Schlick T. Molecular Modeling and Simulation: An Interdisciplinary Guide
Подождите немного. Документ загружается.

472 14. Molecular Dynamics: Further Topics
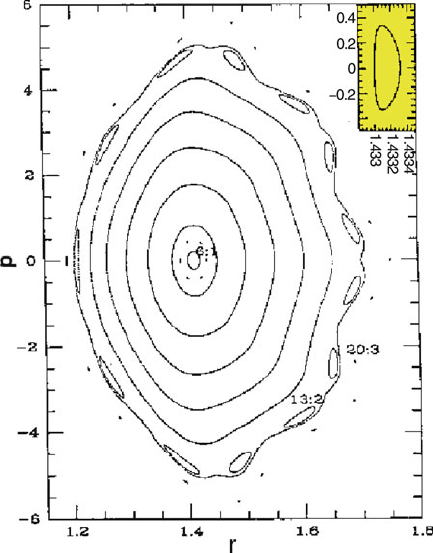
Figure 14.2. Morse oscillator phase diagrams obtained from the leapfrog Verlet integrator
with Δt =2fs at increasing initial energies. Resonances of order 6 and higher can be
noted. One of the inner-most islands (near the 6:1 label) is enlarged in the inset to show
that each corresponds to a closed orbit. See [821] for details.
Section 14.6 presents a resonance analysis for the implicit-midpoint (IM)
scheme, and Box 14.5 derives IM’s resonance condition (based on its phase-space
rotation ω
IM
eff
). Figure 14.16 illustrates resonance for a nonlinear system for both
Verlet and IM.
14.3 Multiple-Timestep (MTS) Methods
14.3.1 Basic Idea
MTS schemes were introduced in the 1970s [1232,1243] in an effort to reduce the
computational cost of molecular simulations. Savings can be realized if the forces
due to distant interactions are held constant over longer intervals than the short-
range forces. Standard integration procedures can then be modified by evaluating
the long-range forces less often than the short-range terms. Between updates, the
slow forces can be incorporated into the discretization as a piecewise constant
14.3. Multiple-Timestep (MTS) Methods 473
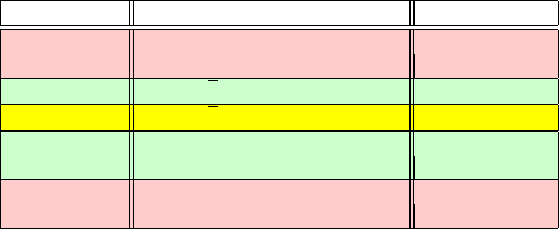
Table 14.2. Stability limit and resonant timestep formulas for the Verlet scheme, with
numerical values for a vibrational mode with period T
p
=10fs.
n, res. order
a
Δt
n:1
(ω)Δt
n:1
(T
p
) Δt
n:1
for MD
b
2 2/ω 0.318 T
p
3.2 fs
[= T
p
/π]
3
√
3/ω 0.276 T
p
2.8 fs
4
√
2/ω 0.225 T
p
2.3 fs
5 1.176/ω 0.188 T
p
1.9 fs
[=2sin(π/5)/ω]
6 1/ω 0.159 T
p
1.6 fs
[= T
p
/2π]
a
For n =2, we have the linear stability condition.
b
To compare resonance in Verlet to implicit-midpoint integration (see end of chapter), consider
the analogous resonant timesteps Δt
n:1
for n =3, 4, 5, 6 of 5.5, 3.2, 2.3, and 1.8 fs, respectively,
for the implicit-midpoint scheme.
function (via extrapolation) or as a sum of delta functions (via impulses). See
Figure 14.4 for a schematic illustration of these two approaches.
To illustrate, in the discussion below we consider below a force splitting scheme
that involves three components for updating the fast, medium, and slow forces:
F
fast
, F
med
,andF
slow
,
with corresponding timesteps
Δτ ≤ Δt
m
≤ Δt.
The timesteps are related via the ratios
k
1
=Δt
m
/Δτ,
k
2
=Δt/Δt
m
, (14.26)
r = k
1
k
2
=Δt/Δτ,
where k
1
and k
2
are integers. Note that when k
1
= k
2
=1we have a single-
timestep (STS) method.
14.3.2 Extrapolation
A simple, Verlet-based, extrapolative force-splitting approach is best formulated
on the basis of position Verlet (eq. (13.15)) since the fast and medium forces
are evaluated at the middle of the corresponding interval rather than at the be-
ginning (and end). In programming style, where new iterates overwrite the old
(i.e., X ← X + ···), this extrapolative force-splitting scheme can be written
as the doubly-nested loop for covering each Δt sweep shown in Figure 14.3,
left-hand side.
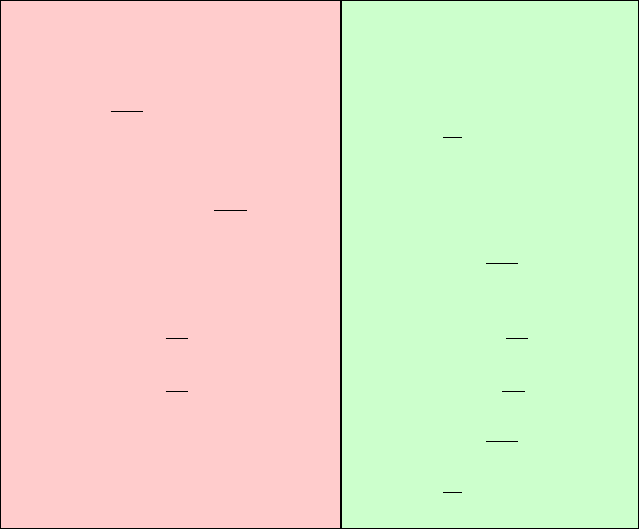
474 14. Molecular Dynamics: Further Topics
Extrapolative MTS Impulse MTS
based on Position Verlet based on Velocity Verlet
X
0
r
≡ X +
Δt
m
2
V
F
slow
≡−M
−1
∇E
slow
(X)
F
slow
≡−M
−1
∇E
slow
(X
r
) V ← V +
Δt
2
F
slow
For j =1 to k
2
For j =0 to k
2
− 1
X
r
≡ X
j
r
← X +
Δt
m
2
V
F
med
≡−M
−1
∇E
med
(X
r
)
F
med
≡−M
−1
∇E
med
(X)
F ←
F
med
+
F
slow
V ← V +
Δt
m
2
F
med
For i =1 to k
1
For i =0 to k
1
− 1
X ← X +
Δτ
2
V V ← V +
Δτ
2
F
fast
V ← V +Δτ (
F +
F
fast
) X ← X +ΔτV
X ← X +
Δτ
2
V V ← V +
Δτ
2
F
fast
End End
V ← V +
Δt
m
2
F
med
End End
V ← V +
Δt
2
F
slow
Figure 14.3. Algorithmic sketches of molecular dynamics integration by two force-splitting
variants: extrapolation (left) versus impulses (right), based on position Verlet (extrapola-
tion) and velocity Verlet (impulses).
Note that in this loop the slow force is evaluated once (at the point X
0
r
), the
medium force is evaluated k
2
times (for each X
j
r
), and the fast force is evalu-
ated k
1
k
2
(or r) times at a corresponding midpoint. If the slow force calculations
take the majority of the CPU time, the MTS approach will result in significant
computational savings.
In practical implementations, the force distance classes are best treated by a
smooth force-switching approach; see Chapter 10, Spherical-Cutoff section.
Simple extrapolation formulations, as above, were first tried for molecu-
lar dynamics [486, 1231, 1232, 1243, 1279]. However, these variants exhibited
systematic energy drifts, a result of their nonsymplecticness.
14.3.3 Impulses
Work continued in the 1980s on MTS methods in a variety of contexts [1276,
1278], leading to the introduction in 1991/1992 (by Schulten and coworkers [486]
and independently by Berne and coworkers [1277]) to an MTS method which

14.3. Multiple-Timestep (MTS) Methods 475
is symplectic and time-reversible. This similar MTS variant, based on impulses
rather than extrapolation, was termed Verlet-I by the former group [486]and
r-Respa [1277] by the latter.
The reversible Respa method was derived from a general Trotter factorization
associated with the Liouville operator L. Liouville operators are fundamental
tools in statistical mechanics for the description of the canonical equations of
motion of Hamiltonian systems. The Liouville operator can be decomposed into
parts corresponding to different components of the energy using the reversible
Trotter factorization. Here, we write the Liouville operator Las a sum of three op-
erators that characterize the scales of motions associated with different potential
components:
L = L
fast
+ L
med
+ L
slow
.
We then use a symmetric factorization of the components to arrive at (recall that
k
1
Δτ =Δt
m
and k
2
Δt
m
=Δt):
exp [iΔtL]=exp[iΔt(L
fast
+ L
med
+ L
slow
)]
=exp
i
Δt
2
L
slow
×
exp
i
Δt
m
2
L
med
(exp [iΔτL
fast
])
k
1
exp
i
Δt
m
2
L
med
k
2
× exp
i
Δt
2
L
slow
+ O(Δt
3
).
This factorization effectively shows that the propagation of the solution can be
approximated by a combination of terms corresponding to several force com-
ponents, each of which is resolved on a suitable timescale (e.g., Δτ/Δt
m
/Δt
for fast/medium/slow components). The middle term, corresponding to the fast
components of the motion, is discretized with the Verlet method at a small
timestep (Δτ).
A sweep over one Δt interval by an impulse-MTS Verlet approach, based on
the leapfrog or velocity Verlet triplet (eq. 13.14), can be written as the doubly-
nested iteration process shown in Figure 14.3, right-hand side.
14.3.4 Vulnerability of Impulse Splitting to Resonance Artifacts
Note that the application of the slow force results in an impulse. The velocities are
modified only outside of the inner loop (i.e., at the onset and at the end of a sweep
covering Δt) by a term proportional to r Δτ, thus r times larger than the changes
made to X and V in the inner loop. This is shown schematically in Figure 14.4
for a dual-timestep method with r =4(see tall spikes at bottom).
Thus, as the time interval between slow-force updates increases, the size of
these “impulses” grows. This causes a resonance artifact when the impulse fre-
quency, or the MTS outer timestep Δt, occurs near a natural frequency of the
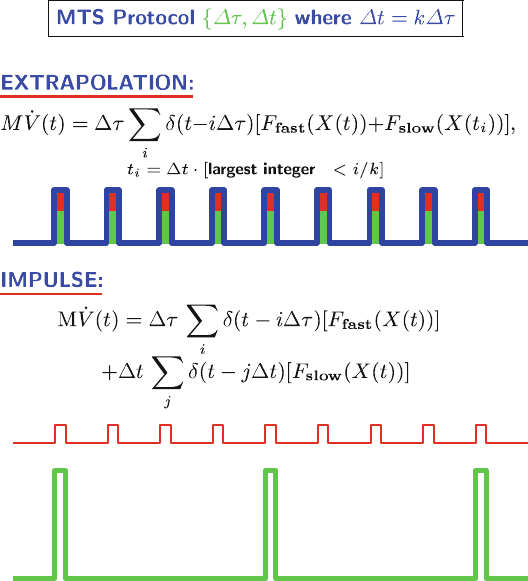
476 14. Molecular Dynamics: Further Topics
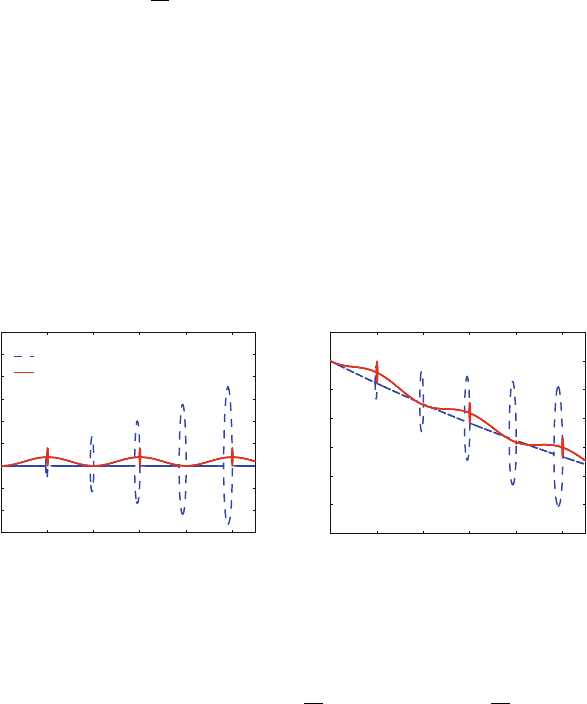
Figure 14.4. Schematic illustration of extrapolative vs. impulse force splitting integration
for a dual-timestep protocol with inner timestep Δτ and outer timestep Δt = kΔτ (k =4
used). In extrapolative splitting, a slow-force contribution (green) is made each time the
fast force (red) is evaluated (i.e., every Δτ interval). In impulse splitting, in contrast, con-
tributions of the slow forces (tall spikes) are considered only at the time of their evaluation
(e.g., every four fast-force evaluations).
system. This artifact results because the long-range forces add energy at pulses
that correspond to the natural frequency of the system. Of course, the true phys-
ical system experiences continuous variation of the long-range forces. Next we
analyze resonance artifacts of MTS methods in more detail for a harmonic model.
14.3.5 Resonance Artifacts in MTS
Analysis of a simple linear system is instructive to illustrate resonance in MTS
schemes [94,446]. Recent works [94,1024,1089] have shown that impulse meth-
ods are generally stable except at integer multiples of half the period of the
fastest motion, with the severity of the instability worsening with the timestep.
14.3. Multiple-Timestep (MTS) Methods 477
Extrapolation methods are generally unstable for the Newtonian model problem,
but the instability is bounded for increasing timesteps. Similar results hold for
stochastic extensions of MTS [94].
Simple Example
Figure 14.5 from [1089] illustrates this behavior as analyzed on a one-dimensional
linear oscillator obeying the equations
˙
X = V,
˙
V = −(λ
1
+ λ
2
)X,
where X and V denote the scalar position and velocity, respectively, and a unit
mass for the particle is assumed. The scalars λ
1
>λ
2
represent two motion com-
ponents differing in timescales. The characteristic angular frequencies associated
with these components and respective periods are
ω
i
=
λ
i
,T
p
i
=2π/ω
i
,i=1, 2 .
For the analysis of Figure 14.5,wesetλ
1
= π
2
and λ
2
=(π/5)
2
to produce
T
p
2
=10=5T
p
1
time units. The slow force component is defined as −λ
1
X and
is updated at timesteps Δt that are k times larger than those (Δτ)usedforthefast
components, −λ
2
X.
In the linear analysis of [1089], eigenvalue magnitudes derived from the prop-
agation matrices associated with the impulse and extrapolation force splitting
schemes are plotted against the outer timestep. A scheme is unstable if the eigen-
value magnitude exceeds unity. Figure 14.5 shows results for both Newtonian
(left) and Langevin (right) dynamics; the latter, stochastic dynamics approach is
described in more detail below. The inner timestep was set to Δτ =0.001 and
the outer timestep to kΔτ (compare to T
p
1
=2and T
p
2
=10).
0 1 2 3 4 5
0.7
1.0
1.3
1.6
Δt
Eig. Magnitude
Impulse Verlet
Constant Ex.
Eig. Magnitude
0 1 2 3 4 5
0.4
0.5
0.6
0.7
0.8
0.9
1
1.1
Δt
Figure 14.5. Propagation-matrix eigenvalue magnitudes for Newtonian (left) and Langevin
(right, γ =0.162) impulse and extrapolative force-splitting methods as a function of
the outer timestep as calculated for a one-dimensional linear test system governed by
¨
X = −(λ
1
+ λ
2
)X with λ
1
= π
2
, λ
2
=(π/5)
2
,whereX denotes the scalar posi-
tion of the particle with unit mass. These settings produce associated characteristic periods
for the fast and slow components of T
p
1
=2π/
√
λ
1
=2and T
p
2
=2π/
√
λ
2
=10time
units. The inner timestep is Δτ =0.001 [1089].
478 14. Molecular Dynamics: Further Topics
We see that the impulse method (dashed line) is unstable at integral multiples
(m) of half the fast period (mT
p
1
/2,whereT
p
1
/2=1here). Furthermore, the
severity of these corruptions increases with k, with the amplitudes of the resonant
spikes increasing linearly with Δt and becoming wider.
While the use of impulses clearly leads to serious artifacts at certain timesteps,
for other timesteps the impulse method is stable for this one-dimensional linear
model. It might seem that avoiding the resonant timesteps is possible in practice.
Unfortunately, experiments on a large nonlinear system show that once a resonant
timestep is reached, good numerical behavior cannot be restored by increasing the
timestep [1126]. See Figure 14.7 for an illustration of resonance on a nonlinear
system.
For the extrapolation method, in contrast, we note in Figure 14.5 (solid
curves) deviations from unit eigenvalues except around isolated outer timesteps.
Since both eigenvalues exceed one in magnitude, the method is not volume-
preserving or symplectic, explaining the energy drift observed in practice for force
splitting by extrapolation. Still, the resonant spikes for constant extrapolation
have magnitudes that are independent of the outer timestep, of values approxi-
mately 1+λ
2
/λ
1
(λ
2
/λ
1
=0.04 here). Therefore, the larger the separation of
frequencies, the more benign the numerical artifacts in practice.
Extensions to Stochasticity and Nonlinearity
These resonance patterns generalize to stochastic dynamics, where friction and
random terms are added (see next section), as seen in the same figure (the eigen-
value magnitudes decrease due to frictional damping). The value of γ used here
ensures that the first spike for the impulse method has magnitude less than unity
[1089]. Behavior is more complicated for a linear three-dimensional test prob-
lem [1089]. Still, applications to a nonlinear potential function for a protein,
asshowninFigure14.7 (discussed below), exhibit similar overall patterns for
impulse versus extrapolative force splitting treatments.
14.3.6 Limitations of Resonance Artifacts
on Speedup; Possible Cures
The original work of Schulten and coworkers [486] expressed reservations regard-
ing force impulses. A subsequent study by Biesiadecki and Skeel [134] discussed
resonance as well as the systematic drift of extrapolative treatment in simple os-
cillator systems, though it conjectured that resonance artifacts were not likely to
be so clear in nonlinear systems. This, unfortunately, turned out not to be true.
Other articles noted a rapid energy growth [1349, 1448] when the interval be-
tween slow-force updates approaches and exceeds 5 fs, half the period of the
fastest oscillations in biomolecules
The presence of these resonances in impulse splitting limits the outermost
timestep, Δt (or the interval of slow-force update), in MTS schemes. In turn,
the overall speedup over a standard Verlet trajectory is capped. The achievable
14.4. Langevin Dynamics 479
speedup factor has been reported to be around 4–6 [1277, 1349, 1448]for
biomolecules. However, this number depends critically on the reference STS
(single-timestep) method to which MTS performanceis compared. Since the inner
timestep in MTS schemes is often small (e.g., 0.5 or even 0.25 fs), such speedup
factors around 5 are obtained with respect to STS trajectories at 0.5 fs (10 with
respect to 0.25 fs!). While accuracy is comparable when this small timestep is
used, typically STS methods use larger timesteps, such as 1 or 2 fs (often with
SHAKE). This means that the actual computational benefit from the use of MTS
schemes in terms of CPU time per nanosecond, for example, is much less. Still,
a careful MTS implementation with inner timestep Δτ =0.5 fs can yield better
accuracy than a STS method using 1 or 2 fs.
Given this resonance limitation on speedup, it is thus of great interest to revise
these methods to yield larger speedups.
The mollified impulse method of Skeel and coworkers [446, 594, 595]has
extended the outer timestep by roughly a factor of 1.5 (e.g., to 8 fs). With addi-
tional Langevin coupling, following the LN approach [95], the Langevin mollified
method (termed LM) can compensate for the inaccuracies due to the use of im-
pulses to approximate a slowly varying force. This is accomplished by substituting
A(X)
T
F
slow
(A(X)) for the slow force term where A(X) is a time-averaging
function.
Another approach altogether is to use extrapolation in the context of a stochas-
tic formulation, as in the LN method [93–95] (see next section). This combination
avoids the systematic energy drift, alleviates severe resonance, and allows much
longer outer timesteps; see also discussion of masking resonances via the
introduction of stochasticity in an editorial overview [665] and review [328].
Though the Newtonian description is naturally altered, the stochastic formu-
lation may be useful for enhanced sampling. The contribution of the stochastic
terms can also be made as small as possible, just to ensure stability [99, 1109].
For example, unlike the predictions in the above review [328], a smaller stochas-
tic contribution (damping constant of 5 to 10 ps
−1
) has been used in the LN
scheme without reducing the outer timestep, and hence without compromising
the speedup [1230].
14.4 Langevin Dynamics
14.4.1 Many Uses
A stochastic alternative to Newtonian dynamics, namely Langevin dynamics, has
been used in a variety of biomolecular simulation contexts for various numeri-
cal and physical reasons. The Langevin model has been employed to eliminate
explicit representation of water molecules [966], treat droplet surface effects
[180,1189], represent hydration shell models in large systems [112–114], enhance
sampling [298, 328, 513, 659, 783, 1036, 1428], and counteract numerical damp-
ing while masking mild instabilities of certain long-timestep approaches [95,992,
480 14. Molecular Dynamics: Further Topics
1132,1435, 1436]. See Pastor’s comprehensive review on the use of the Langevin
equation [966]. The Langevin equation is also discussed in Subsection 10.6.3 of
Chapter 10 in connection with continuum solvation representations.
14.4.2 Phenomenological Heat Bath
The Langevin model is phenomenological [853] — adding friction and random
forces to the systematic forces — but with the physical motivation to represent a
simple heat bath for the macromolecule by accounting for molecular collisions.
The continuous form of the simplest Langevin equation is given by:
M
¨
X(t)=−∇E(X(t)) − γM
˙
X(t)+R(t), (14.27)
where γ is the collision parameter (in reciprocal units of time), also known as the
damping constant. The random-force vector R is a stationary Gaussian process
with statistical properties given by:
R(t) =0, R(t)R(t
)
T
=2γk
B
TM δ(t −t
), (14.28)
where k
B
is the Boltzmann constant, T is the target temperature, and δ is the usual
Dirac symbol.
14.4.3 The Effect of γ
The magnitude of γ determines the relative strength of the inertial forces with
respect to the random (external) forces. Thus, as γ increases, we span the inertial
to the diffusive (Brownian) regime. The Brownian range is used (with suitable
algorithms) to explore configuration spaces of floppy systems efficiently. See
[607, 1321], for instance, for applications to the large-scale opening/closing lid
motion of the enzyme triosephosphate isomerase (TIM), and to the juxtaposition
of linearly-distant segments in long DNA systems, respectively.
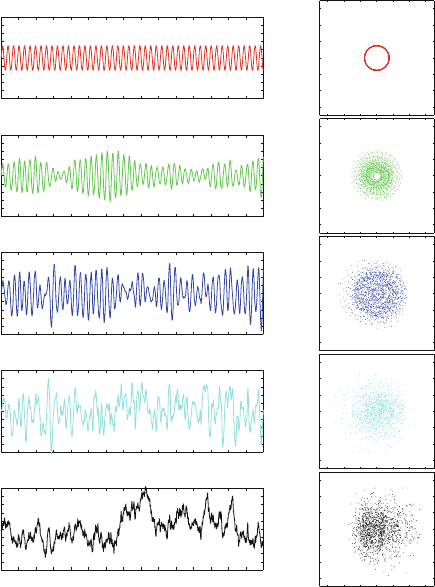
Figure 14.6 illustrates the effects of increasing γ on the trajectories and
phase diagrams of a harmonic oscillator. The systematic harmonic motion and
the closed, circular trajectories characteristic at zero viscosity change as the
relative contribution of the random to systematic forces increases.
Since the stochastic Langevin forces mimic collisions between solvent mole-
cules and the biomolecule (the solute), we see that the characteristic vibrational
frequencies of a molecule in vacuum are damped. In particular, the low-
frequency vibrational modes are overdamped, and various correlation functions
are smoothed (see Case [202] for a review and further references). The magnitude
of such disturbances with respect to Newtonian behavior depends on γ [95].
A physical value for γ for each particle can be chosen according to Stokes’ law
for a hydrodynamic particle of radius a:
γ =6πηa/m , (14.29)
where m is the particle’s mass (not to be confused with the integer m used to
define resonance), and η is the solvent viscosity. For example, γ =50ps
−1
is a
14.4. Langevin Dynamics 481
−.4
−.2
0
.2
.4
−.4
−.2
0
.2
.4
a
b
c
d
e
−.4
−.2
0
.2
.4
−.4
−.2
0
.2
.4
0 1000 2000 3000
−.4
−.2
0
.2
.4
X
γ =0
γ = 0.01
γ = 0.1
γ =1
−.7 0 .7
−.7
0
.7
γ = 10
X
V
Figure 14.6. Langevin trajectories for a harmonic oscillator of angular frequency ω =1
and unit mass simulated by a Verlet extension to Langevin dynamics at a timestep of 0.1
(about 1/60 the period) for various γ. Shown for each γ are plots for position versus time
(left panels) and phase-space diagrams (right squares). Note that points in the phase-space
diagrams are not connected and only appear connected for γ =0because of the closed
orbit.
typical collision frequency for protein atoms exposed to solvent having a viscosity
of 1 cp at room temperature [967]; this value is also in the range of that estimated
for water (γ =54.9 ps
−1
).
It is also possible to choose an appropriate value for γ for a system modeled by
the simple Langevin equation so as to reproduce observed experimental trans-
lation diffusion constants, D
t
. Namely, in the diffusive limit, D
t
is related to
γ by
D
t
= k
B
T/
mγ .
See [966, 1037] for example, for the use of these relations in studies of
supercoiled DNA.
