Schlick T. Molecular Modeling and Simulation: An Interdisciplinary Guide
Подождите немного. Документ загружается.


502 14. Molecular Dynamics: Further Topics
This was verified in an application to a representative nonlinear system, a
blocked alanine model [1126] in Figure 14.16. This figure contrasts the early (i.e.,
with small timestep) instability of Verlet with the resonance of IM around one half
the fastest period, and the absence of resonances in LIM2 (α =1/2).
Unfortunately, the increases of the average energy in LIM2 with the timestep
are unacceptable: they reflect values that are approximately 30% and 100% larger
than the small-timestep value, for Δt = 5 and 9 fs, respectively, for this system.
Part of this behavior is also due to an error constant for LIM2 that is greater than
that of leapfrog/Verlet.
Perspective
In sum, it is difficult to expect reasonable resolution by implicit methods in
the large-timestep regime given the stability limit and resonance problems men-
tioned above. Perhaps semi-implicit [847] or cheaper implementations of implicit
schemes [1440] will better handle this problem: it might be possible to treat the
local terms implicitly and the nonlocal terms explicitly. Exploitation of parallel
machine architecture has potential for further speedup but, if experience to date on
parallelization of linear algebra codes can be considered representative, parallel
computers tend to favor explicit methods.
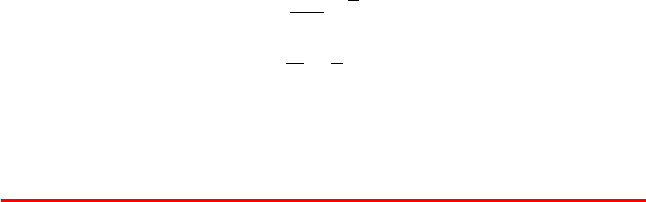
Box 14.6: Implicit Symplectic Integrator Family
The family of implicit symplectic methods parameterized by α can be formulated as
follows [446, 1126]:
X
n+1/2
= X
n
+
Δt
2
V
n
V
n+1/2
= V
n
+
Δt
2
M
−1
F
n+1/2
V
n+1
= V
n+1/2
+
Δt
2
M
−1
F
n+1/2
(14.63)
X
n+1
= X
n+1/2
+
Δt
2
V
n+1
,
where
F
n+1/2
= F (X
n+1/2
+ αΔt
2
M
−1
F
n+1/2
). (14.64)
The values α =0and 1/4 correspond to the Verlet and IM methods, respectively, and
α =1/2 corresponds to the LIM2 method introduced in the text.
The above implicit system can be solved by minimizing the dynamics function Φ to
obtain X and then evaluating F at this minimum point. Here Φ is defined as
Φ(X)=
1
γΔt
2
(X − X
n
0
)
T
M (X − X
n
0
)+αE(X) , (14.65)
with
X
n
0
= X
n
+
Δt
2
V
n
.
14.7. Enhanced Sampling Methods 503
The effective rotation angle for this α-family is [1126]:
θ
α family
eff
=2sin
−1
ωΔt
2
φ
(14.66)
= ωΔt +
1
24
−
α
2
(ωΔt)
3
+ O((ωΔt)
5
)
where
φ =1/ [1 + α(ωΔt)
2
] . (14.67)
Thus the maximal effective rotation can be controlled by the choice of α (see text).
14.7 Enhanced Sampling Methods
14.7.1 Overview
The well known shortcomings of traditional MD have stimulated many innovative
methods for capturing large-scale, long-time configurational changes of biomol-
ecules. Indeed, an enormous range of methods has been developed to enhance
coverage of the conformational space; see [729, 781, 1116, 1117], for example,
for recent reviews. Each method depends on the computation goals and avail-
able computing resources. While many of these methods are suitable for probing
the thermally-accessible configuration space at the cost of altered kinetics, more
sophisticated approaches like transition path sampling or Markov chain models
can yield valuable mechanistic insights, reaction pathways, and reaction rates.
Approaches mentioned already for addressing the sampling problem include
simulating multiple trajectories for improved statistics [69], especially effective
when applied to small systems, as in folding studies of peptides [285] or protein
segments [364, 365]. Other approaches mentioned are Monte Carlo techniques
[509] and variants like simulated annealing, parallel tempering/replica exchange
MC [508, 1237], and hybrid MC methods (Monte Carlo chapter). Implicit solva-
tion (Chapter 10) and Langevin and Brownian dynamics approaches (this chapter)
can also be viewed as approaches for enhancing the sampling because they can
cover longer times and/or reduce computational cost.
Below, additional approaches are described, including methods based on har-
monic analysis and coordinate transformations, coarse-grained models, biasing
approaches and altered MD protocols, and various novel methods for computing
reaction mechanisms and reaction rates.
14.7.2 Harmonic-Analysis Based Techniques
The fundamental oscillations of each molecule about its equilibrium state, termed
normal modes, were described in Chapter 9 in connection to force-field parame-
terization. Each such fundamental mode has a frequency associated with it that

504 14. Molecular Dynamics: Further Topics
is related to a force constant and energy potential that can be propagated to
describe molecular displacements about equilibrium. The same idea based on har-
monic theory has been extended more generally to describe collective molecular
motions, including for complex molecular systems.
In their purest forms, normal mode analysis (NMA) and the related princi-
pal component analysis (PCA) involve propagating the normal modes based on
a spectral decomposition (diagonalization) of a mass-weighted Hessian at ther-
mal equilibrium [535]. Elastic networks [77, 463, 1262] are modern extensions
of these techniques which forgo the computationally-demanding diagonaliza-
tion because the simplified bead/spring coarse-grained models are assumed by
construction to reflect minimum states of the molecular system. Clearly, this har-
monic approximation to describe small-amplitude, high-frequency motions is far
from accurate at ambient temperatures when significant biomolecular fluctua-
tions between minimum-energy regions, as well as rearrangements, occur. Still,
these techniques have provided valuable information on collective motions of
biomolecules.
Besides elastic networks, another successful extension of these harmonic-
analysis-based techniques is called essential dynamics (ED) [36, 37, 650, 1289].
ED attempts to characterize low-frequency, high-amplitude motions modes by
expressing the dynamics in terms of alternative coordinates (principal compo-
nents corresponding to low-frequency motion) which filter out other modes. This
low-dimensional space is constructed from the variance/co-variance matrix of po-
sitional fluctuations obtained from a time series of coordinates, by projecting the
original configurations onto each of the principal components and then following
the principal components of interest in time. ED makes no explicit assumption of
thermal equilibrium.
Specifically, to describe the collective motions, a covariancematrix C of atomic
fluctuations is constructed from a series of M coordinates X
1
, X
2
, ...X
M
where
X
k
is the collective coordinate vector of the system at the kth configuration, with
respect to an average structure X:
X =
1
M
k=1,M
X
k
, (14.68)
from the expression:
C =
1
M
k=1,M
(X
k
−X)(X
k
−X)
T
. (14.69)
Diagonalization of this covariance matrix C produces the eigenvectors {V
n
} and
eigenvalues {λ
n
} as entries of the diagonal matrix Λ =diag(λ
1
,λ
2
,...λ
3N
)
from the spectral decomposition:
V
T
CV = Λ . (14.70)
Thus, the projection
CV
n
= λ
n
V
n
,n=1, 2,...3N, (14.71)
14.7. Enhanced Sampling Methods 505
can be used to visualize and propagate selected motions. In this description, each
eigenvector V
n
defines the direction of motion of the molecular system as a
displacement about the average structure. The normalized magnitude of the cor-
responding eigenvalue λ
n
is a measure of the amplitudes of motion along the
eigenvector V
n
.
The literature is vast with applications of PCA, NMA, and ED with both
all-atom and coarse-grained models in combination with various algorithms,
including molecular, Langevin and Brownian dynamics to biomolecular con-
formational flexibility and dynamics. These approaches have provided valuable
insights into biomolecular flexibility and functional activity. However, the results
depend strongly on the level of convergence of the sampling, which influences the
results and hence the interpretations.
Some examples of using PCA and ED include analyses of protein subdomain
motion correlations in DNA pol β [60] and Dpo4 [1336] and sequence/flexibility
relationships for a series of single-based DNA TATA elements [1026, 1229]
(Fig. 14.17). Other examples include folding dynamics of a β-protein WW do-
main using the coarse-grained protein model UNRES [816], and an investigation
of protein flexibility in water [1075]. Network models have been particularly
effective for applications to molecular machines like the ribosome modeled by
coarse-grained formulations [1248].
14.7.3 Other Coordinate Transformations
This idea in ED of projecting the dynamics onto principal modes is related to vari-
ous variable/coordinate transformation methods in classical statistical mechanical
using configuration partition functions. For example, metadynamics rewrites the
equations of motion in terms of a few collective variables so that key regions
of space are identified and explored [963], and the reference potential spatial
warping algorithm (REPSWA) [860] introduces a variable transformation in the
classical partition function that increases attraction to basins.
Internal or torsion-angle dynamics has long been attempted for the goal of
enhanced sampling, with the rationale that the fewer degrees of freedom com-
pared to Cartesian coordinates will allow longer integration timesteps and hence
greater sampling. Indeed, peptide folding and refinement with dihedral-angle MD
demonstrated a computational advantage of several orders of magnitude over
Cartesian analogues [585] as well as the capturing of folding pathways of he-
lical peptides and local side-chain and domain dynamics [1426]. Dihedral-space
MD has also been combined with PCA in a clever way to systematically construct
the low-dimensional free energy landscape from a classical MD simulation [34].
Though this analysis is interpretive, it shows that major conformational states,
barriers, and reaction pathways for solvated peptides can be visualized from the
constructed energy landscape.
In general, dihedral-angle MD approaches for propagating biomolecular mo-
tion have not caught on at large, perhaps both due to the added cost of the
transformation involved in the Newtonian laws of motion and the fact that
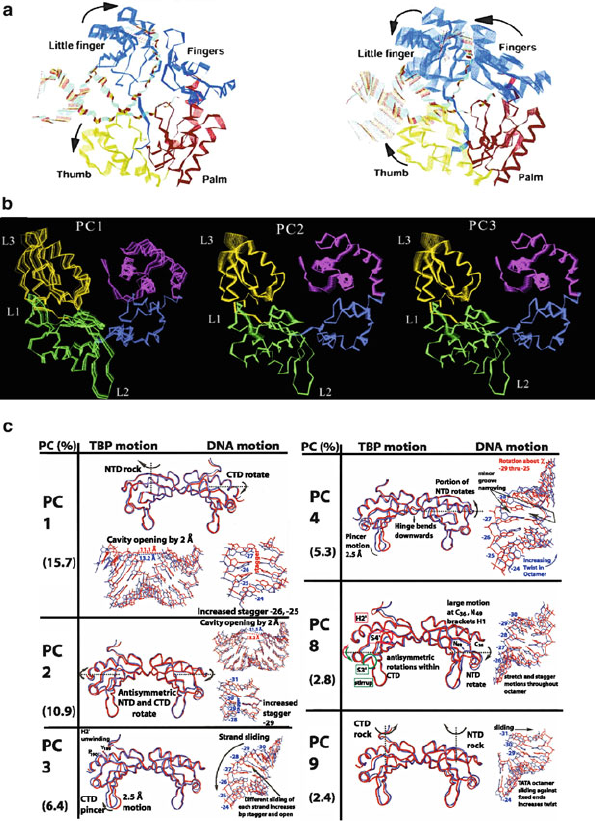
506 14. Molecular Dynamics: Further Topics
Figure 14.17. Examples of PCA and ED applications. (a) A study of the conformational
changes of Dpo4 after chemistry revealed that the little finger and fingers domains may
help translocate the DNA for the next cycle of nucleotide insertion [1336]. (b) A study
of the closing conformational change of DNA polymerase β upon binding the nucleotide
substrate revealed from the top three principal components correlations between the thumb
subdomain and other regions of the protein (palm, 8-kDa) [59]. (c) A study of the motions
of single-base variants of TATA-box DNA sequences bound to the TPB protein helped
explain why these variants revealed a wide range of transcriptional efficiency despite re-
markably similar structures: high-efficiency variants favored complexation motions while
low-efficiency variants tended toward dissociation deformations. The dominant motions
common to all complexes are shown, dissected for the protein and bound TATA-box DNA
separately [1026].
14.7. Enhanced Sampling Methods 507
biomolecular vibrational modes are intricately coupled and hence dynamics can
be critically altered by neglecting the high-frequency bond-length and bond-angle
modes. However, the increased interest in coarse-graining models argues for their
resurgence.
14.7.4 Coarse Graining Models
System specific coarse-grained methods are attractive because they drastically re-
duce the number of degrees of freedom. However, their formulations are highly
system dependent and require as much art as science in constructing, test-
ing/validating, and applying them to appropriately formulated questions. Coarse
graining can involve bead models, implicit solvent approximations, discrete lattice
models, and general multiscale formulations.
The simplest type of coarse graining involves bead models, long used for pro-
teins (e.g., Warshel & Levitt’s united residue model [1344]). Bead models have
also been widely used for supercoiled DNA (wormlike chain model of Allison
and co-workers [32], see Chapter 6) and more recently developed for RNA
(e.g., [315]). Such methods can lead to meaningful insights into large-scale re-
arrangements, including folding, not typically amenable to all-atom simulations.
However, the neglect of many details (e.g., solvent/solute interactions) must be
considered in the biological interpretations.
Lattice models also reduce the conformational degrees of freedom to a discrete
set, therefore allowing in theory exhaustive sampling of the conformational space.
Lattice models of proteins, such as developed by G¯o[463], and by Miyazawa
& Jernigan [869], are associated with ideal funnel energy landscapes: a protein
chain is modeled by attractive interactions between pairs of residues that interact
in the native structures and repulsive interactions of the other pairs, based on
statistical data.
General coarse-grained or multiscale models are most challenging to formu-
late and validate because the various components need to be resolved by different
approaches and combined effectively. For example, the simplified models of the
chromatin fiber developed by the Langowski [1312], Schiessel [883], and Schlick
[64, 1114] groups and others necessarily select the molecular parts to resolve
in detail and those that can be effectively approximated (i.e., coarse grained).
For example, in studies aimed at deducing the architecture of the 30nm chro-
matin fiber [483], the nucleosome core, histone tails, linker DNA, and linker
histones are each modeled differently in a mesoscale model (see Fig. 6.11)and
sampled by MC; parameterization for the core is done by a Poisson Boltzmann
electric field approximation and for the proteins by comparison to atomistic
MD [65,109,1441].
Other examples of general coarse-grained models are innovative models de-
veloped for membrane systems to investigate: the reshaping of the electrostatics-
dominated surface [57,655], pore formation [461], membrane architecture [1150],
and protein/membrane binding interactions [647]. Complex processes involving
508 14. Molecular Dynamics: Further Topics
virus capsid stability [56], GroEL/GroES chaperonin-guided folding [260], and
an ATP-driven molecular motor [667] have also been studied by coarse-graining
approaches.
It is well recognized that rigorous coarse-graining approaches are needed to
ultimately address many levels of biomolecular pathways and reactions to allow a
“telescoping” of views from one level of resolution to another. Some notable work
in that direction has already been reported [506,794,921]. The appropriateness of
the coarse graining has been assessed quantitatively from the collective motions
(ED) compared to atomistic motion [794], by computing solvation free energies
[506], and by a formal statistical mechanical framework [921].
14.7.5 Biasing Approaches
If altering the model — through biasing forces or guiding restraints — is con-
sidered fair game, much room for ingenuity remains, as described by the various
biasing techniques.
The simplest and crudest approach for enhancing the sampling is to manipulate
the energy function used as a basis for MD, by using restrained potentials, as in
targeted MD (TMD) or umbrella sampling, and by using various experimentally-
based biases or guides.
TMD can be used for generating pathways between known endpoints, such
as unfolded and folded state of a peptide [389]. In TMD, an artificial restraining
potential is added like a Lagrange multiplier with parameter λ that is 0 at the initial
state and 1 at the target state. This forces the system to evolve toward a target state
in a specified number of steps [389, 1418]. Despite the fictitious trajectory that
results from such targeted MD, insights into disallowed configurational states can
be obtained, as well as conclusions regarding common pathway themes, though
individual trajectories diverge from one another.
For example, TMD has been used to suggest possible opening pathways be-
tween substrate-free and substrate-bound enzyme states (see Figure 13.1)[1408]
and between closed and open states of a membrane channel [669]. Importantly,
besides suggesting pathways, intermediate pathway configurations generated
in TMD can serve as initial states for regular MD to explore pathways and
conformational transitions.
Many other methods for simulating transitions between defined conformations
have been reported (e.g., [355, 521, 955]). For example, the simplified multiple-
basin Hamiltonian funnel-based potential model [932] was developed for very
large molecular complexes with known endpoint structures to simulate transitions
between ligand-bound and unbound states.
Umbrella sampling (US) allows sampling of specified regions of phase space by
using a similar restraining potential, typically by restraining key conformational
variables to specific regions (e.g., a certain range of sugar puckering for DNA [59,
1030]). It is also possible to employ experimental information to define various
restraints to guide or restrict the pathways. Such experimental information can
14.7. Enhanced Sampling Methods 509
involve distances determined by FRET experiments [859] or the ratio of native
contacts present in protein intermediates relative to those in the native state [956].
US has also been useful in computation of free energies where extensive sampling
is required.
Besides various constraints or restraints, the energy can be alternatively mod-
ified by using other types of biasing terms to facilitate barrier crossing events.
These include a conformation flooding technique in which the bias is determined
based on a coarse-grained simulation for the conformational space density [485],
hyper-MD in which the biasing potential is constructed based on the smallest
eigenvalue of the Hessian matrix [1315], simple local boost method based on
the total potential energy [1332], a bond boost method based on bond-length
deviations [862], and accelerated MD by a potential-energy term “boost” [504].
Umbrella sampling methods can also be very effective in combination with
such biased trajectories (e.g., [130]). For example, diffusion-limited processes as-
sociated with high-energy or entropy barrier can be ‘accelerated’ through biasing
forces in Brownian dynamics simulations, with rate calculations adjusted by as-
sociating lower weights with movements along high biases and vice versa [1458].
Biomolecular systems can be ‘steered’ [592, 593] or ‘guided’ [1393, 1394]—
subjected to time-dependent external forces along certain degrees of freedom or
along a local free-energy gradient — to probe molecular details of certain exper-
iments, such as atomic force microscopy and optical tweezer manipulations (see
citations in [592, 593]), or to study folding/unfolding events [1395]. Interactive
MD [475, 1111] allows such exploration of pathways manually, by a combina-
tion of advanced computer graphics and simulation (VMD and NAMD): the user
steers the system to the desired state by “feeling” the potential force and applying
any desired magnitude (to “pull” the system).
14.7.6 Variations in MD Algorithm and Protocol
The MD simulation protocol rather than the energy can also be manipulated di-
rectly. In this subclass, we have the popular approach LES (locally enhanced
sampling) in which several configurations are generated in a single energy eval-
uation [1063], and Replica exchange MD (REMD) [1237], based on parallel
tempering MC [508] (described in the MC chapter), which itself is based on
Metropolis-coupled Markov chain MC.
The idea in REMD is to simulate the dynamics of multiple, non-interacting
copies (replicas) of identical systems at different temperatures. This requires in-
tegration by using canonical-ensemble algorithms as described in Chapter 13,
such as the the Berendsen weak coupling thermostat (or heat-bath) [123]or
the extended-simple Nos´e-Hoover thermostat approach [566, 924]. Periodically,
the configurations of the different replicas (e.g., replica i at temperature T
i
with replica j at temperature T
j
) are exchanged with a transition probability p
that maintains each temperature’s equilibrium ensemble distribution. Thus, the
510 14. Molecular Dynamics: Further Topics
thermodynamic or temperature states in the canonical ensemble are exchanged,
with the goal of retaining those systems that are making better progress:
p
i→j
=min[1, exp [(β
j
− β
i
)(E
j
− E
i
)] ] , (14.72)
where β
i
is the Boltzmann factor for temperature i: β
i
=1/(k
B
T
i
).
These temperature-ensemble exchanges are intended to accelerate barrier
crossings. Though obeying detailed balance for an extended ensemble of the
canonical states, the state-exchange probability of eq. (14.72) destroys real kinetic
properties. However, REMD has been used at large to simulate folding/unfolding
equilibria of biomolecules. Examples include the folding of a small solvated RNA
hairpin from an extended state [444], folding of a solvated protein A [443], folding
of villin within 1.78
˚
A of the native state [730], and determination of equilibrium
ensembles of proteins in a coarse-grained implicit-solvation model, OPEP [222].
Though REMD has been in wide usage for applications ranging from small
peptides to complex biological systems, its success has largely been empirical and
only recently have some technical issues come to the surface. Indeed, practitioners
of REMD have emphasized the need to formulate careful configurational swap-
ping protocols (temperature ladders and exchange/acceptance ratios) and other
ways to increase the conformational sampling efficiency (see [1064, 1192]for
example) and to use a large number of concurrent processors/replicas for effi-
cient sampling. In fact, a variant distributed replica sampling has been proposed
for serial implementations when a large number of processors is not available
[1060], and a variant for enhanced sampling based on a Tsallis biasing poten-
tial was also proposed [627]. It has also been remarked that REMD may only
be computationally advantageous for systems with relatively high energy barriers
[1459].
More fundamentally, there are issues with REMD because on the inherent
canonical-ensembleintegrators that are used. In their thorough analysis of REMD,
Cooke & Schmidler explain that the formulation of REMD by extension of par-
allel tempering MC to MD has introduced sampling and ergodicity problems
stemming from the failure of the underlying constant-temperature MD integra-
tors to preserve certain variants of REMD [264]. This is because while the MC
analog is based on Markov chains, REMD algorithms cannot use the symplec-
tic leap-frog integrator popular for microcanonical (constant energy) MD and
resort to isothermal integrators as mentioned above. Unfortunately, these meth-
ods are not rigorously ergodic, and this can affect the dynamics of even small
systems, as practitioners noted. As a remedy, combining REMD with hybrid MC
to ensure ergodicity was suggested [264], thereby returning to the original advan-
tages of parallel tempering MC. Other cures, such as using recently developed
entropy-preserving constant-temperature integrators for sampling the canonical
distribution based on a Nos´e-Hoover formulation in the context of Markov chain
Monte Carlo can also solve the ergodicity problem in REMD [732]; systematic
discretization errors are also eliminated by this hybrid MC approach for canonical
sampling.
14.7. Enhanced Sampling Methods 511
14.7.7 Other Rigorous Approaches for Deducing Mechanisms,
Free Energies, and Reaction Rates
Many other innovative methods have been developed by using various constructs
and ideas from physics, mathematics, and engineering such as domain decom-
position, clustering techniques, and Markov models to sample and/or survey
conformational space and dynamics.
For example, for exploring the free energy landscape, the canonical adiabatic
free energy sampling (CAFES) method [1299] propagates the dynamics of decou-
pled solute and solvent systems so that the former follows adiabatic dynamics of
the free energy surface created by the latter at increased temperatures. Metady-
namics explores the free energy surface by following non-Markovian dynamics
determined by rewriting the equations of motion in terms of a few collective
variables so that key regions of space are identified as simulation time increases
[963]. And a sweep method combines temperature-accelerated MD (TAMD) with
a free-energy reconstruction from the mean force using radial basis functions via
optimization [828].
Methods for deducing reaction mechanisms include transition path sampling
(TPS) by Chandler and co-workers which follows a Monte Carlo protocol in MD-
trajectory space to locate key transition states ([147] and recently reviewed in
[294]); the nudged elastic band method (NEB) [543] that optimizes minimum
energy pathways; the max-flux approach [583], which constructs variationally op-
timized reaction pathways based on the max flux for diffusive dynamics; and the
string method [858], which prunes MD trajectories to retain reactive segments.
A recent extension of TPS connects more than two intermediate states in phase
space [1062].
A TPS application to a biomolecular complex, a DNA polymerase β/DNA
complex, combined TPS with an efficient free-energy protocol termed “BOLAS”
[1030] and network models to deduce the mechanism, compute the free energy
profile, and estimate the rate for pol β’s closing conformational change [1031].
The study revealed a complex landscape where sequential, subtle-side chain rear-
rangements lead the enzyme from open to the closed state and where Arg258 is
a “gate keeper” for the reaction (Fig. 14.18); the overall computed rate of 10 per
second corresponds to the 27k
B
T barrier and agrees well with the experimental
value of 3–10 per second for the overall reaction. A similar TPS protocol applied
to the mismatch (G:A instead of G:C) suggested that the higher free energy bar-
rier for the mismatch comes from the instability of the closed mismatched state
compared to the matched base pair system [1032] (Figure 14.19).
More recently, innovative divide and conquer methods have been developed
to compute reaction rates. These include forward flux simulations (FFS) [152],
milestoning [354], and Markov State Models (MSM) [919].
FFS [152] enhances sampling of rare events in stochastic non-equilibrium
systems in which the phase space distribution is not known aprioriby using inter-
faces to partition phase space. Following this partitioning, an adaptive procedure
