems by means
of
387

388
the
mean
entropy
and
the
mean
mutual
entropy
defined
by
the
dynamical
entropy.
The
KS
entropy
14
was
introduced
in classical
systems.
Several
quantum
dynamical
entropy
were
studied
by
Emch
10
,
Connes-Stormer
9,
Connes, Narnhoffer
and
Thirring
8,
Park
29,
.Alicki
and
Fannes
4,
Hudetz
12.
Ohya
21,
Voiculescu
33,
Accardi,
Ohya
and
Watanabe
2,
3,
Kossakowski,
Ohya
and
Watanabe
15,34,
Ohya
and
Petz
23
and
Choda
7,
Bennati
6,
Muraki
and
Ohya
16
and
so on.
In
this
paper,
we briefly
explain
the
mean
entropy
and
the
mean
mutual
entropy
defined
by
Ohya
21.
We
calculate
these
mean
entropies
for some
simple
models
to
discuss
the
complexity
of
information
transmission
for
subsets
of
the
initial
state
space.
2.
Quantum
Channels
The
concept
of
channel
has
been
played
an
important
role in
th
e progress
of
the
quantum
communication
theory. Here we briefly review
the
notion
of
the
quantum
channels.
Let
Hk
(k
=
1,2)
be
complex
separable
Hilbert
spaces. We
denote
the
set
of
all
bounded
linear
operators
on
Hk
by
B(Hd
(k =
1,2)
and
we
express
the
set
of all
density
operators
on
Hk
by
6(Hk)
(k =
1,2).
Let
(B(H
k
),6(H
k
))
(k
=
1,2)
be
input
(k
= 1)
and
output
(k = 2)
quantum
systems,
respectively.
(1) A
mapping
from
6(Hd
to
6(H2)
is called a
quantum
channel
A*.
(2) A * is called a linear
channel
if A * satisfies
the
affine
property
such
as
A*(2:k
AkPk)
= 2:k
Ak
A
*
(Pk)
for
any
Pk
E
6(Hl)
and
any
non-
negative
number
Ak
E [0,1]
with
2:k
Ak
=
1.
For
the
quantum
channel
A *,
the
dual
map
A
of
A * is defined
by
trA*(p)B = trpA(B),
Vp
E
6(H
1
),
VB
E B(H2)'
(3) A * is called a
completely
positive
(CP)
channel
if A * is
linear
channel
and
its
dual
map
A :
B(H
2
)
--+
B(H
1
)
of
A * holds
(
x,t
AiA(A;Aj)AjX);::::: 0
(Vx
E HI)
t,J=1
for
any
n E
N,
any
{Ad
c
B(H
2
)
and
any
{Ai}
C
B(Hd·
Almost
all physical
transform
of
states
can
be
denoted
by
the
CP
chan-
nels
18,19,13,23,27.

389
2.1.
Noisy
optical
channel
Now we explain
the
noisy optical channel such as
an
example
of
the
quan-
tum
communication
channels
19,26.
Let
Kl
and
K2
be
complex separable Hilbert spaces
of
noise
and
loss
systems, respectively. For
an
input
state
p in
6(Hd
and
a noise
state
~
E 6
(Kd,
we defined
in
26
a
mapping
IT*
by
IT*
(p
@~)
==
V
(p
@
~)
V*,
which is called a generalized
beam
splitting, where V is a linear
mapping
from
HI
@
Kl
to
H
2
@K
2
given by
nl+ml
V(lnl;
@
Iml;)
= L
Cj",m"lj;
@ Inl +
ml
-
j;
j=O
for
the
nl,ml,j,(nl+ml-j)
photon
number
state
vectors
Inl;
E
HI,
Iml;
E K
l
, Ij; E
H2,
Inl +
ml
-
j;
E K
2
,
and
K . I , ,
"(
_
')'
Cn1,m1
=
''"'(-It"+
j
-
r
ynl·ml·]·
nl
+ml
].
J
~
r!(nl -
j)!(j
-
r)!(ml
- j + r)!
r=L
xaml-j+2r
(-73)
nl
+j-2r
.
(1)
a
and
{3
are
complex numbers satisfying lal
2
+
1{31
2
=
l.
K
and
L
are
constants
given by K =
min{nl,j},
L =
max{ml
-
j,
O}.
For
the
coherent
input
state
p =
Ie;
(el @
Ih;;
(h;I
E 6
(Hl@Kd,
the
output
state
of
IT*
is
obtained
by
IT*
(Ie; (el @
Ih;;
(h;I)
=
lae
+
{3h;;
(ae
+
{3h;1
@
1-73e
+
(ih;)
\
-73e
+
(ih;I·
IT*
with
the
vacuum
noise
state
~o
=
10;
(01
is called
the
beam
splitter
ITo
given by
ITo
(Ie; (el
@~O)
=
lae;
(ael
@
1-73e)
\-73
e
l
for
the
coherent
input
state
p @
~o
=
Ie;
(el @
10;
(01
E 6
(Hl@Kd·
ITo
was
described by means of
the
lifting
Eo
from 6 (H)
to
6
(H@K)
in
the
sense
of
Accardi
and
Ohya
1 as follows
Eo
(Ie; (el) =
lae;
(ael
@
l{3e)
({3el·
Based
on
the
liftings,
the
beam
splitting
was
studied
by Accardi -
Ohya
and
Fichtner
- Freudenberg - Libsher
11.
Noisy
quantum
channel A *
introduced
in
26
with
a fixed noise
state
~
E 6
(Kd
was defined
by
A*(p)
==
trJC
2
IT*(p
@~)
=
trJC2
V (p
@~)
V*.
(2)
