Voet D., Voet Ju.G. Biochemistry
Подождите немного. Документ загружается.

Cuff, J.A. and Barton, G.J., Evaluation and improvement of multi-
ple sequence methods for protein secondary structure predic-
tion, Proteins 34, 508–519 (1999). [The principles behind
Jpred3.]
Das, R. and Baker, D., Macromolecular modeling with Rosetta,
Annu. Rev. Biochem. 77, 363–382 (2008).
DeGrado W.F., Summa, S.M., Pavone,V., Nastri, F., and Lombardi,
A., De novo design and structural characterization of proteins
and metalloproteins, Annu. Rev. Biochem. 68, 779–819 (1999).
Kuhlman, B., Dantas, G., Ireton, G.C., Varani, G., Stoddard, B.L.,
and Baker, D., Design of a novel protein fold with atomic-
level accuracy, Science 302, 1364–1368 (2003). [The design
of Top7.]
Lesk, A.M., Introduction to Bioinformatics (3rd ed.), pp. 333–358,
Oxford University Press (2008).
Mirny, L. and Shakhnovitch, E., Protein folding theory: From lat-
tice to all-atom models, Annu. Rev. Biophys. Biomol. Struct. 30,
361–396 (2001).
Moult, J., Fidelis, K., Kryshtofovych, A., Rost, B., and Tramon-
tano, A., Critical assessment of methods of protein structure
prediction—Round VIII, Proteins 77 (Issue S9), 1–4 (2009).
[The summary article of the issue of Proteins: Structure,Func-
tion, and Bioinformatics that reports the results of CASP8.]
Rose, G.D., Prediction of chain turns in globular proteins on a hy-
drophobic basis, Nature 272, 586–590 (1978).
Tramontano,A., Protein Structure Prediction. Concepts and Appli-
cations, Wiley-VCH (2006).
Zaki, M.J. and Bystroff, C. (Eds.), Protein Structure Prediction
(2nd ed.), Humana Press (2008).
Protein Dynamics
Henzler-Wildman, K. and Kern, D., Dynamic personalities of pro-
teins, Nature 450, 964–972 (2007).
Karplus, M. and McCammon,A., Molecular dynamics simulations
of biomolecules, Nature Struct. Biol. 9, 646–651 (2002).
Palmer,A.G., III, Probing molecular motion by NMR, Curr. Opin.
Struct. Biol. 7, 732–737 (1997).
Protein dynamics, Science 324, 197–215 (2009). [A special section
containing four articles.]
Rasmussen, B.F., Stock, A.M., Ringe, D., and Petsko, G.A., Crys-
talline ribonuclease A loses function below the dynamical
transition at 220 K, Nature 357, 423–424 (1992).
Ringe, D. and Petsko, G.A., Mapping protein dynamics by X-ray
diffraction, Prog. Biophys. Mol. Biol. 45, 197–235 (1985).
Scheraga, H.A., Khalili, M., and Liwo,A., Protein-folding dynam-
ics: Overview of molecular simulation techniques, Annu. Rev.
Phys. Chem. 58, 57–83 (2007).
Conformational Diseases
Booth, D.R., et al., Instability, unfolding and aggregation of hu-
man lysozyme variants underlying amyloid fibrillogenesis, Na-
ture 385, 787–73 (1997); and Funahashi, J., Takano, K., Ogasa-
hara, K., Yamagata, Y., and Yutani, K., The structure, stability,
and folding process of amyloidogenic mutant lysozyme,
J. Biochem. 120, 1216–1223 (1996).
Büeler, H., Aguzzi, A., Sailer, A., Greiner, R.A., Autenreid, P.,
Aguet, M., and Weissmann, C.,Mice devoid of PrP are resistant
to scrapie, Cell 73, 1339–1347 (1993); and Büeler, H., Fischer,
M., Lang, Y., Bluethmann, H., Lipp, H.-P., DeArmond, S.J.,
Prusiner, S.B., Aguet, M., and Weissmann, C., Normal develop-
ment and behaviour of mice lacking the neuronal cell-surface
PrP protein, Nature 356, 577–582 (1992).
Buxbaum, J.N. and Tagoe, C.E., The genetics of amyloidoses,
Annu. Rev. Med. 51, 543–569 (2000).
Caughey, B., Baron, G.S., Chesebro, B., and Jeffrey, M., Getting a
grip on prions: oligomers, amyloids, and pathological mem-
brane interactions, Annu. Rev. Biochem. 78, 177–204 (2009).
Chien, P.,Weissman, J.S., and DePace, J.H., Emerging principles of
conformation-based inheritance, Annu. Rev. Biochem. 73,
617–656 (2004).
Chiti, F. and Dobson,C.M., Protein misfolding, functional amyloid,
and human disease, Annu. Rev. Biochem. 75, 333–366 (2006).
Collinge, J. and Clarke,A.R.,A general model of prion strains and
their pathogenicity, Science 318, 930–936 (2007).
Deleault, N.R., Harris, B.T., Rees, J.R., and Supattapone, S., For-
mation of native prions from minimal components in vitro,
Proc. Natl. Acad. Sci. 104, 9741–9746 (2007).
Geula, C., Wu, C.-K., Saroff, D., Lorenzo, A., Yuan, M., and
Yankner, B.A., Aging renders the brain vulnerable to amyloid
-protein neurotoxicity, Nature Med. 4, 827–831 (1998).
Goedert, M. and Spillantini, M.G., A century of Alzheimer’s dis-
ease, Science 314, 777–781 (2006).
Gregersen, N., Bross, P., Vang, S., and Christiensen, J.H., Protein
misfolding and human disease, Annu. Rev. Genomics Hum.
Genet. 7, 103–124 (2006).
Hardy, J. and Selkoe, D.J., The amyloid hypothesis of Alzheimer’s
disease: Progress and problems on the road to therapeutics,
Science 297, 353–356 (2002).
Jackson, G.S. and Clarke, A.R., Mammalian prion proteins, Curr.
Opin. Struct. Biol. 10, 69–74 (2000).
Kajava,A., Squire,J.M., and Parry, D.A.D. (Eds.), Fibrous Proteins:
Amyloids, Prions and Beta Proteins, Adv. Prot. Chem. 73
(2006). [The last four chapters are on various aspects of amy-
loids and prions.]
Moore, R.A., Taubner, L.M., and Priola, S.A., Prion misfolding
and disease, Curr. Opin. Struct. Biol. 19, 14–22 (2009).
Pan, K.M., Baldwin, M., Nguyen, J., Gasset, M., Serban, A., Groth,
D., Mehlhorn, I., Huang, Z., Fletterick, R.J., Cohen, F.E., and
Prusiner, S.B., Conversion of ␣-helices into -sheet features in
the formation of the scrapie prion proteins, Proc. Natl. Acad.
Sci. 90, 10962–10966 (1993).
Prusiner, S.B. (Ed.), Prion Biology and Diseases (2nd ed.), Cold
Spring Harbor Laboratory Press (2004); and Prion diseases, in
Valle,D. (Ed.), The Online Metabolic & Molecular Bases of In-
herited Disease, http://www.ommbid.com/, Chap. 224.
Rochet, J.C. and Lansbury, P.T., Jr.,Amyloid fibrillogenesis:Themes
and variations, Curr. Opin. Struct. Biol. 10, 60–68 (2000).
Sawaya, M.R., et al., Atomic structures of amyloid cross- spines
reveal varied steric zippers, Nature 447, 453–457 (2007); and
Nelson, R., Sawaya, M.R., Balbirnie, M., Madsen,A.Ø., Riekel,
C.,Grothe,R.,and Eisenberg,D., Structure of the cross- spine
of amyloid-like fibrils, Nature 435, 773–778 (2005).
Selkoe, D.J., Cell biology of protein misfolding: the examples of
Alzheimer’s and Parkinson’s diseases, Nature Cell Biol. 6,
1054–1061 (2004).
Soto, C., Estrada, L., and Castilla, J., Amyloids, prions and the in-
herent infectious nature of misfolded proteins, Trends
Biochem. Sci. 31, 150–155 (2006).
Sparrer, H.E., Santoso,A., Szoka, F.C., Jr., and Weissman, J.S., Ev-
idence for the prion hypothesis: Induction of the yeast [PSI
⫹
]
factor by in vitro-converted Sup35 protein, Science 289,
595–599 (2000).
Tuite, M.F.,Yeast prions and their prion-forming domain, Cell 100,
289–292 (2000).
Weissmann, C., The state of the prion, Nature Rev. Microbiol. 2,
861–862 (2004).
Wiltzius, J.J.W., Sievers, S.A., Sawaya, M.R., Cascio, D., Popov, D.,
Riekel, C., and Eisenberg, D., Atomic structure of the cross-
References 321
JWCL281_c09_278-322.qxd 6/1/10 7:35 AM Page 321

spine of islet amyloid polypeptide (amylin), Prot. Sci. 17,
1467–1474 (2008).
Zahn, R., Liu, A., Lührs, T., Riek, R., von Schroetter, C., Garcia,
F.L., Billeter, M., Calzolai, L., Wider, G., and Wüthrich, K.,
NMR solution structure of the human prion protein, Proc.
Natl. Acad. Sci. 97, 145–150 (2000); and Liu, H., Farr-Jones, S.,
Ulyanov, N.B., Llinas, M., Marqusee, S., Groth, D., Cohen, F.E.,
Prusiner, S.B., and James, T.L., Solution structure of Syrian
hamster prion protein rPrP(90–231), Biochemistry 38,
5362–5377 (1999).
Structural Evolution
Bajaj, M. and Blundell, T., Evolution and the tertiary structure of
proteins, Annu. Rev. Biophys. Bioeng. 13, 453–492 (1983).
Dickerson, R.E., Timkovitch, R., and Almassy, R.J., The cy-
tochrome fold and the evolution of bacterial energy metabo-
lism, J. Mol. Biol. 100, 473–491 (1976).
Eventhoff, W. and Rossmann, M., The structures of dehydro-
genases, Trends Biochem. Sci. 1, 227–230 (1976).
Lesk, A.M., NAD-binding domains of dehydrogenases, Curr.
Opin. Struct. Biol. 5, 775–783 (1995).
Moore,A.D., Björklund, Å.K., Ekman, D., Bornberg-Baur,E., and
Elofsson, A., Arrangements in the modular evolution of pro-
teins, Trends Biochem. Sci. 33, 444–451 (2008).
Scott, R.A. and Mauk, A.G. (Eds.), Cytochrome c. A Multidisci-
plinary Approach, University Science Books (1996).
322 Chapter 9. Protein Folding, Dynamics, and Structural Evolution
1. How long will it take the polypeptide backbone of a
6-residue folding nucleus to explore all its possible conformations?
Repeat the calculation for 10-, 15-, and 20-residue folding nuclei.
Why, in the classic view of protein folding, are folding nuclei
thought to be no larger than 15 residues?
*2. Consider a protein with 10 Cys residues. On air oxidation,
what fraction of the denatured and reduced protein will randomly
reform the native set of disulfide bonds if: (a) The native protein
has five disulfide bonds? (b) The native protein has three disulfide
bonds?
3. Why are  sheets more commonly found in the hydropho-
bic interiors of proteins than on their surfaces?
4. Under physiological conditions, polylysine assumes a ran-
dom coil conformation. Under what conditions might it form an ␣
helix?
5. Explain how landscape theory is consistent with the obser-
vation that many small proteins appear to fold to their native con-
formations without detectable intermediates, that is, via two-state
mechanisms.
6. Explain why Pro residues can occupy the N-terminal turn
of an ␣ helix.
7. Explain why  sheets are less likely to form than ␣ helices
during the earliest stages of protein folding.
8. Molten globules are thought to be predominantly stabilized
by hydrophobic forces. Why aren’t hydrogen bonding forces im-
plicated in doing so?
*9. The GroEL/ES cycle diagrammed in Fig. 9-25 only circu-
lates in the clockwise direction. Explain the basis for this irre-
versibility in terms of the sequence of structural and binding
changes in the GroEL/ES system.
*10. Predict the secondary structure of the C peptide of proin-
sulin (Fig. 9-4) using the methods of Chou–Fasman and Rose.
11. As Mother Nature’s chief engineer, now certified as a
master helix builder,you are asked to repeat Problem 8-8 with the
stipulation that the ␣ helix really be helical. Use Table 9-1.
*12. Predict the secondary structure of the N-terminal do-
main of yeast protein disulfide isomerase using Jpred3
(http://www.compbio.dundee.ac.uk/www-jpred/). How does this
prediction compare with the observed structure of this domain
(PDBid 2B5E; the a domain in Fig. 9-17)? [To enter the sequence
of this domain into Jpred3, first point your browser at the PDB
(http://www.rcsb.org/pdb), enter the PDBid 2B5E, click on the
“Sequence Details” tab at the top of the resulting page, and deter-
mine the sequence range of the N-terminal domain.Then click on
the UniProt reference (P17967), scroll down to the sequence, copy
the relevant portion to the Jpred3 input box, edit out everything
but the sequence, and click on the “Make Prediction” button. On
the page that comes up indicating that sequence matches were
found in the PDB, click on the “continue” button. When the Re-
sults page appears (you may have to wait some time for it), click
on “View Simple” to see the Jpred3 prediction.The observed sec-
ondary structure of the N-terminal domain is diagrammed on the
foregoing “Sequence Details” page.]
13. Indicate the probable effects of the following mutational
changes on the structure of a protein. Explain your reasoning.
(a) Changing a Leu to a Phe, (b) changing a Lys to a Glu,
(c) changing a Val to a Thr, (d) changing a Gly to an Ala, and
(e) changing a Met to a Pro.
14. Explain why Trp rings are usually completely immobile in
proteins that have rapidly flipping Phe and Tyr rings.
15. Explain why Prn-p
0/0
mice are resistant to scrapie. What
might be the susceptibility of heterozygous Prn-p
⫹/0
mice to
scrapie?
*16. Discuss the merits of the hypothesis that the dinucleotide-
binding domains of the dehydrogenases arose by convergent
evolution.
PROBLEMS
JWCL281_c09_278-322.qxd 6/1/10 7:36 AM Page 322

323
CHAPTER 10
Hemoglobin:
Protein Function
in Microcosm
1 Hemoglobin and Myoglobin Function
A. Heme
B. Oxygen Binding
C. Carbon Dioxide Transport and the Bohr Effect
D. Effect of BPG on O
2
Binding
2 Structure and Mechanism
A. Structure of Myoglobin
B. Structure of Hemoglobin
C. Mechanism of Oxygen-Binding Cooperativity
D. Testing the Perutz Mechanism
E. Origin of the Bohr Effect
F. Structural Basis of BPG Binding
G. Role of the Distal Histidine Residue
3 Abnormal Hemoglobins
A. Molecular Pathology of Hemoglobin
B. Molecular Basis of Sickle-Cell Anemia
4 Allosteric Regulation
A. The Adair Equation
B. The Symmetry Model
C. The Sequential Model
D. Hemoglobin Cooperativity
Appendix: Derivation of Symmetry Model Equations
A. Homotropic Interactions–Equation [10.22]
B. Heterotropic Interactions–Equation [10.23]
also been successful in explaining the control of enzyme ac-
tivity. The first protein X-ray structures to be elucidated
were those of hemoglobin and myoglobin (Greek: myos,
muscle). This central role in the development of protein
chemistry together with its enzymelike O
2
-binding proper-
ties have caused hemoglobin to be dubbed an “honorary
enzyme.”
Hemoglobin is not just a simple oxygen tank. Rather, it
is a sophisticated oxygen delivery system that provides the
proper amount of oxygen to the tissues under a wide vari-
ety of circumstances. In this chapter, we discuss hemoglo-
bin’s properties, structure, and mechanism of action, both
to understand the workings of this physiologically essential
molecule and to illustrate the principles of protein struc-
ture that we have developed in the preceding chapters. We
also consider the properties of abnormal hemoglobins and
their relationship to human disease. Finally, we discuss the-
ories of cooperative interactions among proteins, both to
better understand the properties of hemoglobin and to set
the stage for our later consideration of how enzyme action
is regulated.
1 HEMOGLOBIN AND
MYOGLOBIN FUNCTION
Hemoglobin (Hb), as we have seen in Chapters 7 and 8, is
a 65-kD heterotetramer, ␣
2

2
(alternatively, a dimer of ␣
protomers).The ␣ and  subunits are structurally and evo-
lutionarily related to each other and to myoglobin (Mb),
the 18-kD monomeric oxygen-binding protein of skeletal
and heart muscle (Section 7-3C).
Hemoglobin transports oxygen from the lungs, gills, or
skin of an animal to its capillaries for use in respiration.
Very small organisms do not require such a protein be-
cause their respiratory needs are satisfied by the simple
passive diffusion of O
2
through their bodies. However, since
the transport rate of a diffusing substance varies inversely
with the square of the distance it must diffuse, the O
2
diffu-
sion rate through tissue thicker than ⬃1 mm is too slow to
support life. The evolution of organisms as large and com-
plex as annelids (e.g., earthworms) therefore required the
development of circulatory systems that actively transport
O
2
and nutrients to the tissues.The blood of such organisms
must contain an oxygen transporter such as Hb because the
solubility of O
2
in blood plasma (the fluid component of
The existence of hemoglobin (Greek: haimato, blood), the
red blood pigment, is evident to every child who scrapes a
knee. Its brilliant red color, widespread occurrence, and
ease of isolation have made it an object of inquiry since an-
cient times. Indeed, the early history of protein chemistry is
essentially that of hemoglobin. The observation of crys-
talline hemoglobin was first reported by Friedrich Hüne-
feld in 1840, and by 1909 Edward Reichert and Amos
Brown had published a photographic atlas of hemoglobin
crystals from several hundred species. In contrast, it was
not until 1926 that crystals of an enzyme, those of jack bean
urease, were first reported. Hemoglobin was one of the
first proteins to have its molecular mass accurately deter-
mined, the first protein to be characterized by ultracen-
trifugation, the first to be associated with a specific physio-
logical function (that of oxygen transport), and, in
sickle-cell anemia, the first in which a point mutation was
demonstrated to cause a single amino acid change (Section
7-3Aa). Theories formulated to account for the coopera-
tive binding of oxygen to hemoglobin (Section 10-4) have
JWCL281_c10_323-358.qxd 2/24/10 1:57 PM Page 323
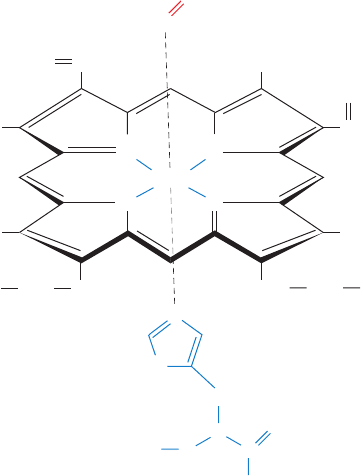
Fe(II) (ferrous) oxidation state whether or not the heme is
oxygenated (binds O
2
).
The Fe atom in deoxygenated Hb and Mb is 5-coordinated
by a square pyramid of N atoms: four from the porphyrin
and one from a His side chain of the protein. On oxygena-
tion, the O
2
binds to the Fe(II) on the opposite side of the
porphyrin ring from the His ligand, so that the Fe(II) is
octahedrally coordinated; that is, the ligands occupy the six
corners of an octahedron centered on the Fe atom (Fig.
10-1). Oxygenation changes the electronic state of the
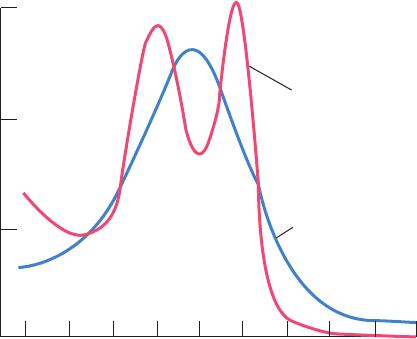
Fe(II)–heme, as is indicated by the color change of blood
from the dark purplish hue characteristic of venous blood to
the brilliant scarlet color of arterial blood and blood from a
cut finger (Fig. 10-2).
Certain small molecules, such as CO, NO, CN
⫺
, and H
2
S,
coordinate to the sixth liganding position of the Fe(II) in
Hb and Mb with much greater affinity than does O
2
. This,
together with their similar binding to the hemes of cy-
tochromes, accounts for the highly toxic properties of these
substances.
The Fe(II) of Hb and Mb can be oxidized to Fe(III) to
form methemoglobin (metHb) and metmyoglobin (metMb).
MetHb and metMb do not bind O
2
; their Fe(III) is already
octahedrally coordinated with an H
2
O molecule in the
sixth liganding position.The brown color of dried blood and
old meat is that of metHb and metMb. Erythrocytes contain
the enzyme methemoglobin reductase, which converts the
blood) is too low (⬃10
⫺4
M under physiological condi-
tions) to carry sufficient amounts of O
2
for metabolic
needs. In contrast, whole blood, which normally contains
⬃150 g of Hb ⴢ L
⫺1
, can carry O
2
at concentrations as high
as 0.01M, about the same as in air.
Although Mb was originally assumed to store oxygen,
it is now clear that this function is significant only in
aquatic mammals such as seals and whales, which have
Mb concentrations in their muscles 10- to 30-fold greater
than those in terrestrial mammals. It would seem more
likely that Mb’s major physiological role in terrestrial
mammals is to facilitate oxygen transport in rapidly
respiring muscle. The rate at which O
2
can diffuse from
the capillaries to the tissues, and thus the level of respira-
tion, is limited by oxygen’s low solubility in aqueous solu-
tion. Mb increases the effective solubility of O
2
in muscle,
the most rapidly respiring tissue under conditions of high
exertion. Hence, in rapidly respiring muscle, Mb functions
as a kind of molecular bucket brigade to facilitate O
2
dif-
fusion. It therefore came as a surprise when knockout
mice for the gene encoding Mb exhibited no obvious ab-
normalities (except for the pale color of their muscles)
and exhibited normal exercise capacity and response to
low oxygen levels. However, further investigations re-
vealed that these knockout mice had compensatory adap-
tations including increases in their hematocrit (the frac-
tion of blood volume occupied by red blood cells;
normally ⬃45%) and the capillary density in their mus-
cles. Moreover, many of the mutant embryos died in utero
due to cardiovascular defects. Additional physiological
functions for Mb have recently been recognized: the
detoxification of the highly reactive biological signaling
molecule nitric oxide (NO) through its conversion to nitrate
ion (NO
⫺
3
) under normal conditions and its synthesis from
nitrite ion (NO
⫺
2
) under hypoxic (having an inadequate
supply of O
2
) conditions (see below).
In this section, we begin our discussions of hemoglobin
by considering its chemical and physical properties and
how they relate to its physiological function. Hemoglobin
structure and the mechanisms by which it carries out these
physiological functions are discussed in Section 10-2.
A. Heme
Myoglobin and each of the four subunits of hemoglobin
noncovalently bind a single heme group (Fig. 10-1; spelled
“haem” in British English). This is the same group that oc-
curs in the cytochromes (Section 9-6A) and in certain re-
dox enzymes such as catalase. Heme is responsible for the
characteristic red color of blood and is the site at which
each globin monomer binds one molecule of O
2
(globins
are the heme-free proteins of Hb and Mb). The hetero-
cyclic ring system of heme is a porphyrin derivative; it con-
sists of four pyrrole rings (labeled A–D in Fig. 10-1) linked
by methene bridges.The porphyrin in heme, with its partic-
ular arrangement of four methyl, two propionate, and two
vinyl substituents, is known as protoporphyrin IX. Heme,
then, is protoporphyrn IX with a centrally bound iron
atom. In Hb and Mb, the iron atom normally remains in the
324 Chapter 10. Hemoglobin: Protein Function in Microcosm
Figure 10-1 The heme group. Fe(II)–heme
(ferroprotoporphyrin IX) is shown liganded to His and O
2
as it is
in oxygenated myoglobin and oxygenated hemoglobin. Note that
the heme is a conjugated system so that, although two of its
Fe¬N bonds are coordinate covalent bonds (bonds in which the
bonding electron pair is formally contributed by only one of the
atoms forming the bond), all of the Fe¬N bonds are equivalent.
The pyrrole ring lettering scheme is shown.
H
2
C
H
3
C
H
3
C
CH
N
N
CH
3
CH
3
CH
CH
2
CH
2
CH
2
COO
–
–
OOC CH
2
CH
2
O
Fe(II)
O
N
CD
BA
N
CH
2
N
HN
C
O
C
N
H
H
JWCL281_c10_323-358.qxd 2/24/10 1:57 PM Page 324
small amount of metHb that spontaneously forms back to
the Fe(II) form.
a. Myoglobin Both Detoxifies and Synthesizes
Nitric Oxide
NO, which is synthesized in many tissues, functions as a
locally active signaling molecule, most notably to induce
vasodilation (Section 19-1L).Once NO has delivered its mes-
sage, it is important that it be rapidly eliminated to prevent
its interference with subsequent NO signals (or lack of
them). Moreover, NO is a highly reactive and hence toxic
substance. In muscle, under normal O
2
concentrations, NO
is detoxified through its reaction with oxygenated myoglo-
bin (oxyMb) to yield nitrate ion and metmyoglobin:
Since the metMb is subsequently reduced to Mb through
the action of an intracellular metmyoglobin reductase,
myoglobin functions as an enzyme in this process. Oxy-
genated hemoglobin (oxyHb) likewise detoxifies the NO
that is present in blood.
Paradoxically, the tissue damage caused by ischemia (in-
adequate blood flow, such as in a heart attack or stroke) is
often exacerbated when blood flow to the deprived tissues
is reestablished. This so-called reperfusion injury is caused,
in part, by the highly destructive reactive oxygen species
(ROS), such as the superoxide ion (O
2
) and the hydroxyl
radical (ⴢOH), initially produced by hypoxic mitochondria
when the blood supply is restored. Nitrite ion is normally
present in blood and tissues at low (micromolar) concen-
trations. Under hypoxic conditions, deoxyMb catalyzes the
formation of NO from nitrite ion:
NO suppresses the ROS-producing mitochondrial electron-
transport chain (Chapter 22) and thereby protects against
reperfusion injury. Indeed, the administration of nitrite ion
NO
⫺
2
⫹ H
⫹
⫹ Mb S NO ⫹ OH
⫺
⫹ metMb
ⴢ
⫺
NO ⫹ MbO
2
S NO
⫺
3
⫹ metMb
to the hearts of normal mice immediately before their
blood supply was cut off for 30 minutes provided a signifi-
cant protective effect against tissue damage, but afforded
no such protection to the hearts of Mb-knockout mice.
b. Other Globins and Oxygen-Transport Proteins
Hb and Mb belong to the globin superfamily, whose
members occur widely in all three kingdoms of life, where
most participate in a variety of enzymatic and O
2
-sensing
functions. Evidently, Hb’s O
2
-transport function is a rela-
tively recent evolutionary adaptation.
Invertebrate hemoglobins vary in quaternary structure
from dimers to as many as 180 subunits. The larger of these,
which are known as erythrocruorins (Latin: cruor, blood),
are extracellular proteins rather than being packaged
within cells; their large sizes (up to 3.5 million kD) permit
them to be retained within their circulatory systems. The
related chlorocruorins (Greek, chloros, pale green), which
occur in the blood of certain annelids, contain a porphyrin
that differs from protoporphyrin IX by the replacement of
its A-ring vinyl group with a formyl group. Consequently,
chlorocruorins are green when deoxygenated and light red
when oxygenated.
Antarctic icefish, the only adult vertebrates that lack
hemoglobin—their blood is colorless—are viable because
of their reduced need for O
2
at low temperatures combined
with the relatively high aqueous solubility of O
2
at the
⫺1.9°C temperature of their environment (recall that the
solubilities of gases increase with decreasing temperature).
Many invertebrates lack Hb but instead produce one of
two alternative types of O
2
-transport proteins:
1. Hemocyanins, which occur in many species of
mollusks and arthropods, form a family of large, multi-
meric, extracellular proteins that differ in their primary
through quaternary structures. However, they have highly
similar O
2
-binding sites that each contain two Cu ions.
Hemocyanins are blue in complex with O
2
and colorless
otherwise.
2. Hemerythrins, which are intracellular proteins, occur
in only a few species of marine worms. Their O
2
-binding
sites contain two non-heme Fe ions. Hemerythrins are
violet-pink in complex with O
2
and colorless otherwise.
Vertebrates also express two recently discovered glo-
bins: neuroglobin, which is present mainly in brain, retina,
and endocrine tissues, and cytoglobin, which occurs in
most tissues. Neuroglobin protects neurons (nerve cells)
from damage under conditions of hypoxia, most likely by
preventing reperfusion injury in much the same way as
does myoglobin in muscle. Cytoglobin may have similar
functions.
B. Oxygen Binding
The binding of O
2
to myoglobin is described by a simple
equilibrium reaction
Mb ⫹ O
2
Δ MbO
2
Section 10-1. Hemoglobin and Myoglobin Function 325
Figure 10-2 The visible absorption spectra of oxygenated and
deoxygenated hemoglobins.
Relative absorbance
Wavelength (nm)
480 500 520 540 560 580 600 620 640 660
5
10
15
Oxyhemoglobin
Deoxyhemoglobin
JWCL281_c10_323-358.qxd 2/24/10 1:57 PM Page 325
with dissociation constant
[10.1]
(biochemists usually express equilibria in terms of dissoci-
ation constants, the reciprocals of the more chemically tra-
ditional association constants). The O
2
dissociation of Mb
may be characterized by its fractional saturation, ,de-
fined as the fraction of O
2
binding sites occupied by O
2
.
[10.2]
Since O
2
is a gas, its concentration is conveniently ex-
pressed by its partial pressure, pO
2
(also called the oxygen
tension). Equation [10.2] may therefore be expressed:
[10.3]
Now define p
50
as the value of pO
2
when that is,
when half of myoglobin’s O
2
-binding sites are occupied.
Substituting this value into Eq. [10.3] and solving for K
yields K ⫽ p
50
. Hence our expression for the fractional sat-
uration of Mb finally becomes
[10.4]
a. Hemoglobin Cooperatively Binds O
2
Myoglobin’s O
2
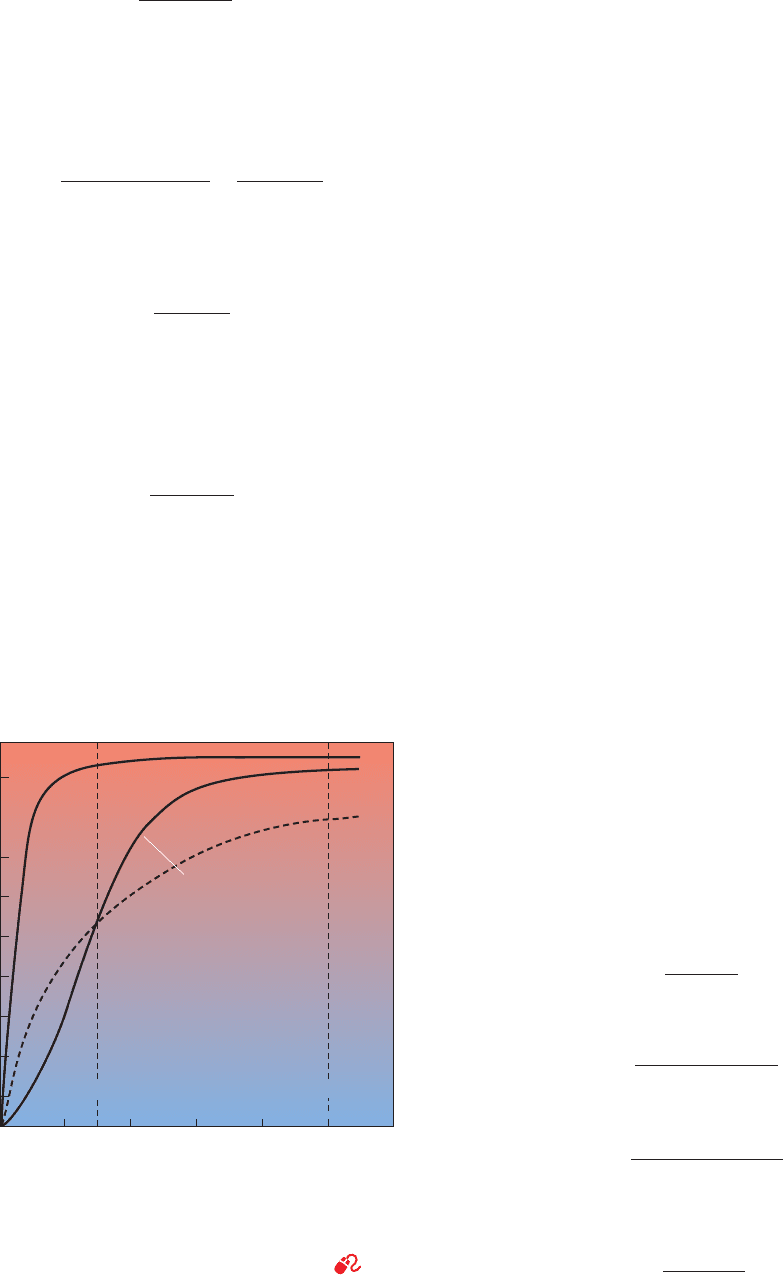
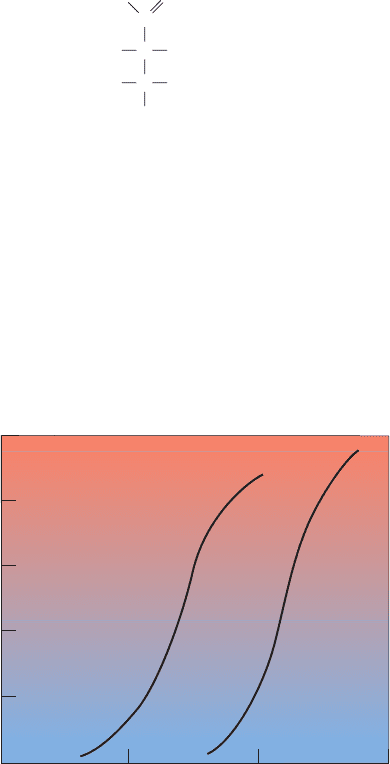
-dissociation curve (Fig. 10-3) closely
follows the hyperbolic curve described by Eq. [10.4]; its
p
50
is 2.8 torr (1 torr ⫽ 1 mm Hg at 0°C ⫽ 0.133 kPa; 760
torr ⫽ 1 atm). Mb therefore gives up little of its bound O
2
Y
O
2
⫽
pO
2
p
50
⫹ pO
2
Y
O
2
⫽ 0.50,
Y
O
2
⫽
pO
2
K ⫹ pO
2
Y
O
2
⫽
[MbO
2
]
[Mb] ⫹ [MbO
2
]
⫽
[O
2
]
K ⫹ [O
2
]
Y
O
2
K ⫽
[Mb][O
2
]
[MbO
2
]
over the normal physiological range of pO
2
in blood (100
torr in arterial blood and 30 torr in venous blood); for ex-
ample, at pO
2
⫽ 100 torr and 0.91 at 30 torr. In
contrast, hemoglobin’s O
2
-dissociation curve (Fig. 10-3),
which has a sigmoidal shape (S shape) that Eq. [10.4] does
not describe, indicates that the amount of O
2
bound by Hb
changes significantly over the normal physiological range
of pO
2
in blood. For example, at 100 torr and
0.55 at 30 torr in whole blood for a difference in of 0.40.
Mb therefore binds O
2
under conditions in which Hb re-
leases it. Thus, the two proteins form a sophisticated O
2
transport system that delivers O
2
from lung to muscle
(where pO
2
may be ⬍20 torr). Hemoglobin’s sigmoidal
O
2
-dissociation curve is of great physiological importance;
it permits the blood to deliver much more O
2
to the tissues
than it could if Hb had a hyperbolic O
2
-dissociation curve
with the same p
50
(26 torr; dashed curve in Fig. 10-3). Such a
hyperbolic curve has at 100 torr and 0.54 at 30
torr for a difference in of only 0.25.
A sigmoidal dissociation curve is diagnostic of a cooper-
ative interaction between a protein’s small molecule bind-
ing sites; that is, the binding of one small molecule affects
the binding of others. In this case, the binding of O
2
in-
creases the affinity of Hb for binding additional O
2
.The
structural mechanism of hemoglobin cooperativity is dis-
cussed in Section 10-2C.
b. The Hill Equation Phenomenologically Describes
Hemoglobin’s O
2
-Binding Curve
The earliest attempt to analyze hemoglobin’s sigmoidal
O
2
-dissociation curve was formulated by Archibald Hill in
1910. We shall follow his analysis in general form because
it is useful for characterizing the cooperative behavior of
oligomeric enzymes as well as that of hemoglobin.
Consider a protein E consisting of n subunits that can
each bind a molecule S, which, in analogy with the sub-
stituents of metal ion complexes, is known as a ligand. As-
sume that the ligand binds with infinite cooperativity,
that is, the protein either has all or none of its ligand-
binding sites occupied, so that there are no observable in-
termediates ES
1
,ES
2
, etc.The dissociation constant for this
reaction is
[10.5]
and, as before, its fractional saturation is expressed:
[10.6]
Combining Eqs. [10.5] and [10.6] yields
which on algebraic rearrangement and cancellation of
terms becomes the Hill equation:
[10.7]Y
S
⫽
[S]
n
K ⫹ [S]
n
Y
S
⫽
[E][S]
n
>K
[E](1 ⫹ [S]
n
>K)
Y
S
⫽
n[ES
n
]
n([E] ⫹ [ES
n
])
K ⫽
[E][S]
n
[ES
n
]
E ⫹ nS Δ ES
n
Y
O
2
Y
O
2
⫽ 0.79
Y
O
2
Y
O
2
⫽ 0.95
Y
O
2
⫽ 0.97
326 Chapter 10. Hemoglobin: Protein Function in Microcosm
Figure 10-3 Oxygen-dissociation curves of Mb and of Hb in
whole blood. The normal sea level values of human arterial and
venous pO
2
values are indicated.The dashed line is a hyperbolic
O
2
-dissociation curve with the same p
50
as Hb (26 torr). See
the Animated Figures
1.00
0.80
0.60
0.40
0.20
0.0
0 20 40 60 80 100 120
Hemoglobin
in whole blood
Myoglobin
Venous pressure
Arterial pressure
pO
2
(torr)
Y
O
2
JWCL281_c10_323-358.qxd 2/24/10 1:57 PM Page 326

which, in a manner analogous to Eq. [10.4], describes the
degree of saturation of a multisubunit protein as a function
of ligand concentration.
Infinite ligand-binding cooperativity (n equal to the
number of protein subunits), as assumed in deriving the
Hill equation, is a physical impossibility. Nevertheless, n
may be taken to be a noninteger parameter related to the
degree of cooperativity among interacting ligand-binding
sites rather than the number of subunits per protein. The
Hill equation then becomes a useful empirical curve-fitting
relationship rather than an indicator of a particular model
of ligand binding. The quantity n, the Hill constant, in-
creases with the degree of cooperativity of a reaction and
thereby provides a convenient, although simplistic, charac-
terization of a ligand-binding reaction. If n ⫽ 1, Eq. [10.7]
describes a hyperbola, as do Eqs. [10.3] and [10.4] for Mb,
and the ligand-binding reaction is said to be noncoopera-
tive. A reaction with n ⬎ 1 is described as positively coop-
erative: Ligand binding increases the affinity of E for fur-
ther ligand binding (cooperativity is infinite in the limit
that n is equal to the number of ligand-binding sites in E).
Conversely, if n ⬍ 1, the reaction is termed negatively co-
operative: Ligand binding reduces the affinity of E for sub-
sequent ligand binding.
c. Hill Equation Parameters May Be
Graphically Evaluated
The Hill constant, n, and the dissociation constant, K,
that best describe a saturation curve can be graphically de-
termined by rearranging Eq. [10.7] as follows:
Y
S
1 ⫺ Y
S
⫽
[S]
n
K ⫹ [S]
n
1 ⫺
[S]
n
K ⫹ [S]
n
⫽
[S]
n
K
and then taking the log of both sides to yield a linear
equation:
[10.8]
The linear plot of log[Y
S
/(1 ⫺ Y
S
)] versus log[S], the Hill
plot, has a slope of n and an intercept on the log[S] axis of
(log K)/n (recall that the linear equation y ⫽ mx ⫹ b de-
scribes a line with a slope of m and an x intercept of ⫺b/m).
For Hb, if we substitute pO
2
for [S] as was done for Mb,
the Hill equation becomes:
[10.9]
As in Eq. [10.4], let us define p
50
as the value of pO
2
at
. Then, substituting this value into Eq. [10.9],
so that
[10.10]
Substituting this result back into Eq. [10.9] yields
[10.11]
(Note: Equation [10.4] is a special case of Eq. [10.11] with
n ⫽ 1). Equation [10.8] for the Hill plot of Hb therefore
takes the form
[10.12]
so that this plot has a slope of n and an intercept on the log
pO
2
axis of log p
50
.
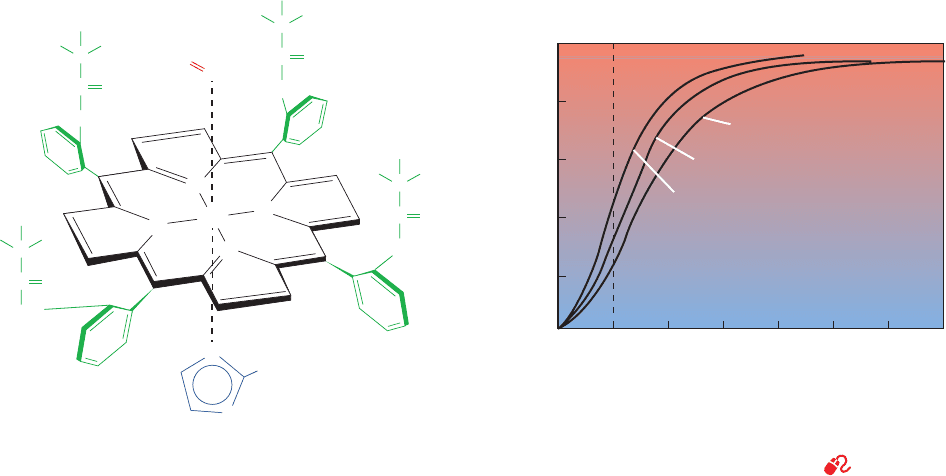
Figure 10-4 shows the Hill plots for Mb and Hb. For Mb
it is linear with a slope of 1, as expected.Although Hb does
log
a
Y
O
2
1 ⫺ Y
O
2
b⫽ n log pO
2
⫺ n log p
50
Y
O
2
⫽
(pO
2
)
n
(p
50
)
n
⫹ (pO
2
)
n
K ⫽ (p
50
)
n
0.50 ⫽
(p
50
)
n
K ⫹ (p
50
)
n
Y
O
2
⫽ 0.50
Y
O
2
⫽
(pO
2
)
n
K ⫹ (pO
2
)
n
log a
Y
S
1 ⫺ Y
S
b⫽ n log[S] ⫺ log K
Section 10-1. Hemoglobin and Myoglobin Function 327
Figure 10-4 Hill plots for Mb and
purified (“stripped”) Hb. Note that this is
a log–log plot. Hence the horizontal axis,
occurs where
(and pO
2
⫽ p
50
).Y
O
2
>(1 ⫺ Y
O
2
) ⫽ 1
log[Y
O
2
>(1 ⫺ Y
O
2
)] ⫽ 0,
100
10
1
0.1
0.01
0.99
0.5
10 100 100010.10.01
1 –
p
50
of
the last
oxygen
bound
p
50
of
the first
oxygen
bound
Myoglobin
slope = 1
Upper asymptote
Y
O
2
Y
O
2
Y
O
2
Hemoglobin
slope 3.0
~
~
Lower asymptote
pO
2
0.9
0.1
0.01
pO
2
(torr)
JWCL281_c10_323-358.qxd 2/24/10 1:57 PM Page 327
not bind O
2
in a single step as is assumed in deriving the
Hill equation, its Hill plot is essentially linear for values of
between 0.1 and 0.9. Its maximum slope, which occurs
near pO
2
⫽ p
50
is nor-
mally taken to be the Hill constant. For normal human Hb,
the Hill constant is between 2.8 and 3.0; that is, hemoglobin
oxygen binding is highly, but not infinitely, cooperative.
Many abnormal hemoglobins exhibit smaller Hill con-
stants (Section 10-3A), indicating that they have a less than
normal degree of cooperativity. At values near 0, when
few Hb molecules have bound even one O
2
molecule, the
Hill plot of Hb assumes a slope of 1 (Fig. 10-4, lower
asymptote) because the Hb subunits independently com-
pete for O
2
as do molecules of Mb. At values near 1,
when at least three of each of hemoglobin’s four O
2
-binding
sites are occupied, the Hill plot also assumes a slope of 1
(Fig. 10-4, upper asymptote) because the few remaining un-
occupied sites are on different molecules and therefore
bind O
2
independently.
Extrapolating the lower asymptote in Fig. 10-4 to the
horizontal axis indicates, according to Eq. [10.11], that
p
50
⫽ 30 torr for binding the first O
2
to Hb. Likewise, ex-
trapolating the upper asymptote yields p
50
⫽ 0.3 torr for
binding hemoglobin’s fourth O
2
.Thus the fourth O
2
to bind
to Hb does so with 100-fold greater affinity than the first.
This difference, as we shall see in Section 10-2C, is entirely
due to the influence of the globin on the O
2
affinity of heme.
It corresponds to a free energy difference of 11.4 kJ ⴢ mol
⫺1
between binding the first and binding the last O
2
to Hb
(Section 3-4A).
More sophisticated mathematical models than the Hill
equation have been developed for analyzing the coopera-
tive binding of ligands to proteins. We examine some of
them in Section 10-4.
Y
O
2
Y
O
2
[(Y
O
2
⫽ 0.5; Y
O
2
>(1 ⫺ Y
O
2
) ⫽ 1],
Y
O
2
d. Globin Prevents Oxyheme from Autooxidizing
Globin not only modulates the O
2
-binding affinity of
heme, but makes reversible O
2
binding possible. Fe(II)–heme
by itself is incapable of binding O
2
reversibly. Rather,in the
presence of O
2
it autooxidizes irreversibly to the Fe(III)
form through the intermediate formation of a complex
consisting of an O
2
bridging the Fe atoms of two hemes.
This reaction can be inhibited by derivatizing the heme
with bulky groups that sterically prevent the close face-
to-face approach of two hemes. Such picket-fence
Fe(II)–porphyrin complexes (Fig. 10-5), which James Coll-
man first synthesized, bind O
2
reversibly. The back side of
this porphyrin is unhindered and is complexed with a sub-
stituted imidazole in a manner similar to that in Mb and
Hb. In fact, the O
2
affinity of the picket-fence complex is
similar to that of Mb.Thus, the globins of Mb and Hb func-
tion to prevent the autooxidation of oxyheme by surround-
ing it, rather like a hamburger bun surrounds a hamburger,
so that only its propionate side chains are exposed to the
aqueous solvent (Section 10-2B).
C. Carbon Dioxide Transport and the Bohr Effect
In addition to being an O
2
carrier, Hb plays an important
role in the transport of CO
2
by the blood. When Hb (but
not Mb) binds O
2
at physiological pH’s, it undergoes a
conformational change (Section 10-2Ba) that makes it a
slightly stronger acid. It therefore releases protons on
binding O
2
:
where n ⫽ 0, 1, 2, or 3 and x 0.6 under physiological con-
ditions. Conversely, increasing the pH, that is, removing pro-
tons, stimulates Hb to bind O
2
(Fig. 10-6). This phenomenon,
whose molecular basis is discussed in Section 10-2E, is
known as the Bohr effect after Christian Bohr (the father
⬇
Hb(O
2
)
n
⫹ O
2
Δ Hb(O
2
)
n⫹1
⫹ xH
⫹
328 Chapter 10. Hemoglobin: Protein Function in Microcosm
N
N
CH
3
CH
3
CH
3
H
3
C
C
CO
NH
CH
3
CH
3
H
3
C
C
NH
CH
3
CH
3
H
3
C
C
HN
O
O
CH
CH
3
H
3
C
C
NH
N
N
N
N
Fe
CO
CO
CO
0 20 40 60 80 100 120 140
1.0
0.8
0.6
0.4
0.2
0
pO
2
(torr)
pH 7.6
pH 7.2
pH 7.4
Y
O
2
Figure 10-5 A picket-fence Fe(II)–porphyrin complex with
bound O
2
. [After Collman, J.P., Brauman, J.I., Rose, E., and
Suslick, K.S., Proc. Natl. Acad. Sci. 75, 1053 (1978).]
Figure 10-6 Effect of pH on the O
2
-dissociation curve of Hb:
the Bohr effect. The vertical dashed line indicates the pO
2
in
actively respiring muscle tissue. [After Benesch, R.E. and
Benesch, R., Adv. Prot. Chem. 28, 212 (1974).]
See the
Animated Figures
JWCL281_c10_323-358.qxd 2/25/10 10:51 AM Page 328
of the pioneering atomic physicist Niels Bohr), who first
reported it in 1904.
a. The Bohr Effect Facilitates O
2
Transport
The ⬃0.8 molecules of CO
2
formed per molecule of O
2
consumed by respiration diffuse from the tissues to the
capillaries largely as dissolved CO
2
as a result of the slow-
ness of the reaction forming bicarbonate:
This reaction, however, is catalyzed in the erythrocyte by
carbonic anhydrase (Fig. 8-41). Accordingly, most of the
CO
2
in the blood is carried in the form of bicarbonate (in
the absence of carbonic anhydrase, the hydration of CO
2
would equilibrate 100-fold more slowly, so bubbles of the
only slightly soluble CO
2
would form in the blood and
tissues).
In the capillaries, where pO
2
is low, the H
⫹
generated by
bicarbonate formation is taken up by Hb, which is thereby
induced to unload its bound O
2
. This H
⫹
uptake, moreover,
facilitates CO
2
transport by stimulating bicarbonate forma-
tion. Conversely, in the lungs, where pO
2
is high, O
2
binding
by Hb releases the Bohr protons, which drive off the CO
2
.
These reactions are closely matched, so they cause very lit-
tle change in blood pH.
The Bohr effect provides a mechanism whereby addi-
tional O
2
can be supplied to highly active muscles. Such
muscles generate acid (Section 17-3A) so fast that they
lower the pH of the blood passing through them from 7.4
to 7.2. At pH 7.2, Hb releases ⬃10% more O
2
at the ⬍20
torr pO
2
in these muscles than it does at pH 7.4 (Fig. 10-6).
b. CO
2
and Cl
⫺
Modulate Hemoglobin’s O
2
Affinity
CO
2
modulates O
2
binding directly and by combining re-
versibly with the N-terminal amino groups of blood proteins
to form carbamates:
The conformation of deoxygenated Hb (deoxyHb), as we
shall see in Section 10-2Ba, is significantly different from
that of oxygenated Hb (oxyHb). Consequently, deoxyHb
binds more CO
2
as carbamate than does oxyHb. CO
2
like
H
⫹
, is therefore a modulator of hemoglobin’s O
2
affinity:A
high CO
2
concentration, as occurs in the capillaries, stimu-
lates Hb to release its bound O
2
. Note the complexity of
this Hb⫺O
2
⫺CO
2
⫺H
⫹
equilibrium: The protons released
by carbamate formation are, in part, taken up through
the Bohr effect, thereby increasing the amount of O
2
that
Hb would otherwise release. Although the difference in
CO
2
binding between the oxy and deoxy states of hemoglo-
bin accounts for only ⬃5% of the total blood CO
2
, it is nev-
ertheless responsible for around half of the CO
2
trans-
ported by blood. This is because only ⬃10% of the total
blood CO
2
turns over in each circulatory cycle.
Cl
⫺
is also bound more tightly to deoxyHb than to
oxyHb (Section 10-2E). Accordingly, hemoglobin’s O
2
affinity also varies with [Cl
⫺
]. freely permeates theHCO
⫺
3
R¬NH
2
⫹ CO
2
Δ R¬NH¬ COO
⫺
⫹ H
⫹
CO
2
⫹ H
2
O Δ H
⫹
⫹ HCO
⫺
3
erythrocyte membrane (Section 12-3D), so that once
formed, it equilibrates with the surrounding plasma. The
need for charge neutrality on both sides of the membrane,
however,requires that Cl
⫺
, which also freely permeates the
membrane, replace the that leaves the erythrocyte
(the erythrocyte membrane is impermeable to cations).
Consequently, [Cl
⫺
] in the erythrocyte is greater in the ve-
nous blood than it is in the arterial blood. Cl
⫺
is therefore
also a modulator of hemoglobin’s O
2
affinity.
D. Effect of BPG on O
2
Binding
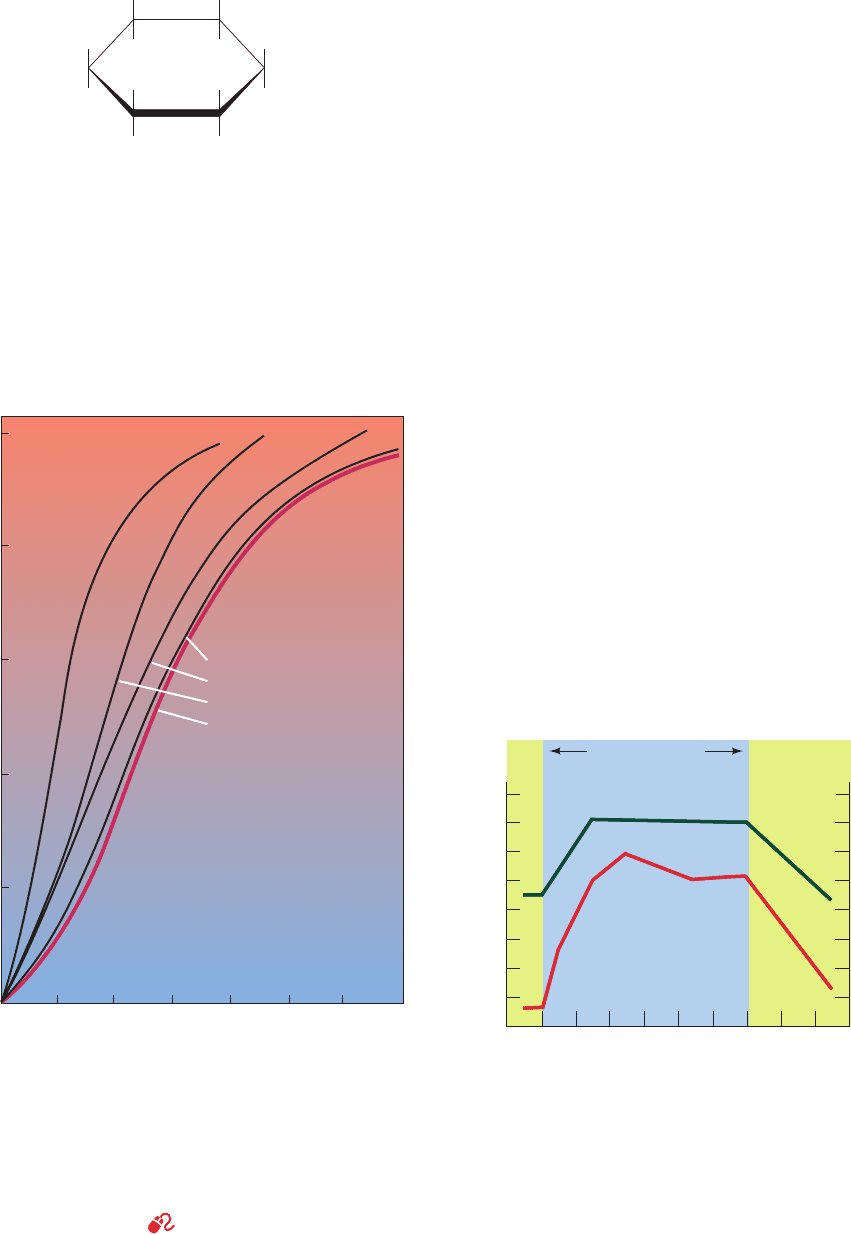
Purified (stripped) hemoglobin has a much greater O
2
affinity than does hemoglobin in whole blood (Fig. 10-7).
This observation led Joseph Barcroft, in 1921, to speculate
that blood contains some other substance that complexes
with Hb so as to reduce its O
2
affinity. In 1967, Reinhold
and Ruth Benesch demonstrated that this substance is
D-2,3-bisphosphoglycerate (BPG)
[previously known as 2,3-diphosphoglycerate (DPG)].
BPG binds tightly to deoxyHb in a 1:1 mole ratio (K ⫽
1.5 ⫻ 10
⫺5
M) but only weakly to oxyHb. The presence
of BPG therefore decreases hemoglobin’s oxygen affinity
by keeping it in the deoxy conformation; for example, the
p
50
of stripped hemoglobin is increased from 12 to 22 torr
by 4.7 mM BPG, its normal concentration in erythrocytes
OO
–
C
CH
C
H
H
D-2,3-Bisphosphoglycerate (BPG)
OPO
3
2⫺
OPO
3
2⫺
HCO
⫺
3
Section 10-1. Hemoglobin and Myoglobin Function 329
Figure 10-7 Comparison of the O
2
-dissociation curves of
“stripped” Hb and whole blood in 0.01M NaCl at pH 7.0. [After
Benesch, R.E. and Benesch, R., Adv. Prot. Chem. 28, 217 (1974).]
1.0
0.8
0.6
0.4
0.2
–1
0
012
log
pO
2
(torr)
Stripped
hemoglobin
Whole
blood
Y
O
2
JWCL281_c10_323-358.qxd 2/25/10 10:52 AM Page 329
(similar to that of Hb). Organic polyphosphates, such as
inositol hexaphosphate (IHP)
and ATP, also have this effect on Hb. In fact, in birds, IHP
functionally replaces BPG and ATP does so in fish and in
most amphibians. The ⬃2 mM ATP normally present in
Inositol hexaphosphate (IHP)
⫺2
O
3
PO
⫺2
O
3
PO
H
H
H
H
H
OPO
3
2⫺
OPO
3
2⫺
OPO
3
2⫺
HOPO
3
2⫺
mammalian erythrocytes is prevented from binding to Hb
by its complexation with Mg
2⫹
.
BPG has an indispensable physiological function: In arte-
rial blood, where pO
2
is ⬃100 torr, Hb is ⬃95% saturated
with O
2
,but in venous blood, where pO
2
is ⬃30 torr,it is only
⬃55% saturated (Fig. 10-3). Consequently, in passing
through the capillaries, Hb unloads ⬃40% of its O
2
. In the
absence of BPG, little of this O
2
is released since hemoglo-
bin’s O
2
affinity is increased, thus shifting the O
2
-dissociation
curve significantly toward lower pO
2
(Fig. 10-8, left).
CO
2
and BPG independently modulate hemoglobin’s
O
2
affinity. Figure 10-8 indicates that stripped Hb can be
made to have the same oxygen-dissociation curve as the
Hb in whole blood by adding CO
2
and BPG in the concen-
trations found in erythrocytes (the pH and [Cl
⫺
] are also
the same). Hence, the presence of these four substances in
whole blood—BPG, CO
2
,H
⫹
, and Cl
⫺
—accounts for the
O
2
-binding properties of Hb.
a. Increased BPG Levels Are Partially Responsible
for High-Altitude Adaptation
High-altitude adaptation is a complex physiological
process that involves an increase in the amount of hemo-
globin per erythrocyte and in the number of erythrocytes.
It normally requires several weeks to complete. Yet, as is
clear to anyone who has quickly climbed to high altitude,
even a 1-day stay there results in a noticeable degree of
adaptation.This effect results from a rapid increase in the
erythrocyte BPG concentration (Fig. 10-9; BPG, which
cannot pass through the erythrocyte membrane, is synthe-
sized in the erythrocyte; Section 17-2Hb).The consequent
decrease in O
2
-binding affinity, as indicated by its elevated
330 Chapter 10. Hemoglobin: Protein Function in Microcosm
Figure 10-8 The effects of BPG and CO
2
, both separately and
combined, on hemoglobin’s O
2
-dissociation curve compared with
that of whole blood (red curve). In the Hb solutions, which were
0.1M KCl and pH 7.22, pCO
2
⫽ 40 torr and the BPG concentration
was 1.2 times that of Hb. The blood had pCO
2
⫽ 40 torr and a
plasma pH of 7.40, which corresponds to a pH of 7.22 inside the
erythrocyte. [After Kilmartin, J.V. and Rossi-Bernardi, L.,
Physiol. Rev. 53, 884 (1973).]
See the Animated Figures
1.0
0.8
0.6
0.4
0.2
0
0 10203040506070
Stripped Hb
Hb + BPG +
CO
2
Hb + BPG
Hb + CO
2
Whole blood
pO
2
(torr)
Y
O
2
p
50
34
33
32
31
30
29
28
27
26
160
140
120
100
80
60
40
20
0
–1 0 1 2 3 4 5 6/0 1 2 3
BPG concentration
( g phosphorus
.
mL
–1
blood) μ
Time (days)
(torr)
Sea
level
Sea
level
4530 m above
sea level
p
50
BPG concentration
Figure 10-9 The effect of high-altitude exposure on the p
50
and the BPG concentration of blood in sea level–adapted
individuals. The region on the right marked “Sea level” indicates
the effects of exposure to sea level on high altitude–adapted
individuals. [After Lenfant, C.,Torrance, J.D., English, E., Finch,
C.A., Reynafarje, C., Ramos, J., and Faura, J., J. Clin. Invest. 47,
2653 (1968).]
JWCL281_c10_323-358.qxd 2/24/10 1:57 PM Page 330
