Wilkinson D.J. Stochastic Modelling for Systems Biology
Подождите немного. Документ загружается.


234
INFERENCE FOR STOCHASTIC KINETIC MODELS
advantages
of
using a sequential algorithm is that it is very convenient to use in the
context
of
multiple data sets from different cells
or
experiments, possibly measuring
different species in each experiment. A discussion
of
this issue in the context
of
an
illustrative example is given
in
Golightly & Wilkinson (2006).
Given this discussion, it is clearly possible to develop a generic piece of software
for inferring the rate parameters
of
the CLE for realistic experimental data (at the
single-cell level) using MCMC techniques. At the time
of
writing, I am currently
developing such an application
(stochinf2),
and a link to it will be posted on the
website for this book when it becomes available.
10.5 Network inference
At
this point it is worth saying a few words regarding network inference.
It
is cur-
rently fashionable in some parts
of
the literature to attempt to deduce the structure of
biochemical networks
ab initio from routinely available experimental data. While it
is clearly possible to develop computational algorithms to do this, the utility
of
doing
so is not at all clear due to the fact that there will typically be a very large number
of
distinct network structures all
of
which are consistent with the available experimen-
tal data (large numbers
of
these will be biologically implausible, but a large number
will also
be
quite plausible). In this case the "best fitting" network is almost certainly
incorrect. For the time being it seems more prudent to restrict attention to the less
ambitious goal
of
comparing the experimental support for a small number
of
com-
peting network structures. Typically this will concern a relatively well-characterised
biochemical network where there is some uncertainty
as
to whether one or two
of
the potential reaction steps actually take place. In this case, deciding whether or not
a given reaction is present is a problem
of
discrimination between two competing
network structures. There are a number
of
ways that this problem could be tack-
led. The simplest approach would be to fit all competing models using the MCMC
techniques outlined in the previous sections and compute the marginal likelihoods
associated with each (Chib 1995) in order to compute Bayes factors. More sophis-
ticated strategies might adopt reversible jump MCMC techniques (Green 1995) in
order to simultaneously infer parameters and structure. In principle, the reversible
jump techniques could also be used for
ab initio network inference, but note well
the previous caveat. In particular, note that inferences are sensitive not only
to
the
prior adopted over the set
of
models to
be
considered, but also to the prior structure
adopted for the rate constants conditional on the model. Simultaneous inference for
parameters and network structure is currently an active research area.
10.6 Exercises
1.
Pick a simple model (say, from Chapter 7) and simulate some complete data from
it using the Gillespie algorithm.
(a) Use these data to form the complete-data likelihood
(10.2).
(b) Maximise the complete data likelihood using (10.4).




238
CONCLUSIONS
possible to extend model calibration technology for the fitting
of
stochastic models,
it
is not straightforward
to
do so in practice. The development
of
fast and approx-
imate techniques for calibrating stochastic simulation models is currently an active
res.earch area.
In
conclusion, this text has covered only the essential concepts required for begin-
ning to
think seriously about systems biology from a stochastic viewpoint. The really
interesting statistical problems concerned with marrying stochastic systems biology
models to routinely available high-throughput experimental data remain largely un-
solved and the subject
of
a great deal
of
ongoing research.


240
</math>
<listOfParameters>
<parameter
id="kl
11
value=-
11
1''
/>
</listOfPararneters>
</kineticLaw>
</reaction>
SBMLMODELS
<reaction
id="ReverseRepressionBinding"
name="Reverse
repression
binding"
reversible=
11
false
11
>
<listOfReactants>
<SpeciesReference
species="P2Gene"/>
</listOfReactants>
<listOfProducts>
<speciesReference
species=
11
Gene
11
/>
<SpeciesReference
species=
11
P2
11
/>
</listOfProducts>
<kineticLaw>
<math
xmlns="http://www.w3.org/1998/Math/MathML">
<apply>
<times/>
<ci>
klr
</ci>
<Ci>
P2Gene
</Ci>
</apply>
</math>
<listOfParameters>
<parameter
id=
11
klr
11
value="10''/>
</listOfParameters>
</kineticLaw>
</reaction>
,
<reaction
id="Transcription
11
reversible=
11
false
11
>
<listOfReactants>
<SpeciesReference
species="Gene"/>
</listOfReactants>
<listOfProducts>
<speciesReference
species="Gene"/>
<SpeciesReference
species="Rna"/>
</listOfProducts>
<kineticLaw>
<math
xmlns="http://www.w3.org/1998/Math/MathML">
<apply>
<times/>
<Ci> k2
</Ci>
<Ci>
Gene
</Ci>
</apply>
</math>
<listOfParameters>
<parameter
id=
11
k2
11
value=
11
0.01"/>
</listOfParameters>
</kineticLaw>
</reaction>
<reaction
id=
11
Translation"
reversible="false">
<listOfReactants>
<speciesReference
species="Rna
11
/>
</listOfReactants>
<listOfProducts>
<speciesReference
species=
11
Rna"/>
<speciesReference
species="P"/>
</listOfProducts>
<kineticLaw>
<math
xmlns="http://www.w3.org/1998/Math/MathML">
<apply>
<times/>
<Ci> k3
</Ci>
<Ci>
Rna
</Ci>
</apply>
</math>
<listOfParameters>
<parameter
id="k3"
value=''10''
/>

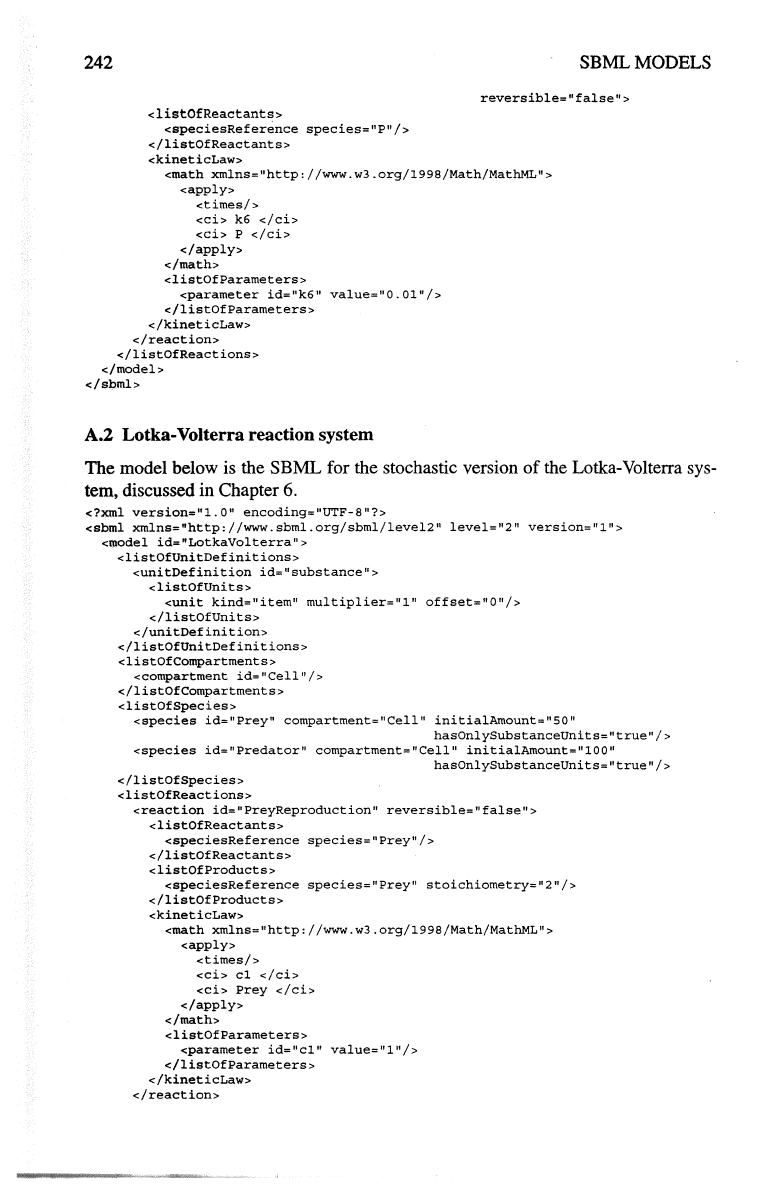
242
<listOfReactants>
<SpeciesReference
species="P"/>
</listOfReactants>
<kineticLaw>
SBMLMODELS
reversible="false''
>
<math
xmlns="http://www.w3.org/1998/Math/MathML">
<apply>
<times/>
<Ci> k6
</Ci>
<Ci>
P
</Ci>
</apply>
</math>
<listOfParameters>
<parameter
id="k6"
value="O.Ol"/>
</listOfParameters>
</kineticLaw>
</reaction>
</listOfReactions>
</model>
</sbml>
A.2 Lotka-Volterra reaction system
The model below is the SBML for the stochastic version
of
the Lotka-Volterra sys-
tem, discussed in Chapter 6.
<?xml
version=nl.O"
encoding=
11
UTF-8"?>
<Sbml
xmlns="http://www.sbml.org/sbml/level2
11
level="2"
version="l">
<model
id="LotkaVolterra
11
>
<listOfUnitDefinitions>
<unitDefinition
id="substance
11
>
<listOfUnits>
<unit
kind=nitemn
multiplier="l"
offset="0
11
/>
</listOfUnits>
</unitDefinition>
</listOfUnitDefinitions>
<listOfCompartrnents>
<compartment
id="Cell"/>
</listOfCompartments>
<listOfSpecies>
<species
id=
11
Prey"
compartment="Cell
11
initia1Amount="50
11
hasOnlySubstanceUnits=
11
true
11
/>
<species
id=''Predator
11
compartment="Cell"
initia1Amount=
11
100"
hasOnlySubstanceUnitS=
11
true"/>
</listOfSpecies>
<listOfReactions>
<reaction
id=
11
PreyReproduction"
reversible="false">
<listOfReactants>
<speciesReference
species="Prey"/>
</listOfReactants>
<listOfProducts>
<speciesReference
species=
"Preyu
stoichiometry="2
''I>
</listOfProducts>
<kineticLaw>
<math
xmlns="http://www.w3.org/1998/Math/MathML">
<apply>
<times/>
<Ci>
Cl
</Ci>
<Ci>
Prey
</ci>
</apply>
</math>
<listOfParameters>
<parameter
id="cl"
value="l"/>
</listOfParameters>
</kineticLaw>
</reaction>

