Voet D., Voet Ju.G. Biochemistry
Подождите немного. Документ загружается.

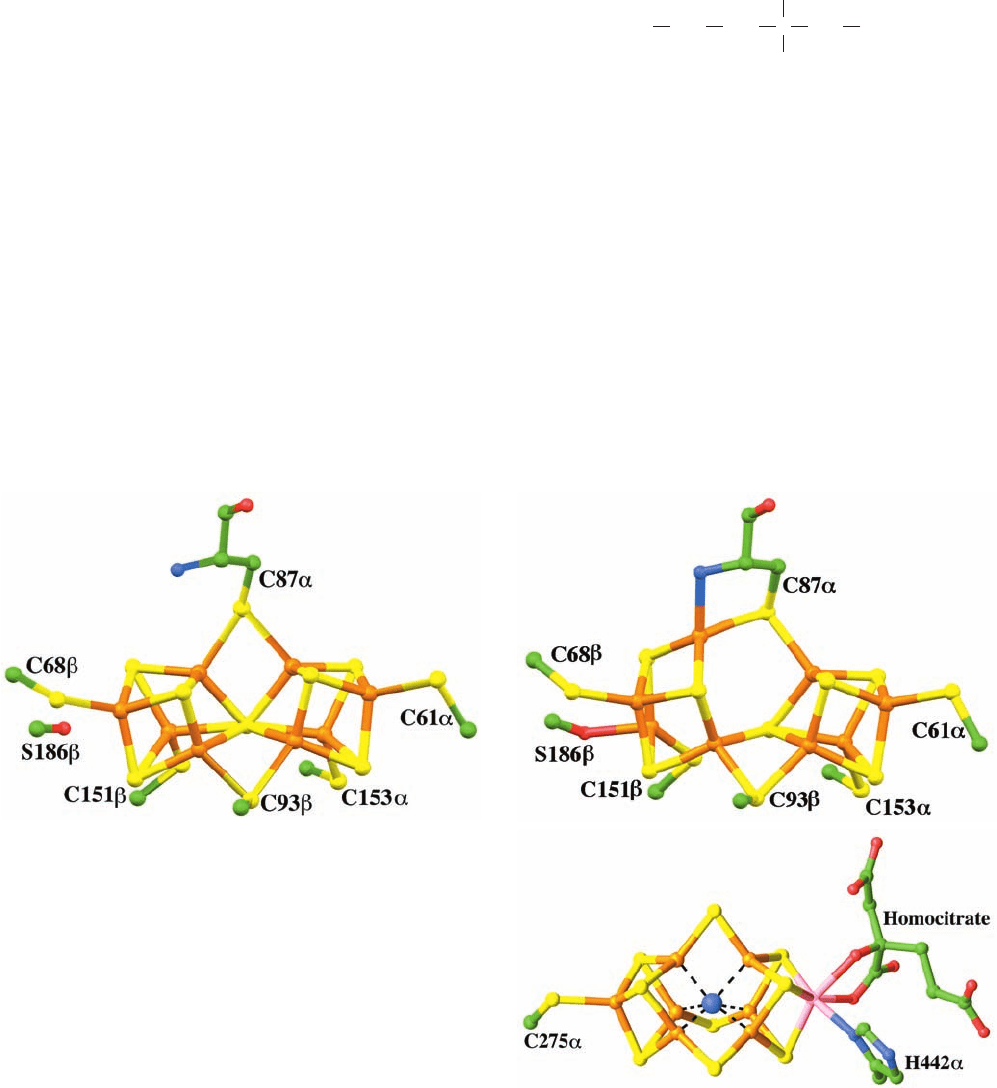
and is symmetrically linked to Cys 97 and Cys 132 from
both subunits such that an Fe-protein resembles an “iron
butterfly” with the [4Fe–4S] cluster at its head. Its two
identical nucleotide binding sites are located at the inter-
face between its two subunits. The [4Fe–4S] cluster cycles
between its ⫹1 and ⫹2 oxidation states.
The MoFe-protein’s ␣ and  subunits assume similar
folds and extensively associate to form a pseudo-2-fold
symmetric ␣ dimer, two of which more loosely associate
to form the 2-fold symmetric ␣
2

2
tetramer (Fig. 26-67).
Each ␣ dimer has two bound redox centers:
1. The P-cluster (Fig. 26-68a,b), which consists of two
[4Fe–3S] clusters linked through an additional sulfide ion
forming the eighth corner of each of the clusters to make
cubane-like structures, and bridged by two Cys thiol lig-
ands, each coordinating one Fe from each cluster. Four ad-
ditional Cys thiols coordinate the remaining four Fe atoms.
The positions of two of the Fe atoms in one of the [4Fe–3S]
clusters change on oxidation, rupturing the bonds from
these Fe atoms to the linking sulfide ion. These bonds are
replaced in the oxidized state by a Ser oxygen ligand to one
of the Fe atoms, and by a bond to the amide N of a Cys
from the other Fe atom.
2. The FeMo-cofactor (Fig. 26-68c), which consists of a
[4Fe–3S] cluster and a [1Mo–3Fe–3S] cluster bridged by
three sulfide ions.The FeMo-cofactor’s Mo atom is approx-
imately octahedrally coordinated by three cofactor sulfide
ions, a His imidazole nitrogen, and two oxygens from a
bound homocitrate ion:
(an essential component of the FeMo-cofactor). The
FeMo-cofactor contains a central cavity that a high resolu-
tion (1.16 Å) X-ray structure of A. vinelandii MoFe-
protein, also determined by Rees, reveals contains what
most probably is a nitrogen atom (although a C or an O atom
cannot be ruled out). This putative N atom is liganded to
the FeMo-cofactor’s central six Fe atoms such that it com-
pletes the approximate tetrahedral coordination environ-
ment of each of these Fe atoms.
The FeMo-cofactor is located ⬃10 Å below the ␣ subunit
surface, and hence the N
2
is thought to gain access to its
binding site through conformational fluctuations of the
protein (recall that myoglobin and hemoglobin likewise
have no clear path for O
2
to approach its heme binding
CH
2
C
CH
2
–
COO COO
–
COO
–
Homocitrate
OH
CH
2
Section 26-6. Nitrogen Fixation 1081
Figure 26-68 The prosthetic groups of the nitrogenase
MoFe-protein. The molecules are drawn in ball-and-stick form
with C green, N blue, O red, S yellow, Fe orange, and Mo pink.
(a) The reduced Klebsiella pneumoniae P-cluster. It consists of
two [4Fe–3S] complexes linked by an additional sulfide ion
forming the eighth corner of each cubane-like structure, and
bridged by two Cys thiol ligands, each coordinating one Fe from
each cluster. Four additional Cys thiols coordinate the remaining
4 Fe atoms. (b) The 2-electron-oxidized K. pneumoniae P-cluster.
In comparison with the reduced complex in Part a, two of the
Fe¬S bonds from the centrally located sulfide ion that bridges
the two [4Fe–3S] clusters have been replaced by ligands from the
Cys 87␣ amide N and the Ser 186 side chain O yielding a
[4Fe–3S] cluster (left) and a [4Fe–4S] cluster (right) that remain
linked by a direct Fe¬S bond and two bridging Cys thiols.
(c) The A. vinelandi FeMo-cofactor. It consists of a [4Fe–3S] cluster
and a [1Mo–3Fe–3S] cluster that are bridged by three sulfide
ions.The FeMo-cofactor is linked to the protein by only two
ligands at its opposite ends, one from His 442␣ to the Mo atom
and the other from Cys 275␣ to an Fe atom. The Mo atom is
additionally doubly liganded by homocitrate. What is most likely
an N atom (blue sphere) is liganded to the FeMo-cluster’s six
central Fe atoms (dashed black lines). [Parts a and b based on
X-ray structures by David Lawson, John Innes Centre, Norwich,
U.K. Part c based on an X-ray structure by Douglas Rees,
California Institute of Technology. PDBids (a) 1QGU, (b) 1QH1,
and (c) 1M1N.]
(a) (b)
(c)
JWCL281_c26_1019-1087.qxd 4/20/10 9:26 AM Page 1081

sites in these proteins; Section 10-2).The P-cluster, which is
also ⬃10 Å below the protein surface,is at the interface be-
tween the ␣ and  subunits on the pseudo-2-fold axis that
roughly relates these two subunits. The 2-fold axis of the
Fe-protein and the pseudo-2-fold axis of the MoFe-pro-
teins coincide in their complex.
The Fe-protein hydrolyzes two ATP molecules for each
electron it transfers from its [4Fe–4S]
⫹1
cluster to the
P-cluster. Since the nucleotide binding sites and the [4Fe–4S]
cluster on the Fe-protein are separated by ⬃20 Å, a distance
too large for direct coupling between electron transfer and
ATP hydrolysis, it appears that these processes are alloster-
ically coupled through conformational changes at the sub-
unit interface. Indeed, portions of the Fe-protein resemble
those of G-proteins, in which nucleotide hydrolysis is cou-
pled to conformational changes controlling the protein’s ac-
tions (Sections 19-2Cb and 19-3Cf).Specifically,two regions
of the Fe-protein, designated Switch I and Switch II (Fig.26-
67),are homologous with those of Ras (Section 19-3Cf).The
binding of ADP ⴢ AIF
⫺
4
to Fe-protein induces conforma-
tional changes in Switch I that affect the interactions be-
tween the Fe-protein and the MoFe-protein,and in Switch II
that affect the environment of the [4Fe–4S] cluster.
In nitrogenase, the [4Fe–4S] cluster of the Fe-protein ap-
proaches within ⬃14 Å of the P-cluster in the MoFe-protein,
whereas the P-cluster and the FeMo-cofactor are ⬃13 Å
apart. Hence, the sequence of the electron-transfer steps in
the nitrogenase reaction appears to be
It therefore seems that the role of ATP hydrolysis is to sta-
bilize a conformation in the Fe-protein that it cannot
achieve on its own and which facilitates electron transfer
from the [4Fe–4S] cluster on the Fe-protein to the P-cluster
on the MoFe-protein.
b. N
2
Reduction Is Energetically Costly
Nitrogen fixation requires two participants in addition
to N
2
and nitrogenase: (1) a source of electrons and (2) ATP.
Electrons are generated either oxidatively or photosyn-
thetically, depending on the organism. These electrons are
transferred to ferredoxin (Section 22-2C1a), a [4Fe–4S]-
containing electron carrier that transfers an electron to the
Fe-protein of nitrogenase, beginning the nitrogen fixation
process (Fig. 26-69). Two molecules of ATP bind to the
FeMo-cofactor
¡
N
2
[4Fe–4s] cluster
¡
P-cluster
¡
reduced Fe-protein and are hydrolyzed as each electron is
passed from the Fe-protein to the MoFe-protein.The ATP
hydrolysis-induced conformational change in the Fe-protein
alters its redox potential from ⫺0.29 to ⫺0.40 V, making
the electron capable of N
2
reduction ( for
the half-cell N
2
⫹ 6H
⫹
⫹ 6e
⫺
34 2NH
3
).
The actual reduction of N
2
occurs on the MoFe-protein
in three discrete steps, each involving an electron pair:
An electron transfer must occur six times per N
2
molecule
fixed so that a total of 12 ATPs are required to fix one N
2
molecule. Although the N
2
binding site is almost certainly
the FeMo-cofactor, exactly how the N
2
is bound and re-
duced are largely a matter of speculation.Theoretical stud-
ies suggest that the FeMo-cofactor’s prismatically arranged
Fe atoms provide favorable interaction sites for N
2
and its
reduction products. Indeed, it seems highly likely that the
putative N atom that is liganded to the FeMo-cofactor
(Fig. 26-68c) participates in N
2
reduction.
Nitrogenase also reduces H
2
O to H
2
, which in turn re-
acts with diimine to reform N
2
:
The resulting futile cycle is favored when the ATP level is
low and/or the reduction of the Fe-protein is sluggish. Even
when ATP is plentiful, however, the cycle cannot be sup-
pressed beyond about one H
2
molecule produced per N
2
reduced and hence appears to be a requirement of the ni-
trogenase reaction. The total cost of N
2
reduction is there-
fore 8 electrons transferred and 16 ATPs hydrolyzed (phys-
iologically, 20–30 ATPs). Hence nitrogen fixation is an
energetically expensive process; indeed, the nitrogen-fixing
bacteria in the root nodules of pea plants consume nearly
20% of the ATP that the plant produces.
c. Leghemoglobin Protects Nitrogenase
from Oxygen Inactivation
Nitrogenase is rapidly inactivated by O
2
,so the enzyme must
be protected from this reactive substance. Cyanobacteria
HN“NH ⫹ H
2
¡
N
2
⫹ 2H
2
2H
+
+ 2e
–
2H
+
+ 2e
–
2H
+
+ 2e
–
NN
NHH
HH
HH
N N 2NH
3
HydrazineDiimine
N
e°¿ ⫽⫺0.34 V
1082 Chapter 26. Amino Acid Metabolism
2P
(Ferredoxin)
red
(Ferredoxin)
ox
photosynthesis
or oxidative
electron
transport
(Fe-protein)
ox
(Fe-protein)
red
(MoFe-protein)
red
(MoFe-protein)
ox
8H
+
N
2
+
H
N
3
+2 H
2
+2ADP
i
2ATP
8 times
Figure 26-69 The flow of electrons in
the nitrogenase-catalyzed reduction of N
2
.
JWCL281_c26_1019-1087.qxd 4/20/10 9:26 AM Page 1082
fix their own nitrogen, a complex undertaking in which the
plant must be made to provide a hospitable environment
for nitrogen fixation as well as to acquire the enzymatic
machinery to do so. This would free farmers, particularly
those in developing countries, from the need for either pur-
chasing fertilizers, periodically letting their fields lie fallow
(giving legumes the opportunity to grow), or following the
slash-and-burn techniques that are rapidly destroying the
world’s tropical forests and contributing significantly to the
greenhouse effect (atmospheric CO
2
pollution causing
long-term global warming).
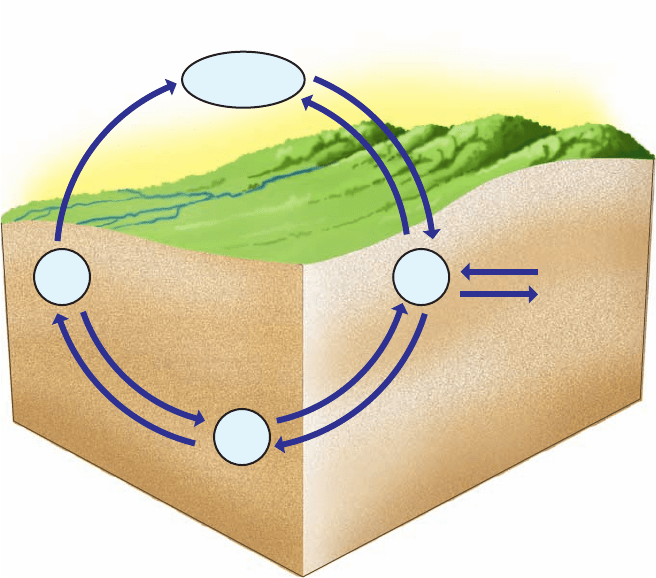
e. The Nitrogen Cycle Describes the Interconversion
of Nitrogen in the Biosphere
The ammonia produced by the nitrogenase reaction and
incorporated into amino acids is eventually recycled in the
biosphere as described by the nitrogen cycle (Fig. 26-70).
Nitrate is produced by certain bacteria that oxidize NH
3
to
NO
2
⫺
and then NO
3
⫺
, a process called nitrification. Still
other organisms convert nitrate back to N
2
, which is known
as denitrification. In addition, nitrate is reduced to NH
3
by
plants, fungi, and many bacteria, a process called ammoni-
fication in which nitrate reductase catalyzes the two-electron
reduction of nitrate to nitrite (NO
2
⫺
):
and then nitrite reductase converts nitrite to ammonia,
The direct anaerobic oxidation of NH
3
back to N
2
without
the intermediacy of nitrate, the reverse of nitrogen fixa-
tion, has recently been discovered in certain bacteria.
NO
2
⫺
⫹ 7H
⫹
⫹ 6e
⫺
¡
NH
3
⫹ 2H
2
O
NO
3
⫺
⫹ 2H
⫹
⫹ 2e
⫺
¡
NO
2
⫺
⫹ H
2
O
Section 26-6. Nitrogen Fixation 1083
Atmospheric
N
2
NO
2
–
NO
3
–
NH
3
Nitrite
Nitrication
Ammonication
Nitrogen xation
Nitrate
Ammonia
Assimilation
N-containing
biomolecules
Decomposition
nitrogenase
nitrite
reductase
nitrate
reductase
nitrogenase
nitrite
reductase
nitrate
reductase
Dentrication
Figure 26-70 The nitrogen cycle. Nitrogen
fixation by nitrogenase converts N
2
to the biologically
useful ammonia. Nitrate can also be converted to ammonia
by the sequential actions of nitrate reductase and nitrite reductase.
Ammonia is transformed to N
2
by nitrification followed by
denitrification.Ammonia may be assimilated into nitrogen-containing
biomolecules, which may be decomposed back to ammonia.
(photosynthetic oxygen-evolving bacteria; Section 1-1Ab)
provide protection by carrying out nitrogen fixation in spe-
cialized nonphotosynthetic cells called heterocysts, which
have Photosystem I but lack Photosystem II (Section 24-
2Ca). In the root nodules of legumes (Fig. 26-66), however,
protection is afforded by the symbiotic synthesis of leghemo-
globin. The globin portion of this ⬃145-residue monomeric
oxygen-binding protein is synthesized by the plant (an evolu-
tionary curiosity since globins are otherwise known to occur
only in animals), whereas the heme is synthesized by the Rhi-
zobium. Leghemoglobin has a very high O
2
affinity, thus
keeping the pO
2
low enough to protect the nitrogenase while
providing passive O
2
transport for the aerobic bacterium.
d. Installing the Nitrogen Fixation Machinery in
Nonleguminous Plants Would Revolutionize
Agriculture
Although atmospheric N
2
is the ultimate nitrogen source
for all living things, most plants do not support the symbi-
otic growth of nitrogen-fixing bacteria.They must therefore
depend on a source of “prefixed” nitrogen such as nitrate or
ammonia. These nutrients come from lightning discharges
(the source of ⬃10% of naturally fixed N
2
), decaying or-
ganic matter in the soil, or from fertilizer applied to it. The
Haber process, which was invented by Fritz Haber in 1910,
is a chemical process for N
2
fixation that is still widely used
in fertilizer manufacture. This direct reduction of N
2
by H
2
to form NH
3
requires temperatures of 300 to 500°C, pres-
sures of ⬎300 atm, and an Fe catalyst. Intriguingly, the spac-
ing of the Fe atoms on the surface of this catalyst resembles
that of the FeMo-cofactor’s central Fe atoms (Fig. 26-68c).
One of the major long-term goals of genetic engineering
is to induce agriculturally useful nonleguminous plants to
JWCL281_c26_1019-1087.qxd 6/8/10 9:38 AM Page 1083

1084 Chapter 26. Amino Acid Metabolism
1 Amino Acid Deamination Amino acids are the precur-
sors for numerous nitrogen-containing compounds such as
heme, physiologically active amines, and glutathione. Excess
amino acids are converted to common metabolic intermedi-
ates for use as fuels. The first step in amino acid breakdown is
removal of the ␣-amino group by transamination. Transami-
nases require pyridoxal phosphate (PLP) and convert amino
acids to their corresponding ␣-keto acids. The amino group is
transferred to ␣-ketoglutarate to form glutamate, oxaloac-
etate to form aspartate, or pyruvate to form alanine. Gluta-
mate is subsequently oxidatively deaminated by glutamate de-
hydrogenase (GDH) to form ammonia and regenerate
␣-ketoglutarate. Hyperinsulinism/hyperammonemia (HI/HA),
a genetic disease, is caused by a mutation of the GDH gene
that decreases GTP’s ability to inhibit GDH.
2 The Urea Cycle In the urea cycle, amino groups from
NH
3
and aspartate combine with HCO
⫺
3
to form urea. This
pathway takes place in the liver,partially in the mitochondrion
and partially in the cytosol. It begins with the ATP-dependent
condensation of NH
3
and HCO
3
⫺
by carbamoyl phosphate syn-
thetase, an enzyme with a 96-Å-long tunnel connecting its
three active sites through which its highly reactive intermedi-
ate products are channeled. The resulting carbamoyl phos-
phate then combines with ornithine to yield citrulline, which
combines with aspartate to form argininosuccinate, which in
turn is cleaved to fumarate and arginine. The arginine is then
hydrolyzed to urea, which is excreted, and ornithine, which
reenters the urea cycle. N-Acetylglutamate regulates the
urea cycle by activating carbamoyl phosphate synthetase
allosterically.
3 Metabolic Breakdown of Individual Amino Acids The
␣-keto acid products of transamination reactions are de-
graded to citric acid cycle intermediates or their precursors.
The amino acids leucine and lysine are ketogenic in that they
are converted only to the ketone body precursors acetyl-CoA
and acetoacetate. The remaining amino acids are, at least in
part, glucogenic in that they are converted to the glucose pre-
cursors pyruvate, oxaloacetate, ␣-ketoglutarate, succinyl-CoA,
or fumarate. Alanine, cysteine, glycine, serine, and threonine
are converted to pyruvate. Serine hydroxymethyltransferase
catalyzes the PLP-dependent C
␣
¬C

bond cleavage of serine
to form glycine. This reaction requires the transfer of a meth-
ylene group from N
5
,N
10
-methylene-tetrahydrofolate, which
the tetrahydrofolate (THF) obtains from the glycine cleavage
system, a multienzyme system. Asparagine and aspartate are
converted to oxaloacetate. ␣-Ketoglutarate is a product of
arginine, glutamate, glutamine, histidine, and proline degrada-
tion. Methionine, isoleucine, and valine are degraded to suc-
cinyl-CoA. Methionine breakdown involves the synthesis of
S-adenosylmethionine (SAM), a sulfonium ion that acts as a
methyl donor in many biosynthetic reactions. Hyperhomocys-
teinemia, a risk factor for cardiovascular disease, cognitive im-
pairment, and neural tube defects, is caused by a deficiency in
its folate-dependent degradation. Maple syrup urine disease
(MSUD) is caused by an inherited defect in branched-chain
amino acid degradation. Branched-chain amino acid degra-
dation pathways contain reactions common to all acyl-CoA
oxidations. Tryptophan is degraded to alanine and acetoac-
etate. Phenylalanine and tyrosine are degraded to fumarate
and acetoacetate. Most individuals with the hereditary disease
phenylketonuria lack phenylalanine hydroxylase (PAH), which
converts phenylalanine to tyrosine.
4 Amino Acids as Biosynthetic Precursors Heme is syn-
thesized from glycine and succinyl-CoA. These precursors
condense to form ␦-aminolevulinic acid (ALA), which cyclizes
to form the pyrrole porphobilinogen (PBG). Four molecules
of PBG condense to form uroporphyrinogen III, which then
goes on to form heme, with the final reaction, the insertion of
Fe(II) into protoporphyrin IX, catalyzed by ferrochelatase.
Defects in heme biosynthesis, which are known as porphyrias,
have a variety of bizarre symptoms. Heme is degraded to form
linear tetrapyrroles, which are subsequently excreted as bile pig-
ments.The hormones and neurotransmitters
L-DOPA, epineph-
rine, norepinephrine, serotonin, ␥-aminobutyric acid (GABA),
and histamine are all synthesized from amino acid precursors.
Glutathione, a tripeptide that is synthesized from glutamate,
cysteine, and glycine, is involved in a variety of protective,
transport, and metabolic processes.Tetrahydrofolate is a coen-
zyme that participates in the transfer of C
1
units.
5 Amino Acid Biosynthesis Amino acids are required
for many vital functions of an organism. Those amino acids
that mammals can synthesize from common ␣-keto acid car-
bon skeletons and preformed ␣-amino nitrogen such as that of
glutamate are known as nonessential amino acids; those that
mammals must obtain from their diets are called essential
amino acids. The biosynthesis of nonessential amino acids in-
volves relatively simple pathways, whereas those forming the
essential amino acids are generally more complex.
6 Nitrogen Fixation Although the ultimate source of ni-
trogen for amino acid biosynthesis is atmospheric N
2
, this
nearly inert gas must first be reduced to a metabolically useful
form, NH
3
, by nitrogen fixation. This process occurs only in
certain types of bacteria, one genus of which occurs in symbi-
otic relationship with legumes. N
2
is fixed in these organisms
by an oxygen-sensitive enzyme, nitrogenase, that consists of
two proteins: the Fe-protein dimer, which contains one
[4Fe–4S] cluster and two ATP binding sites, and the MoFe-
protein ␣
2

2
tetramer, which contains one P-cluster (consist-
ing of two [4Fe–3S] clusters linked by a sulfide ion) and one
FeMo-cofactor (a [4Fe–3S] cluster and a [1Mo–3Fe–3S] clus-
ter bridged by three sulfide ions and coordinated with homo-
citrate) in each ␣ dimer.These cofactors each function as two-
electron carriers for the ATP-driven reduction of N
2
to NH
3
.
CHAPTER SUMMARY
JWCL281_c26_1019-1087.qxd 6/8/10 9:38 AM Page 1084

References 1085
General
Bender, D.A., Amino Acid Metabolism, Wiley (1985).
Frey, P.A. and Hegeman, A.D., Enzymatic Reaction Mechanisms,
Oxford University Press (2007). [Describes the mechanisms of
many of the enzymes discussed in this chapter.]
Valle,D. (Ed.), The Online Metabolic & Molecular Bases of Inher-
ited Disease, http://www.ommbid.com/. [Part 8 contains numer-
ous chapters on defects in amino acid metabolism.]
Amino Acid Deamination and the Urea Cycle
Eliot, A.C. and Kirsch, J.F., Pyridoxal phosphate enzymes, Annu.
Rev. Biochem. 73, 383–415 (2004).
Holden, H.M., Thoden, J.B., and Raushel, F.M., Carbamoyl phos-
phate synthetase: a tunnel runs through it, Curr. Opin. Struct.
Biol. 8, 679–685 (1998); and Thoden, J.B., Holden, H.M.,
Wesenberg, G., Raushel, F.M., and Rayment, I., Structure of
carbamoyl phosphate synthetase:A journey of 96 Å from sub-
strate to product, Biochemistry 36, 6305–6316 (1997).
Jansonius, J.N., Structure, evolution and action of vitamin B
6
-
dependent enzymes, Curr. Opin. Struct. Biol. 8, 759–769 (1998).
Jungas, R.L., Halperin, M.L., and Brosnan, J.T., Quantitative
analysis of amino acid oxidation and related gluconeogenesis
in humans, Physiol. Rev. 72, 419–448 (1992).
Miles, E.W., Rhee, S., and Davies, D.R., The molecular basis of
substrate channeling, Biochemistry 274, 12193–12196 (1999).
Saeed-Kothe, A. and Powers-Lee, S.G., Specificity determining
residues in ammonia- and glutamine-dependent carbamoyl
phosphate synthetases, J. Biol. Chem. 277, 7231–7238 (2002).
Smith, T.J., Schmidt, T., Fang, J., Wu, J., Siuzdak, G., and Stanley,
C.A., The structure of apo human glutamate dehydrogenase
details subunit communication and allostery, J. Mol. Biol. 318,
765–777 (2002).
Smith, T.J. and Stanley, C.A., Untangling the glutamate dehydro-
genase allosteric nightmare, Trends Biochem. Sci. 33, 557–564
(2008).
Stipanuk, M.H. and Watford, M., Amino acid metabolism, Chap.
11, in Stipanuk,M.H. (Ed.), Biochemical and Physiological Ba-
sis of Nutrition, Saunders (2000).
Torchinsky, Yu.M., Transamination: its discovery, biological and
chemical aspects (1937–1987), Trends Biochem. Sci. 12,
115–117 (1987).
Metabolic Breakdown of Individual Amino Acids
Anderson, O.A., Flatmark, T., and Hough, E., High resolution
crystal structures of the catalytic domain of human phenylala-
nine hydroxylase in its catalytically active Fe(II) form and bi-
nary complex with tetrahydrobiopterin, J. Mol. Biol. 314,
279–291 (2001).
Ævarsson, A., Chuang, J.L., Wynn, R.M., Turley, S., Chuang, D.T.,
and Hol, W.G.J., Crystal structure of human branched-chain
␣-ketoacid dehydrogenase and the molecular basis of multien-
zyme complex deficiency in maple syrup urine disease, Struc-
ture 8, 277–291 (2000); and Wynn, R.M., Davie, J.R., Chuang,
J.L., Cote, C.D., and Chuang, D.T., Impaired assembly of E1 de-
carboxylase of the branched-chain ␣-ketoacid dehydrogenase
complex in type IA maple syrup urine disease, J. Biol. Chem.
273, 13110–13118 (1998).
Binda, C., Bossi, R.T., Wakatsuki, S., Arzt, S., Coda, A., Curti, B.,
Vanoni, M.A., and Mattevi, A., Cross-talk and ammonia chan-
neling between active centers in the unexpected domain
arrangement of glutamate synthase, Structure 8, 1299–1308
(2000).
Douce, R., Bourguignon, J., Neuburger, M., and Rébeillé, F., The
glycine decarboxylase system: a fascinating complex, Trends
Plant Sci. 6, 167–176 (2001).
Drennen,C.L., Huang,S., Drumond,J.T., Matthews, R.,and Ludwig,
M.L., How a protein binds B
12
: A 3.0 Å X-ray structure of
B
12
-binding domains of methionine synthase, Science 266,
1669–1674 (1994).
Faure, M., Rourguignon, J., Neuburger, M., Macherel, D., Sieker,
L., Ober, R., Kahn, R., Cohen-Addad, C., and Douce, R., Inter-
action between the lipoamide-containing H-protein and the
lipoamide dehydrogenase (L-protein) of the glycine decar-
boxylase multienzyme system, 2. Crystal structures of H- and
L-proteins, Eur. J. Biochem. 267, 2890–2898 (2000).
Guenther, B.D., Sheppard, C.A., Tran, P., Rozen, R., Matthews,
R.G., and Ludwig, M.L.,The structure and properties of meth-
ylenetetrahydrofolate reductase from Escherichia coli suggest
how folate ameliorates human hyperhomocysteinemia, Nature
Struct. Biol. 6, 359–365 (1999).
Guilhaudis, L., Simorre, J.-P., Blackledge, M., Marion, D., Gans, P.,
Neuburger, M., and Douce, R., Combined structural and bio-
chemical analysis of the H–T complex in the glycine decar-
boxylase cycle: evidence for a destabilization mechanism of
the H-protein, Biochemistry 39, 4259–4266 (2000).
Huang, X., Holden, H.M., and Raushel, F.M., Channeling of sub-
strates and intermediates in enzyme-catalyzed reactions,
Annu. Rev. Biochem. 70, 149–180 (2001).
Kelly, A. and Stanley, C.A., Disorders of glutamate metabolism,
Mental Retard. Devel. Dis. Res. Rev. 7, 287–295 (2001).
Ludwig, M.L. and Matthews, R.G., Structure-based perspectives
on B
12
-dependent enzymes, Annu. Rev. Biochem. 66, 269–313
(1997).
Matthews, R.G., Koutmos,M., and Datta, S., Cobalamin-dependent
and cobamide-dependent methyltransferases,Curr. Opin.Struct.
Biol. 18, 658–666 (2008).
Medina, M.Á., Urdiales, J.L., and Amores-Sánchez, M.I., Roles of
homocysteine in cell metabolism: Old and new functions, Eur.
J. Biochem. 268, 3871–3882 (2001).
Spiro, T.G. and Kozlowski, P.M., Is the CO adduct of myoglobin
bent, and does it matter? Acc. Chem. Res. 34, 137–144 (2001).
Swain,A.L., Jaskólski, M., Housset, D., Rao, J.K.M., and Wladower,
A., Crystal structure of Escherichia coli
L-asparaginase, an
enzyme used in cancer therapy, Proc. Natl. Acad. Sci. 90,
1474–1478 (1993).
Varughese, K.I., Skinner, M.M., Whiteley, J.M., Matthews, D.A.,
and Xuong, N.H., Crystal structure of rat liver dihydropteri-
dine reductase, Proc. Natl. Acad. Sci. 89, 6080–6084 (1992).
Zalkin, H. and Smith, J.L. Enzymes utilizing glutamine as an
amide donor, Adv. Enzymol. 72, 87–144 (1998).
Amino Acids as Biosynthetic Precursors
Ajioka, R.S., Phillips, J.D., and Kushner, J.P., Biosynthesis of heme
in mammals, Biochim. Biophys. Acta 1763, 723–736 (2006).
Al-Karadaghi, S., Franco, R., Hansson, M.,Shelnutt, J.A., Isaya, G.,
and Ferreira, G.C., Chelatases: distort to select, Trends
Biochem. Sci. 31, 135–142 (2006).
Battersby,A.R.,Tetrapyrroles: the pigments of life,Nat.Prod. Rep.
17, 507–526 (2000).
Erskine, P.T., Newbold, R., Brindley, A.A., Wood, S.P., Shoolingin-
Jordan, P.M., Warren, M.J., and Cooper J.B., The X-ray struc-
ture of yeast 5-aminolaevulinic acid dehydratase complexed
with substrate and three inhibitors, J. Mol. Biol. 312, 133–141
(2001).
REFERENCES
JWCL281_c26_1019-1087.qxd 6/8/10 9:38 AM Page 1085
Fitzpatrick, P.F., Tetrahydropterin-dependent amino acid hydrox-
ylases, Annu. Rev. Biochem. 68, 355–381 (1999).
Jaffe, E.K., Martins, J., Li, J., Kervinen, J., and Dunbrack, R.L., Jr.,
The molecular mechanism of lead inhibition of human por-
phobilinogen synthase, J. Biol. Chem. 276, 1531–1537 (2001).
Jaffe, E.K., Morpheeins—a new structural paradigm for allosteric
regulation, Trends Biochem. Sci. 39, 490–497 (2005); and
Lawrence, S.H.and Jaffe, E.K.,Expanding the concepts in protein
structure-function relationships and enzyme kinetics: Teaching
using morpheeins, Biochem. Mol. Biol. Educ. 36, 274–283 (2008).
Kauppinen, R., Porphyrias, Lancet 365, 241–252 (2005).
Louie, G.V., Brownlie, P.D., Lambert, R., Cooper, J.B., Blundell,
T.L., Wood, S.P., Warren, M.J., Woodcock, S.C., and Jordan,
P.M., Structure of porphobilinogen deaminase reveals a flexi-
ble multidomain polymerase with a single catalytic site, Nature
359, 33–39 (1992).
Medlock, A.E., Dailey, T.A., Ross, T.A., Dailey, H.A., and
Lanzilotta, W.N., A -helix switch selective for porphyrin de-
protonation and product release in human ferrochelatase,
J. Mol. Biol. 373, 1006–1016 (2007).
Pagola, S., Stephens, P.W., Bohle, D.S., Kosar, A.D., and Madsen,
S.K.,The structure of malaria pigment -haematin, Nature 404,
307–310 (2000).
Schneider-Yin, X., Gouya, L., Dorsey, M., Rüfenacht, U., Deybach,
J.-C., and Ferreira, G.C., Mutations in the iron-sulfur cluster
ligands of the human ferrochelatase lead to erythropoietic pro-
toporphyria, Blood 96, 1545–1549 (2000).
Sellers, V.M., Wu, C.-K., Dailey, T.A., and Dailey, H.A., Human
ferrochelatase: Characterization of substrate-iron binding and
proton-abstracting residues, Biochemistry 40, 9821–9827 (2001).
Shoolingin-Jordan, P.M., Warren, M.J., and Awan, S.J., Discovery
that the assembly of the dipyrromethane cofactor of porpho-
bilinogen deaminase holoenzyme proceeds initially by the re-
action of preuroporphyrinogen with the apoenzyme, Biochem.
J. 316, 373–376 (1996).
Song,G., Li,Y., Cheng, C.,Zhao,Y., Gao,A., Zhang, R., Joachimiak,
A., Shaw, N., and Li,Z.-J., Structural insight into acute intermit-
tent porphyria, FASEB J. 23, 396–404 (2009). [Reports the
X-ray structure of human porphobilinogen deaminase.]
Thunell, S., Porphyrins, porphyrin metabolism and porphyrias. I.
Update, Scand. J. Clin. Lab. Invest. 60, 509–540 (2000).
Wellems,T.E., How chloroquine works, Nature 355, 108–109 (1992).
Wu, C.-K., Dailey, H.A., Rose, J.P., Burden, A., Sellers, V.M., and
Wang, B.-C., The 2.0 Å structure of human ferrochelatase, the
terminal enzyme of heme biosynthesis, Nature Struct. Biol. 8,
156–160 (2001).
Amino Acids Biosynthesis
Chaudhuri, B.N., Lange, S.C., Myers, R.S., Chittur, S.V., Davisson,
V.J., and Smith, J.L., Crystal structure of imidazole glycerol
phosphate synthase: a tunnel through a (/␣)
8
barrel joins two
active sites, Structure 9, 987–997 (2001).
Dunn, M.F., Niks, D., Ngo, H., Barends,T.R.M., and Schlichting, I.,
Tryptophan synthase: the workings of a channeling nano-
machine, Trends Biochem. Sci. 33, 254–264 (2008).
Eisenberg,D., Gill, H.S.,Pfluegl,M.U., and Rotstein, S.H., Structure–
function relationships of glutamine synthetases, Biochim.
Biophys.Acta 1477, 122–145 (2000);and Gill,H.S. and Eisenberg,
D., The crystal structure of phosphinothricin in the active site
of glutamine synthetase illuminates the mechanism of enzy-
matic inhibition, Biochemistry 40, 1903–1912 (2001).
Hyde, C.C., Ahmed, S.A., Padlan, E.A., Miles, E.W., and Davies,
D.R., Three-dimensional structure of the tryptophan synthase
␣
2

2
multienzyme complex from Salmonella typhimurium,
J. Biol. Chem. 263, 17857–17871 (1988).
Katagiri, M. and Nakamura, M., Animals are dependent on pre-
formed ␣-amino nitrogen as an essential nutrient, Life 53,
125–129 (2002).
Kishore, G.M. and Shah, D.M.,Amino acid biosynthesis inhibitors
as herbicides, Annu. Rev. Biochem. 57, 627–663 (1988). [Dis-
cusses the biosynthesis of the essential amino acids.]
Larsen, T.M., Boehlein, S.K., Schuster, S.M., Richards, N.G.J.,
Thoden, J.B.,Holden,H.M.,and Rayment,I.,Three-dimensional
structure of Escherichia coli asparagine synthetase B: a short
journey from substrate to product, Biochemistry 38, 16146–
16167 (1999).
Stadtman, E.R., The story of glutamine synthetase regulation,
J. Biol. Chem. 276, 44357–44364 (2001).
Stallings, W.C., Abdel-Meguid, S.S., Lim, L.W., Shieh, H.-S.,
Dayringer, H.E., Leimgruber, N.K., Stegeman, R.A.,Anderson,
K.S., Sikorski, J.A., Padgette, S.R., and Kishore, G.M., Structure
and topological symmetry of the glyphosate target 5-enol-
pyruvylshikimate-3-phosphate synthase: A distinctive protein
fold, Proc. Natl. Acad. Sci. 88, 5046–5050 (1991). [The enzyme
that catalyzes Reaction 6 of Fig. 26-62 in complex with
glyphosate, an inhibitor that is a broad-spectrum herbicide.]
Weeks, A., Lund, L., and Raushel, F.M., Tunneling of intermedi-
ates in enzyme-catalyzed reactions, Curr. Opin. Chem. Biol. 10,
465–472 (2006).
Nitrogen Fixation
Einsle, O., Tezcan, F.A., Andrade, A.L.A., Schmidt, B., Yoshida,
M., Howard, J.B., and Rees, D.C., Nitrogense MoFe-protein at
1.16 Å resolution: A central ligand in the FeMo-cofactor, Sci-
ence 297, 1696–1700 (2002).
Fisher, R.F. and Long, S.R., Rhizobium–plant signal exchange,Na-
ture 357, 655–660 (1992). [Discusses the signals through which
Rhizobiaceae and legumes communicate to symbiotically gen-
erate the root nodules in which nitrogen fixation occurs.]
Jang, S.B., Seefeldt, L.C., and Peters, J.W., Insights into nucleotide
signal transduction in nitrogenase: Structure of an iron protein
with MgADP bound, Biochemistry 39, 14745–14752 (2000).
Lawson, D.M. and Smith, B.E., Molybdenum nitrogenases: a crys-
tallographic and mechanistic view, Metal Ions Biol. Sys. 39,
75–120 (2002).
Mayer, S.M., Lawson, D.M., Gormal, C.A., Roe, S.M., and Smith,
B.E., New insights into structure-function relationships in ni-
trogenase: A 1.6 Å resolution X-ray crystallographic study of
Klebsiella pneumoniae MoFe-protein, J. Mol. Biol. 292,
871–891 (1999).
Peters, J.W., Stowell, M.H.B., Soltis, S.M., Finnegan, M.G., Johnson,
M.K., and Rees, D.C., Redox-dependent structural changes
in the nitrogenase P-cluster, Biochemistry 36, 1181–1187
(1997).
Peters, J.W. and Szilagyi, R.K., Exploring new frontiers of nitroge-
nase structure and mechanism, Curr. Opin. Chem. Biol. 10,
101–108 (2006).
Rees, D.C.,Tezcan, F.A., Haynes, C.A., Walton, M.Y., Andrade, S.,
Einsle, O., and Howard, J.B., Structural basis of biological
nitrogen fixation, Philos.Trans. Roy. Soc.A 363, 971–984 (2005);
and Howard, J.B. and Rees, D.C., How many metals does it
take to fix N
2
? A mechanistic overview of biological nitrogen
fixation, Proc. Natl.Acad. Sci. 103, 17088–17093 (2006).
Schindelin, H., Kisker, C., Schlessman, J.L., Howard, J.B., and
Rees., D.C., Structure of ADP ⴢ AIF
⫺
4
stabilized nitrogenase
complex and its implications for signal transduction, Nature
387, 370–376 (1997).
Seefeldt, L.C.,Hoffman,B.M., and Dean, D.R., Mechanism of Mo-
dependent nitrogenase, Annu. Rev. Biochem. 78, 701–722
(2009).
1086 Chapter 26. Amino Acid Metabolism
JWCL281_c26_1019-1087.qxd 6/8/10 9:38 AM Page 1086

Problems 1087
1. Write the reaction for the transamination of an amino acid
in terms of Cleland notation (Section 14-5A).
2. Explain why the symptoms of the partial deficiency of a
urea cycle enzyme may be attenuated by a low-protein diet.
3. Why are people on a high-protein diet instructed to drink
lots of water?
4. A student on a particular diet expends 10,000 kJ ⴢ day
⫺1
while excreting 40 g of urea. Assuming that protein is 16% N by
weight and that its metabolism yields 18 kJ ⴢ g
⫺1
, what percentage
of the student’s energy requirement is met by protein?
5. Production of the enzymes that catalyze the reactions of the
urea cycle can increase or decrease according to the metabolic
needs of the organism. High levels of these enzymes are associ-
ated with high-protein diets as well as starvation. Explain this ap-
parent paradox.
6. Helicobacter pylori, the bacterium responsible for gastric
ulcers, can survive in the stomach (where the pH is as low as 1.5)
in part because it synthesizes large amounts of the enzyme urease.
(a) Write the reaction for urea hydrolysis by urease. (b) Explain
why this reaction could help establish a more hospitable environ-
ment for H. pylori, which tolerates acid but prefers to grow at
near-neutral pH.
7. Why are phenylketonurics warned against eating products
containing the artificial sweetener aspartame (NutraSweet; chem-
ical name
L-aspartyl-L-phenylalanine methyl ester)?
8. Demonstrate that the synthesis of heme from PBG as la-
beled in Fig. 26-35 results in the heme-labeling pattern given in
Fig. 26-32.
9. Explain why certain drugs and other chemicals can precipi-
tate an attack of acute intermittent porphyria.
10. Heterozygotes for erythropoietic protoporphyria show
only 20 to 30% residual ferrochelatase activity rather than the
50% that is normally expected for an autosomal dominant inher-
ited disease. Provide a plausible explanation for this observation.
11. One of the symptoms of kwashiorkor, the dietary protein
deficiency disease in children, is the depigmentation of the skin
and hair. Explain the biochemical basis of this symptom.
12. What are the metabolic consequences of a defective uridylyl-
removing enzyme in E. coli?
13. Figure 26-60, Reaction 9, indicates that methionine is syn-
thesized in microorganisms by the methylation of homocysteine
in a reaction in which N
5
-methyl-THF is the methyl donor.Yet, in
the breakdown of methionine (Fig. 26-18), its demethylation oc-
curs in three steps in which SAM is an intermediate. Discuss why
this reaction does not occur via the simpler one-step reversal of
the methylation reaction.
*14. In the glucose–alanine cycle (Fig. 26-3), glycolytically de-
rived pyruvate is transaminated to alanine and exported to the
liver for conversion to glucose and return to the cell. Explain how
a muscle cell is able to participate in this cycle under anaerobic
(vigorously contracting) conditions. (Hint: The breakdown of
many amino acids yields NH
3
.)
15. Draw the activated intermediates involved in (a) glutamine
and (b) asparagine biosynthesis from glutamate and aspartate, re-
spectively. (c) Provide an example of another metabolic activation
of a carboxylic acid group analogous to each of these reactions.
16. The ␣
2

2
tetramer of tryptophan synthase catalyzes the
PLP-dependent reaction of indole-3-glycerol phosphate and ser-
ine to form tryptophan (Fig. 26-63, Reactions 5 and 6). Draw the
chemical reactions involved in this synthesis, including the partic-
ipation of PLP, and use curved arrows to show the flow of elec-
trons.What role does PLP play in the reaction?
17. Suggest a reason why the nitrogen-fixing heterocysts of
cyanobacteria have lost Photosystem II but retain Photosystem I.
PROBLEMS
JWCL281_c26_1019-1087.qxd 4/20/10 9:26 AM Page 1087
1088
NPY/
AgRP
POMC/
CART
–
+
–
–
CHAPTER 27
Energy Metabolism:
Integration and Organ
Specialization
1 Major Pathways and Strategies of Energy
Metabolism: A Summary
2 Organ Specialization
A. Brain
B. Muscle
C. Adipose Tissue
D. Liver
E. Kidney
3 Metabolic Homeostasis: Regulation of Appetite,
Energy Expenditure, and Body Weight
A. AMP-Dependent Protein Kinase Is the Cell’s Fuel Gauge
B. Adiponectin Regulates AMPK Activity
C. Leptin
D. Insulin
E. Ghrelin and PYY
3–36
F. Hypothalamic Integration of Hormonal Signals
G. Control of Energy Expenditure by Adaptive Thermogenesis
H. Did Leptin Evolve as a Thrifty Gene?
4 Metabolic Adaptation
A. Starvation
B. Diabetes Mellitus
At this point in our narrative we have studied all of the ma-
jor pathways of energy metabolism. Consequently, we are
now in a position to consider how organisms, mammals in
particular, orchestrate the metabolic symphony to meet
their energy needs. This chapter therefore begins with a re-
capitulation of the major metabolic pathways and their
control systems, then considers how these processes are ap-
portioned among the various organs of the body, and ends
with a discussion of metabolic adaptation, including how
the body maintains energy balance (homeostasis), how it
deals with the metabolic challenges of starvation and obe-
sity, and how it responds to the loss of control resulting
from diabetes mellitus.
1 MAJOR PATHWAYS AND
STRATEGIES OF ENERGY
METABOLISM: A SUMMARY
Figure 27-1 indicates the interrelationships among the ma-
jor pathways involved in energy metabolism. Let us review
these pathways and their control mechanisms.
1. Glycolysis (Chapter 17) The metabolic degrada-
tion of glucose begins with its conversion to two molecules
of pyruvate with the net generation of two molecules each
of ATP and NADH. Under anaerobic conditions, pyruvate
is converted to lactate (or, in yeast, to ethanol) so as to re-
cycle the NADH. Under aerobic conditions, however,
when glycolysis serves to prepare glucose for further oxi-
dation, the NAD
⫹
is regenerated through oxidative phos-
phorylation (see below). The flow of metabolites through
the glycolytic pathway is largely controlled by the activity
of phosphofructokinase (PFK).This enzyme is activated by
AMP and ADP, whose concentrations rise as the need for
energy metabolism increases, and is inhibited by ATP and
citrate, whose concentrations increase when the demand
for energy metabolism has slackened. Citrate, a citric acid
cycle intermediate, also inhibits PFK and glycolysis when
aerobic metabolism takes over from anaerobic metabo-
lism, making glucose oxidation more efficient (the Pasteur
effect; Section 22-4C), and when fatty acid and/or ketone
body oxidation (which are also aerobic pathways) are
providing for energy needs (the glucose–fatty acid or
Randle cycle; Section 22-4Bb). PFK is also activated by
fructose-2,6-bisphosphate, whose concentration is regulated
by the levels of glucagon, epinephrine, and norepinephrine
through the intermediacy of cAMP (Section 18-3Fc). Liver
and heart muscle F2,6P levels are regulated oppositely: A
[cAMP] increase causes an [F2,6P] decrease in liver and an
[F2,6P] increase in heart muscle. However, skeletal muscle
[F2,6P] does not respond to changes in [cAMP].
2. Gluconeogenesis (Section 23-1) Mammals can syn-
thesize glucose from a variety of precursors, including
pyruvate, lactate, glycerol, and glucogenic amino acids (but
not fatty acids), through pathways that occur mainly in
liver and kidney. Many of these precursors are converted to
oxaloacetate which, in turn, is converted to phospho-
enolpyruvate and then, through a series of reactions that
largely reverse the path of glycolysis, to glucose. The irre-
versible steps of glycolysis, those catalyzed by PFK and
hexokinase, are bypassed in gluconeogenesis by hydrolytic
reactions catalyzed, respectively, by fructose-1,6-bisphos-
phatase (FBPase) and glucose-6-phosphatase. FBPase and
PFK may both be at least partially active simultaneously,
creating a substrate cycle.This cycle, and the reciprocal reg-
ulation of PFK and FBPase, are important in regulating
both the rate and direction of flux through glycolysis and
JWCL281_c27_1088-1106.qxd 4/22/10 9:58 AM Page 1088
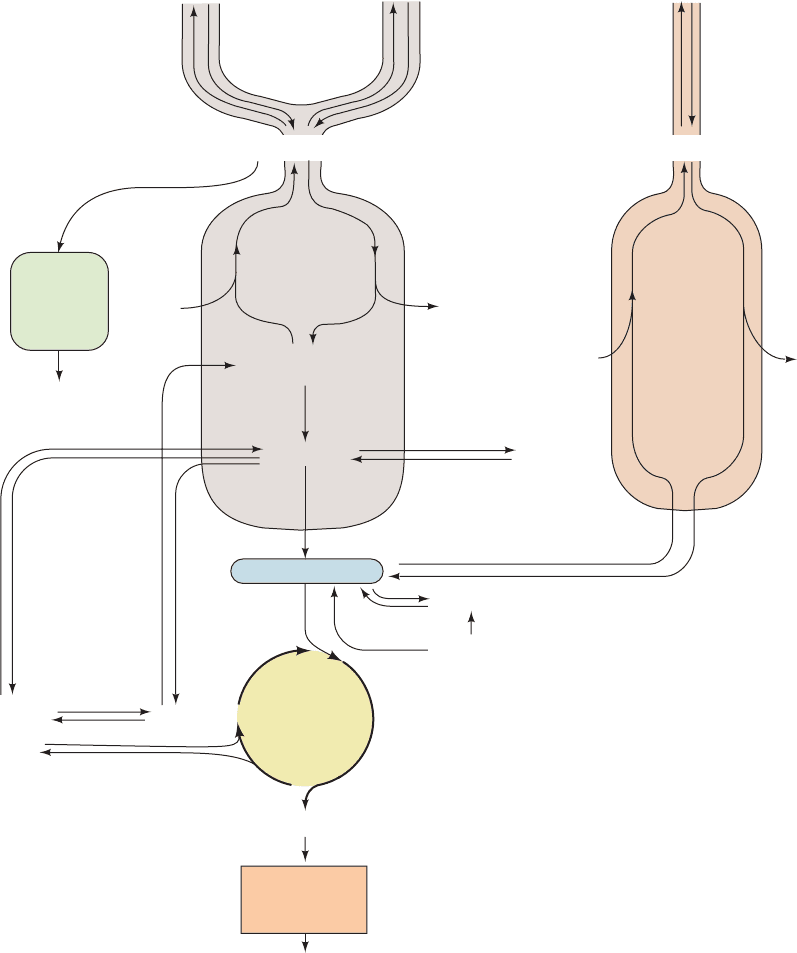
gluconeogenesis (Sections 17-4F and 23-1B). Fatty acid and
ketone body oxidation can increase the rate of gluconeogen-
esis in liver by decreasing the concentration of F2,6P
(Section 18-3Fc). This occurs because the increased citrate
concentration accompanying activation of the citric acid
cycle during fatty acid oxidation inhibits PFK-2 as well as
PFK (Table 23-1). Phosphoenolpyruvate carboxykinase
(PEPCK) bypasses the third irreversible reaction of glycol-
ysis, that catalyzed by pyruvate kinase (PK), and is con-
trolled exclusively by long-term transcriptional regulation.
3. Glycogen degradation and synthesis (Chapter 18)
Glycogen, the storage form of glucose in animals, occurs
mostly in liver and muscle. Its conversion to glucose-6-
phosphate (G6P) for entry into glycolysis in muscle and its
conversion to glucose in liver is catalyzed, in part, by glyco-
gen phosphorylase, whereas the opposing synthetic path-
way is mediated by glycogen synthase. These enzymes are
reciprocally regulated through phosphorylation/dephos-
phorylation reactions as catalyzed by amplifying cascades
that respond to the levels of the hormones glucagon and
epinephrine through the intermediacy of cAMP, and by
insulin (Sections 18-3E and 19-4F). The glucagon–insulin
ratio is therefore a crucial factor in determining the rate and
direction of glycogen metabolism.
4. Fatty acid degradation and synthesis (Sections 25-1
through 25-5) Fatty acids are broken down in increments
of C
2
units through  oxidation to form acetyl-CoA. They
are synthesized from this compound via a separate path-
way.The activity of the -oxidation pathway varies with the
fatty acid concentration.This, in turn, depends on the activ-
ity of “hormone-sensitive” triacylglycerol lipase in adipose
tissue that is stimulated, through cAMP-regulated phos-
phorylation/dephosphorylation reactions, by glucagon and
epinephrine but inhibited by insulin.The fatty acid synthe-
sis rate varies with the activity of acetyl-CoA carboxylase,
Section 27-1. Major Pathways and Strategies of Energy Metabolism: A Summary 1089
glucose-6-
phosphatase
fructose-1,6-
bisphosphatase
phosphofructokinase
acetyl-CoA carboxylase
Glucose
Glycogen
hexo-
kinase
glycogen
synthase
glycogen
phosphorylase
Glucose-6-phosphate
pentose
phosphate
pathway
NADPH
+
Ribose-5-phosphate
gluconeo-
genesis
glycolysis
Phosphoenol-
pyruvate
pyruvate
kinase
Pyruvate
pyruvate
dehydro-
genase
Acetyl-CoA
Citric
acid
cycle
Glucogenic
amino
acids
Oxaloacetate
NADH + FADH
2
ATP
Oxidative
phosphorylation
ATP
Ketone bodies
Ketogenic amino acids
pyruvate
carboxylase
phosphoenol-
pyruvate
carboxykinase
Lactate
NADH
NAPDH
lactate
dehydrogenase
Triacylglycerols
Fatty acids
hormone-
sensitive
triacylglycerol
lipase
triacylglycerol
synthesis
oxidation
β
fatty acid
synthesis
+
2
FADH
ATP
Figure 27-1 The major energy metabolism pathways.
JWCL281_c27_1088-1106.qxd 6/8/10 8:48 AM Page 1089
which is activated by citrate and insulin-dependent de-
phosphorylation, and inhibited by the pathway product
palmitoyl-CoA and by cAMP- and AMP-dependent phos-
phorylation. Fatty acid synthesis is also subject to long-
term regulation through alterations in the rates of synthe-
sis of the enzymes mediating this process as stimulated by
insulin and inhibited by fasting. The glucagon–insulin ratio
is therefore of prime importance in determining the rate and
direction of fatty acid metabolism.
5. Citric acid cycle (Chapter 21) The citric acid cycle
oxidizes acetyl-CoA, the common degradation product of
glucose, fatty acids, ketone bodies, and ketogenic amino
acids, to CO
2
and H
2
O with the concomitant production of
NADH and FADH
2
. Many glucogenic amino acids can also
be oxidized via the citric acid cycle through their break-
down, ultimately to pyruvate and then to acetyl-CoA,
sometimes via the cataplerosis (using up) of a citric acid cy-
cle intermediate (Section 21-5). The activities of the citric
acid cycle regulatory enzymes citrate synthase, isocitrate
dehydrogenase, and ␣-ketoglutarate dehydrogenase are
controlled by substrate availability and feedback inhibition
by cycle intermediates, NADH, and ATP.
6. Oxidative phosphorylation (Chapter 22) This mito-
chondrial pathway oxidizes NADH and FADH
2
to NAD
⫹
and FAD with the coupled synthesis of ATP. The rate of
oxidative phosphorylation, which is tightly coordinated
with the metabolic fluxes through glycolysis and the citric
acid cycle, is largely dependent on the concentrations of
ATP, ADP, and P
i
, as well as O
2
.
7. Pentose phosphate pathway (Section 23-4) This
pathway functions to generate NADPH for use in reductive
biosynthesis, as well as the nucleotide precursor ribose-5-
phosphate, through the oxidation of G6P. Its flux-generating
step is catalyzed by glucose-6-phosphate dehydrogenase,
which is controlled by the level of NADP
⫹
. The ability of
enzymes to distinguish between NADH, which is mainly
utilized in energy metabolism, and NADPH permits energy
metabolism and biosynthesis to be regulated independently.
8. Amino acid degradation and synthesis (Sections 26-1
through 26-5) Excess amino acids may be degraded to com-
mon metabolic intermediates. Most of these pathways begin
with an amino acid’s transamination to its corresponding ␣-
keto acid with the eventual transfer of the amino group to
urea via the urea cycle. Leucine and lysine are ketogenic
amino acids in that they can be converted only to acetyl-CoA
or acetoacetate and hence cannot be glucose precursors. The
other amino acids are glucogenic in that they may be, at least
in part, converted to one of the glucose precursors pyruvate,
oxaloacetate, ␣-ketoglutarate, succinyl-CoA, or fumarate.
Five amino acids are both ketogenic and glucogenic. Essen-
tial amino acids are those that an animal cannot synthesize it-
self; they must be obtained from plant and microbial sources.
Nonessential amino acids can be synthesized by animals uti-
lizing preformed amino groups via pathways that are gener-
ally simpler than those synthesizing essential amino acids.
Two compounds lie at the crossroads of the foregoing
metabolic pathways: acetyl-CoA and pyruvate (Fig. 27-1).
Acetyl-CoA is the common degradation product of most
metabolic fuels, including polysaccharides, lipids, and pro-
teins. Its acetyl group may be oxidized to CO
2
and H
2
O via
the citric acid cycle and oxidative phosphorylation or used
to synthesize fatty acids. Pyruvate is the product of glycoly-
sis, the dehydrogenation of lactate, and the breakdown of
certain glucogenic amino acids. It may be oxidatively decar-
boxylated to yield acetyl-CoA, thereby committing its
atoms either to oxidation or to the biosynthesis of fatty
acids.Alternatively, it may be carboxylated via the pyruvate
carboxylase reaction to form oxaloacetate, which, in turn,
either replenishes citric acid cycle intermediates or enters
gluconeogenesis via phosphoenolpyruvate, thereby bypass-
ing an irreversible step in glycolysis. Pyruvate is therefore a
precursor of several amino acids as well as of glucose.
The foregoing pathways occur in specific cellular com-
partments. Glycolysis, glycogen synthesis and degradation,
fatty acid synthesis, and the pentose phosphate pathway
are largely or entirely cytosolically based, whereas fatty
acid degradation, the citric acid cycle, and oxidative phos-
phorylation occur in the mitochondrion. Different phases
of gluconeogenesis and amino acid degradation occur in
each of these compartments. The flow of metabolites across
compartment membranes is mediated, in most cases, by spe-
cific carriers that are also subject to regulation.
The enormous number of enzymatic reactions that simul-
taneously occur in every cell (Fig. 16-1) must be coordinated
and strictly controlled to meet the cell’s needs. Such regula-
tion occurs on many levels. Intercellular communications
regulating metabolism occur via certain hormones, including
epinephrine, norepinephrine, glucagon, and insulin, as well as
through a series of steroid hormones known as glucocorti-
coids (whose effects are discussed in Section 19-1Ga). These
hormonal signals trigger a variety of cellular responses, in-
cluding the synthesis of second messengers such as cAMP in
the short term and the modulation of protein synthesis rates
in the long term. On the molecular level, enzymatic reaction
rates are controlled by phosphorylation/dephosphorylation
via amplifying reaction cascades, by allosteric responses to
the presence of effectors, which are usually precursors or
products of the reaction pathway being controlled, and by
substrate availability. The regulatory machinery of opposing
catabolic and anabolic pathways is generally arranged such
that these pathways are reciprocally regulated.
2 ORGAN SPECIALIZATION
Different organs have different metabolic functions and
capabilities. In this section we consider how the special
needs of the mammalian body organs are met and how
their metabolic capabilities are coordinated to meet these
needs. In particular, we discuss brain, muscle, adipose tis-
sue, liver, and kidney (Fig. 27-2).
A. Brain
Brain tissue has a remarkably high respiration rate. For in-
stance, the human brain constitutes only ⬃2% of the adult
1090 Chapter 27. Energy Metabolism: Integration and Organ Specialization
JWCL281_c27_1088-1106.qxd 4/21/10 9:41 AM Page 1090
