Schlick T. Molecular Modeling and Simulation: An Interdisciplinary Guide
Подождите немного. Документ загружается.

482 14. Molecular Dynamics: Further Topics
14.4.4 Generalized Verlet for Langevin Dynamics
The Verlet algorithm can easily be generalized to include the friction and stochas-
tic terms above. A common discretization is that described by Brooks, Br¨unger
and Karplus, known as BBK [180, 967]:
Generalized Verlet Algorithm for Langevin Dynamics
V
n+1/2
= V
n
+ M
−1
Δt
2
[−∇E(X
n
) − γMV
n
+ R
n
]
X
n+1
= X
n
+ΔtV
n+1/2
(14.30)
V
n+1
= V
n+1/2
+ M
−1
Δt
2
−∇E(X
n+1
) − γMV
n+1
+ R
n+1
.
This Langevin scheme reduces to velocity Verlet (triplet eq. (13.14)) when γ
and hence R
n
are zero.
Note that the third equation above defines V
n+1
implicitly; the linear depen-
dency, however, allows solution for V
n+1
in closed form (i.e., explicitly). The
superscript used for R has little significance, as the random force is chosen
independent at each step.
When the Dirac delta function of eq. (14.28) is discretized, δ(t −t
) is replaced
by δ
nm
/Δt.
The BBK method is only appropriate for the small-γ regime. A more ver-
satile algorithm was derived in [1293]. See also [1200] for a related Langevin
discretization.
14.4.5 The LN Method
The idea of combining force splitting via extrapolation with Langevin dynamics
can alleviate severe resonance effects — as discussed in the MTS section (and
shown in Figure 14.5 for a simple harmonic model) — and allow larger outer
timesteps to be used than impulse-based MTS schemes [93–95].
The LN algorithm based on this combination (see background in Box 14.1)
is sketched in Figure 14.8 in the same notation used for the MTS schemes. The
direct-force algorithms on the left forms the basis for the algorithms implemented
in the CHARMM [95]andAMBER[89,97,1025] programs.
Note that LN is based on position Verlet rather than velocity Verlet. If a
constrained dynamics formulation is used (e.g., SHAKE), this splitting version
requires on average two SHAKE iterations per inner loop rather than the one
required by velocity Verlet. However, we have found stability advantageous in
practice for the position Verlet scheme over velocity Verlet in the unusual limit of
moderate to large timesteps and large timescale separations [99].
14.4. Langevin Dynamics 483
0 20 40
−4
−2
0
2
Outer Timestep [fs]
Energy [Kcal/mol]
LN
Langevin Impulse
0 5 10
−5
0
2
5
10
x 10
4
x 10
4
Outer Timestep [fs]
Energy [Kcal/mol]
Newtonian Extrapolation
Newtonian Impulse
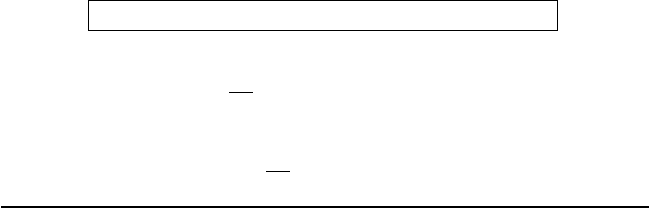
Figure 14.7. Energy means and deviations as computed over 5-ps Newtonian and Langevin
impulse and extrapolative force splitting schemes for the protein BPTI with the damping
constant γ =20ps
−1
, as functions of the outer timestep for a fixed inner timestep of 0.5 fs
and medium timestep of 1 fs [1089].
Resonance Alleviation
Applications to the solvated model of the protein BPTI in Figure 14.7 show
how the Langevin extrapolative treatment alleviates severe resonances present
in impulse treatments for both Newtonian and Langevin dynamics. Newto-
nian extrapolation yields large energy increases as the timestep increases. Both
Newtonian and Langevin impulse splitting exhibit resonant spikes: beyond half
the fastest period for Newtonian impulse, and beyond the fastest period for
Langevin impulse splitting. Unfortunately, once such resonance is encountered,
good behavior cannot be restored after the first resonant timestep. The LN com-
bination, in contrast, does not exhibit resonant spikes for this value of damping
constant, 20 ps
−1
. It stabilizes the energy growth seen for Newtonian extrapola-
tion. See also [1134] for detailed performance evaluations for a large protein/DNA
complex.
The exchange of Hamiltonian dynamics for stochastic dynamics can guarantee
better numerical behavior, but the resulting dynamics are altered. Stochastic dy-
namics are, however, suitable for many thermodynamic and sampling questions.
Small-timestep dynamics simulations can always be performed in tandem once
an interesting region of conformation space is identified.

484 14. Molecular Dynamics: Further Topics
LN Algorithm
(Direct Force Version) (Linearization Variant changes)
X
0
r
≡ X X
r
≡ X +
Δt
m
2
V
F
slow
≡−∇E
slow
(X
r
)
For j =1 to k
2
X
r
≡ X
j
r
← X +
Δt
m
2
V X
r
← X +
Δt
m
2
V ;
H
r
≡
H(X
r
)
F
med
≡−∇E
med
(X
r
) F
med
≡−∇E
med
(X
r
) −∇E
fast
(X
r
)
F ← F
med
+ F
slow
For i =1 to k
1
Evaluate Gaussian force R
X ← X +
Δτ
2
V
F
tot
← F + F
fast
(X)+R F
tot
← F −
H
r
· (X −X
r
)+R
V ←
V + M
−1
ΔτF
tot
/γ
X ← X +
Δτ
2
V
End
End [γ =1+γΔτ] [
H
r
= local Hessian approx.]
Figure 14.8. Algorithmic sketch of the LN scheme [95] for Langevin dynamics by extrap-
olative force-splitting based on position Verlet. The version on the left uses direct fast-force
evaluations, while the reference version on the right (only alternative statements shown)
uses linearization to approximate the fast forces over a Δt
m
interval. The latter requires a
local Hessian formulation,
H
r
=
H(X
r
), at point X
r
every time the medium forces are
evaluated. See Box 14.1 for background details.
Skeel, Izaguirre, and coworkers have also adopted the stochastic coupling [594,
595] in the context of the mollified impulse method [1133] to extend the timestep
beyond half the fast period. Because their γ is yet smaller, the agreement with
Hamiltonian dynamics is better, but the largest possible outer timesteps and hence
speedup factors are reduced.
An interesting recent variation called “NML” [1242] is an extension of a LIN
[1435,1436] developed as a way to increase the timestep in standard MD integra-
tion. The idea in LIN was to use implicit integration for the low-frequency modes
and normal-mode analysis (NMA) for the high-frequency modes; Langevin rather
than Newtonian dynamics was also used to dampen resonance effects, which
limit the timestep to around 3.3 fs when all light-atom motions are consid-
ered. In NML, the costly NMA of LIN is replaced by Brownian dynamics
for the high-frequency modes to keep the fast oscillations around their equilib-
rium values; the low-frequency modes are propagated by Langevin dynamics as
in LIN.

14.4. Langevin Dynamics 485
Box 14.1: LN Background and Assessment
Background. LN arose fortuitously [93] upon analysis of the range of harmonic validity
of the Langevin/Normal-mode method LIN [1435,1436]. Essentially, in LIN the equations
of motion are linearized using an approximate Hessian and solved for the harmonic com-
ponent of the motion; an implicit integration step with a large timestep then resolves the
residual motion (see subsection on implicit methods). Approaches based on linearization
of the equations of motion have been attempted for MD [67, 603, 1209, 1280], but compu-
tational issues ruled out general macromolecular applications. Indeed, the LIN method is
stable over large timesteps such as 15 fs but the speedup is modest due to the cost of the
minimization subproblem involved in the implicit discretization. The discarding of LIN’s
implicit-discretization phase — while reducing the frequency of the linearization — in
combination with a force splitting strategy forms the basis of the LN approach [95].
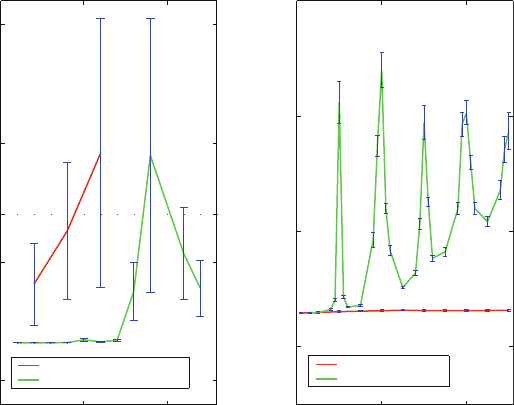
Performance: Energetics and Speedup. Performance of MTS schemes can be analyzed
by ‘Manhattan plots’, as shown in Figure 14.9 for a large polymerase/DNA system of
41,973 atoms (illustrated in Figure 14.10)[1408]; that is, differences of mean energy
components are reported as a function of the outer timestep Δt relative to STS Langevin
simulations. For three LN protocols — using different combinations of Δτ , Δt
m
, γ,and
bond constraints (SHAKE on or off) — these plots, along with corresponding CPU times
and speedup, show that the first protocol has the optimal combination of low relative
error in all energy components (below 3%) and low CPU time per physical time unit. The
computational speedup factor is 4 or more in all cases.
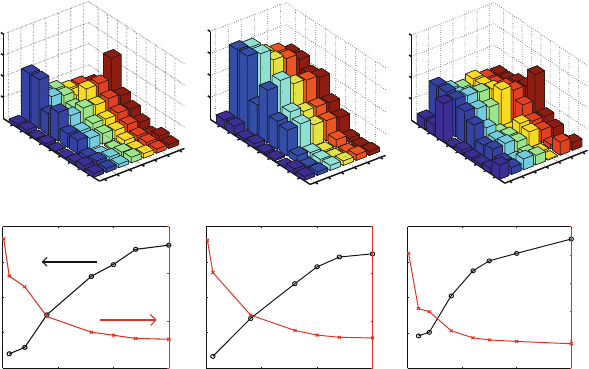
Performance: Dynamics. The assignment of the Langevin parameter γ in the LN scheme
ensures numerical stability on one hand and minimizes the perturbations to Hamiltonian
dynamics on the other; we have used γ =10ps
−1
or smaller in biomolecular simulations.
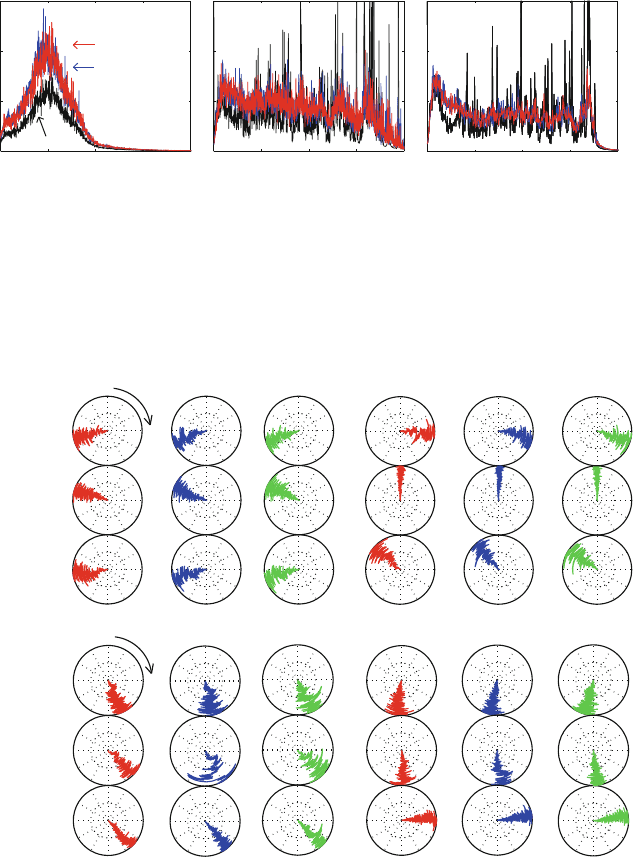
To assess the effect of γ of dynamic properties, the protocol-sensitive spectral density
functions computed from various trajectories can be analyzed (see Figure 14.11 caption).
The densities for solvated BPTI in Figure 14.11 show how the characteristic frequencies
can be more closely approximated as γ is decreased; the densities for the large polymerase
system in Figure 14.12 show, in addition, the good agreement between the STS Langevin
and LN-computed frequencies for the same γ. This emphasizes the success of MTS in-
tegrators as long as the inner timestep is small. (Recall another illustration of this point
for butane in Figure 13.5, where the average butane end-to-end distance is shown for STS
versus MTS protocols).
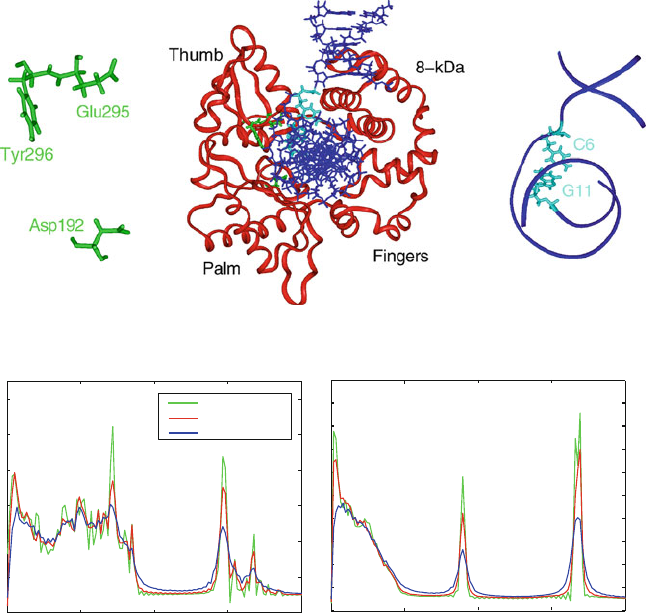
Detailed comparisons of the evolution of various geometric variables (Figure 14.13)re-
flect the agreement between LN and the reference Langevin simulation as well [1408]. As
expected, individual trajectories diverge, but the angular fluctuations are all in reasonable
ranges. The flexibility of the DNA backbone angles is expected at the base pair near the
kink induced by the polymerase [1408].
486 14. Molecular Dynamics: Further Topics
6
40
80
100
120
150
Ek
Temp
Evdw
Etot
Ep
Eelec
Etors
Eangle
Ebond
1
2
3
4
6
20
40
80
100
120
150
Ek
Temp
Evdw
Etot
Ep
Eelec
Etors
Eangle
Ebond
1
2
3
4
0 50 100 150
1
2
3
4
5
0
50
100
150
6
20
40
80
100
120
150
Ek
Temp
Ebond
Etot
Evdw
Ep
Eelec
Etors
Eangle
1
2
3
4
abc{Δτ, Δt
m
} = {1, 2} fs
No SHAKE
SHAKE
SHAKE
LN speedup factor
CPU time (days/ns)
Relative error [%]
Energy [kcal/mol] Outer timestep [fs]
{Δτ, Δt
m
} = {1, 2} fs {Δτ, Δt
m
} = {0.5, 1} fs
γ = 10 ps
−1
γ = 5 ps
−1
γ = 10 ps
−1
Outer timestep [fs]
0 50 100 150
1
2
3
4
5
0
50
100
150
0 50 100 150
1
2
3
4
5
0
50
100
150
200
250
CPU time
Speedup
Figure 14.9. ‘Manhattan plots’ for a solvated polymerase/DNA system showing the LN
relative errors in different energy components (bond, bond angle, torsion, electrostatic, po-
tential, total, van der Waals) and the kinetic temperature as a function of the outer timestep
Δt compared to single-timestep Langevin trajectories at the same Δτ,aswellascorre-
sponding speedups (scale at left) and CPU times per nanosecond (scale at right). ‘SHAKE’
refers to constraining all bonds involving hydrogen. The data for three parameter com-
bination in (a–c) are based on simulations of length 12 ps started from the intermediate
polymerase/DNA complex [1408], pictured in Figure 14.10. See also Box 14.1.
Testing and Application
Results have shown that good parameter choices for a 3-class LN scheme are
Δτ =0.5 fs, Δt
m
=1fs, and Δt up to 150 fs. If constrained dynamics
for the light atom bonds are used, the inner timestep can be increased to 1 fs
and the medium timestep to around 2 fs. Various biomolecular applications have
shown good agreement of LN trajectories to small-timestep Langevin simulations
and significant speedup factors [95, 1230, 1408]. See Box 14.1 for performance
assessment of energetics, dynamics, and speedup on a biomolecule.
Recent work has tailored elements of the LN integrator to particle-mesh Ewald
protocols [97, 99, 1024, 1025]. Challenges remain regarding the most effective
MTS splitting procedures for Ewald formulations, given the fast terms present
in the Ewald reciprocal component [89, 97, 98, 1025, 1110, 1236, 1449]and
the numerical artifact stemming from subtraction of the term accounting for
excluded-nonbonded atoms pairs [89,1019,1025].
14.5. Brownian Dynamics (BD) 487
Figure 14.10. Solvated polymerase/DNA system used for evaluating LN [1408].
0 1000 2000 3000 4000
Protein
Velocity Verlet
γ
= 5 ps
-1
γ
=20 ps
−1
0 1000 2000 3000 4000
Water
Wave number [cm
−1
] Wave number [cm
−1
]
Figure 14.11. Spectral density functions calculated by Verlet and LN over 5 ps runs for a
solvated system of the protein BPTI (protein and water frequencies shown separately) at
three γ values: 0 (by the velocity Verlet scheme), 5 and 20 ps
−1
by the LN scheme [1089].
The functions are computed by Fourier transforming the velocity autocorrelation time se-
ries for each atom in the system to obtain a power spectrum for each atom. These spectra
are averaged over the water and biomolecule atoms separately for global characterization
of the motion. See [1089] for the detailed protocol. Note that the characteristic frequencies
obtained by this procedure reflect the force field constants rather than physical frequencies
per se.
14.5 Brownian Dynamics (BD)
14.5.1 Brownian Motion
The mathematical theory of Brownian motion is rich and subtle, involving high-
level physics and mathematics. Important contributors to the theory include
Einstein (who explained Brownian motion in 1905) and Planck. Here we present
488 14. Molecular Dynamics: Further Topics
0 500 1000 1500 2000 0 500 1000 1500 20000 500 1000 1500 2000
Wave number [cm
−1
]
Water
DNA
Protein
LN 1/2/150 fs
BBK 1 fs
γ = 10 ps
−1
VV 1 fs
Figure 14.12. Spectral density functions calculated by the STS Newtonian (velocity Verlet)
and STS Langevin (BBK, γ =10ps
−1
) schemes versus the MTS LN scheme at
γ =10ps
−1
(triple timestep protocol 1/2/150 fs) for a solvated polymerase/DNA sys-
tem simulated over 12 ps and sampled every 2 fs. Spectral densities for the protein, DNA,
and water atoms are shown separately. See also caption to Figure 14.11.
Asp192
Glu295
Tyr296
Protein φ
Protein ϕ
LN BBK VV LN BBK VV
DNA C6 DNA G11
P
β
δ
0
90
270
0
90
270
Figure 14.13. Comparisons of the evolution of representative dihedral angles (from protein
φ and ψ and DNA sugar and backbone near one CG base pair) over 12 ps for the STS veloc-
ity Verlet, STS Langevin (BBK), and the MTS LN scheme for a solvated polymerase/DNA
system. The LN protocol used is Δτ =1fs, Δt
m
=2fs, and Δt = 150 fs, with SHAKE.
The polar vector coordinates correspond to time and angle. See Figure 14.10 for location
of residues.

14.5. Brownian Dynamics (BD) 489
the BD framework as an extension to the Langevin model presented above and
focus on numerical algorithms for Brownian dynamics. See statistical mechanics
texts, such as [853], for more comprehensive presentations.
The term Brownian is credited to the botanist Robert Brown, who in 1827 ob-
served that fine particles — like pollen grains, dust, and soot — immersed in a
fluid undergo a continuous irregular motion due to collisions of the particles with
the solvent molecules. Dutch physician Jan Ingenhausz was actually the first, in
1785, to report this motion for powdered charcoal on an alcohol surface. The ef-
fective force on such particles originates from friction, as governed by Stokes’
law, and a fluctuating random force.
14.5.2 Brownian Framework
Generalized Friction
Generalized frictional interactions among the particles can be incorporated into
the Langevin equation introduced in Section 14.4 using a friction tensor Z.
This matrix replaces the single parameter γ, so as to describe the action of the
solvent by:
M
¨
X(t)=−∇E(X(t)) − Z
˙
X(t)+R(t), (14.31)
where the mean and covariance of the random force R are given by:
R(t) =0, R(t)R(t
)
T
=2k
B
TZ δ(t − t
) . (14.32)
The above relation is based on the fluctuation/dissipation theorem, a fundamental
result showing how friction is related to fluctuations of the random force, as-
suming the Brownian particle is randomly moving about thermal equilibrium.
This description ensures that the ensembles of the trajectories generated from
eq. (14.31) are governed by the Fokker-Planck equation, a partial-differential
equation describing the density function in phase space of a particle undergoing
diffusive motion.
The random force R is white noise and has no natural timescale. Thus, the
inertial relaxation times given by the inverses of the eigenvalues of the matrix
M
−1
Z define the characteristic timescale of the thermal motion in eq. (14.31).
Neglect of Inertia
When the inertial relaxation times are short compared to the timescale of interest,
it is often possible to ignore inertia in the governing equation, that is, discard the
momentum variables, assuming M
¨
X(t)=0.Fromeq.(14.31), we have:
˙
X(t)=− Z
−1
∇E(X(t)) + Z
−1
R(t), (14.33)
and this can be written in the Brownian dynamics form:
˙
X(t)=−
D
k
B
T
∇E(X(t)) + R
B
(t); (14.34)
490 14. Molecular Dynamics: Further Topics
here D is the diffusion tensor
D = k
B
TZ
−1
, (14.35)
and the mean and covariance of the random force R
B
depend on D as:
R
B
(t) =0, R
B
(t)R
B
(t
)
T
=2D δ(t − t
) . (14.36)
Thus, solvent effects are sufficiently large to make inertial forces negligible,
and the motion is overall Brownian and random in character. This description
is effective for very large, dense systems whose conformations in solution are
continuously and significantly altered by the fluid flow in their environment.
Transport Properties
Brownian theory allows us to determine average behavior, such as transport prop-
erties, for systems governed by such diffusional motion. For example, a molecule
modeled as a free Brownian particle that moves a net distance x over a time in-
terval Δt has an expected mean square distance x
2
(analogous to the mean
square end-to-end distance in a polymer chain) proportional to Δt: x
2
∝Δt.
Einstein showed that the proportionality constant is 2D,whereD is the diffusion
coefficient:
x
2
=2D Δt.
Algorithms
An appropriate simulation algorithm for following Brownian motion can be
defined by assuming that the timescales for momentum and position relaxation
are well separated, with the former occurring much faster. BD algorithms then
prescribe recursion recipes that displace each current position by a random force
— similar in flavor to the Langevin random force — and additional terms that
depend on the diffusion tensor.
It is not always possible to neglect inertial contributions. For example, it was
shown that inertial contributions can affect long-time processes in long DNA due
to mode coupling [107]. An “inertial BD” algorithm termed IBD has been devel-
oped, tested on simple systems [106], and applied to bead models of long DNA
[107]. IBD more accurately approximates long-time kinetic processes that occur
within equilibrium ensembles, as long as the timestep is not too small.
In practice, the IBD scheme adds a mass-dependent correction term to the usual
BD propagation scheme (see below). Though IBD has an additional computa-
tional cost of a factor of two over BD, the computational complexity is the same
as for the usual BD scheme based on the Cholesky factorization (consult a numer-
ical methods textbook like [280] for the Cholesky factorization, and see Box 14.4
and below for IBD details).

14.5. Brownian Dynamics (BD) 491
14.5.3 General Propagation Framework
In general, larger timesteps are used for Brownian dynamics simulations than for
molecular and Langevin dynamics simulations. As a first example, consider a free
particle in one dimension whose diffusion constant D is (by definition) the mean
square displacement divided by 2t:
2tD ≈|x(t) −x(0)|
2
over sufficiently long times. This diffusional motion can be simulated by the
simple scheme:
x
n+1
= x
n
+ R
n
, (14.37)
where the random force R is related to D by:
R
n
=0, (R
n
)
2
=2DΔt. (14.38)
This propagation scheme reproduces D over long times t; see homework
assignment 12.
Ermak/McCammon
Ermak and McCammon [367] derived a basic BD propagation scheme from the
generalized Langevin equation in the high-friction limit, where it is assumed
that momentum relaxation occurs much faster than position relaxation. For a
three-dimensional particle diffusing relative to other particles and subject to a
force F (X), the derived BD scheme becomes:
X
n+1
= X
n
+
Δt
k
B
T
D
n
F (X
n
)+R
n
, (14.39)
R
n
=0, R
n
(R
m
)
T
=2D
n
Δtδ
nm
. (14.40)
For reference, the IBD algorithm developed in [106] has the following mass-
dependent correction term:
X
n+1
= X
n
+
Δt
k
B
T
D
n
F (X
n
)+
1
(k
B
T)
2
D
n
MD
n
[F (X
n−1
)−F (X
n
)]+R
n
,
(14.41)
with the same random-force properties for R as above (eq. (14.40)).
14.5.4 Hydrodynamic Interactions
Various approaches have been developed to define the diffusion tensor D in these
equations. Recall from Subsection 14.2.5 that the simple Langevin formulation
offers only a simple isotropic description of viscous effects: frictional effects
are taken to be isotropic, so γ is a scalar. Furthermore, the random force R
acts independently on each particle, thereby ignoring changes in force due to the
solvent-mediated dynamic interparticle interactions.
